Deep Learning Analysis for Predicting Tumor Spread through Air Space in Early-Stage Lung Adenocarcinoma Pathology Images
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Pathological Data Review
2.3. Pathological Spread through Air Space (STAS) Prediction Model Development
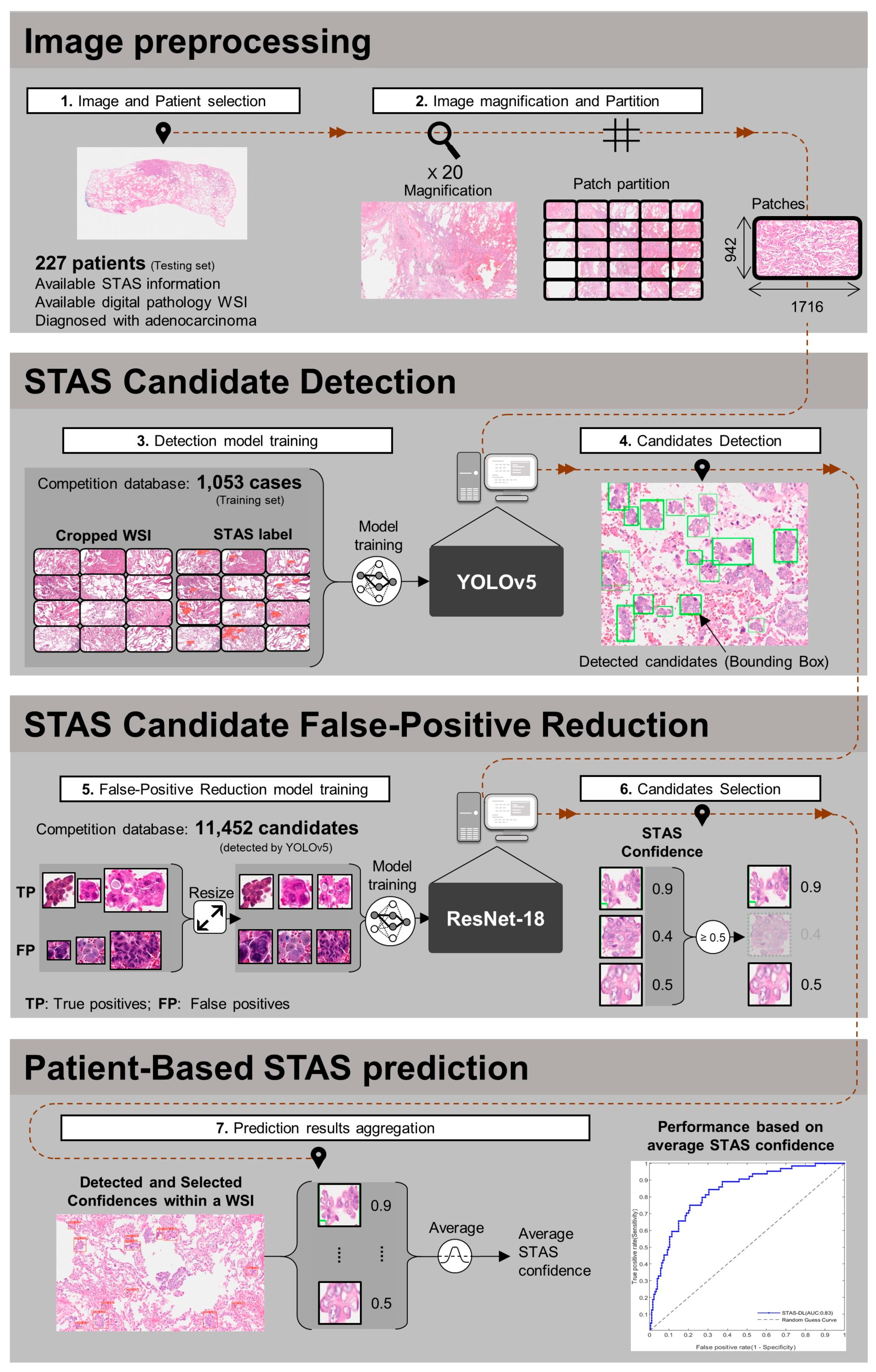
2.3.1. Image Preprocessing
2.3.2. Spead through Air Space (STAS) Candidate Detection
2.3.3. False-Positive Reduction
2.3.4. Patient-Based Spread through Air Space (STAS) Prediction
2.4. Correlation Analysis between Histological Grades and Model Prediction
2.5. Statistical Analyses
3. Results
3.1. Patient Demographics and Clinicopathological Characteristics
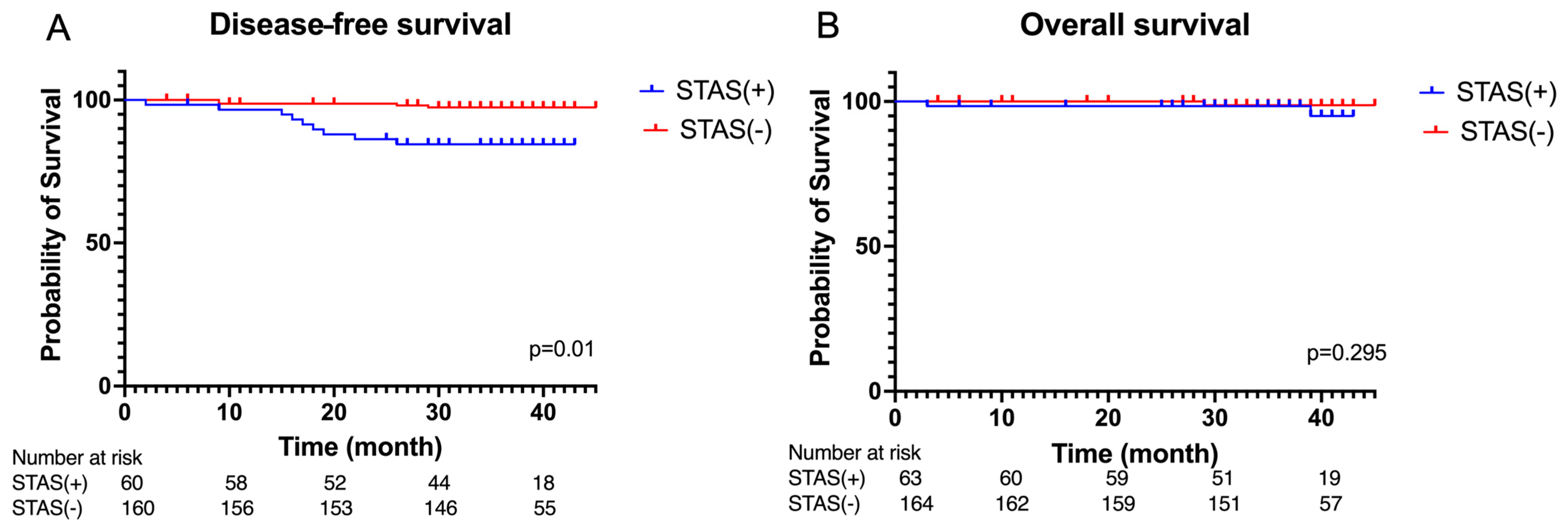
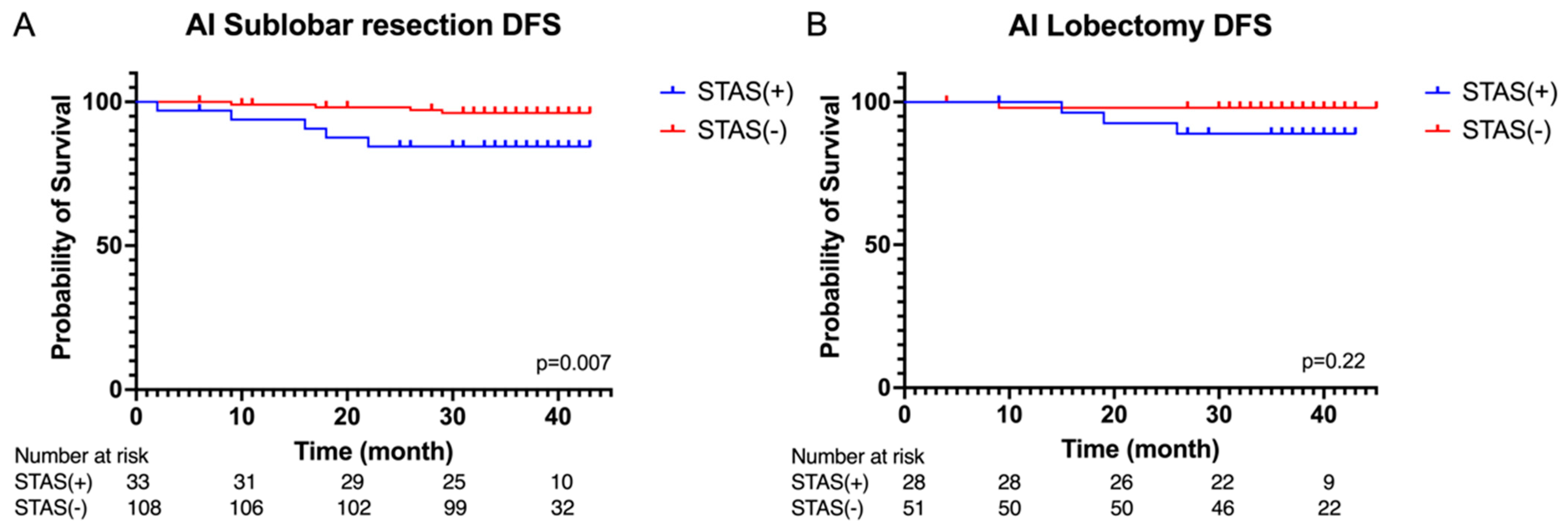
3.2. Peri-Operative Outcomes and Survival Analysis
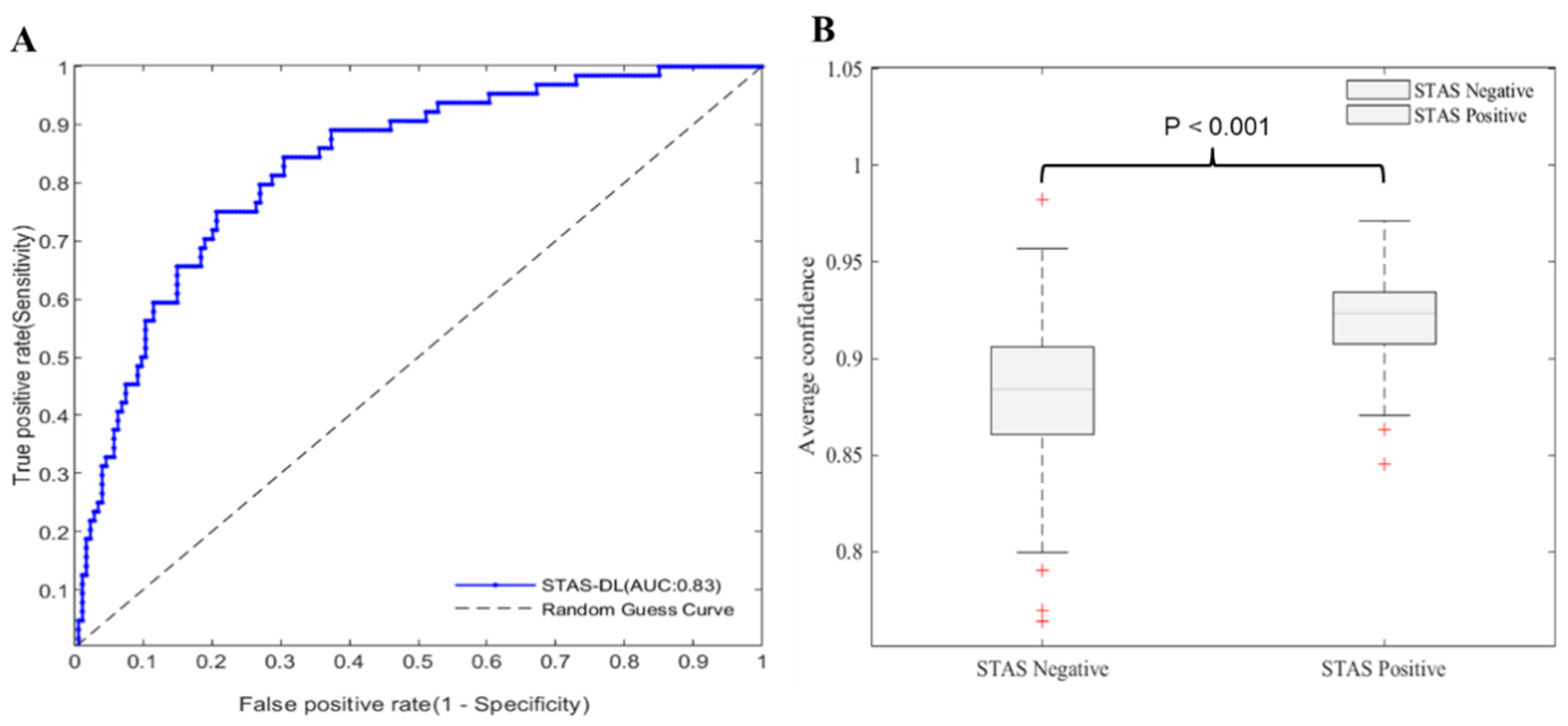
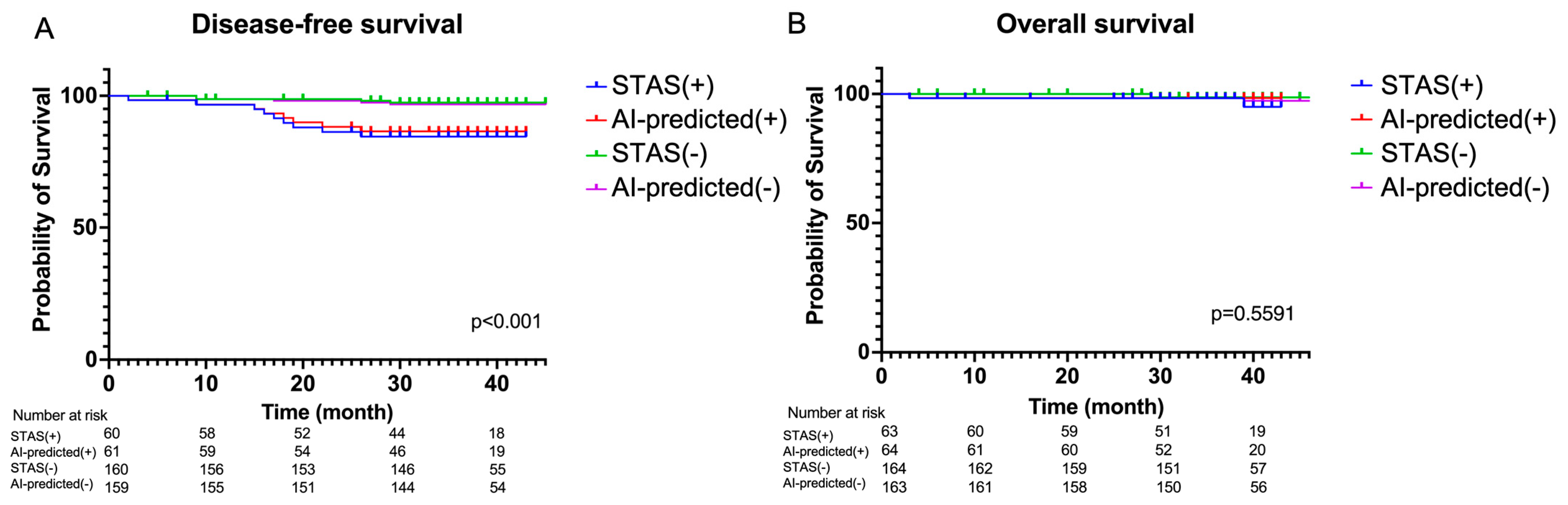
3.3. Performance of Pathological Spread through Air Space (STAS) Prediction Model and Correlation Results between Different Histological Grades
Confidence and Histological Grades
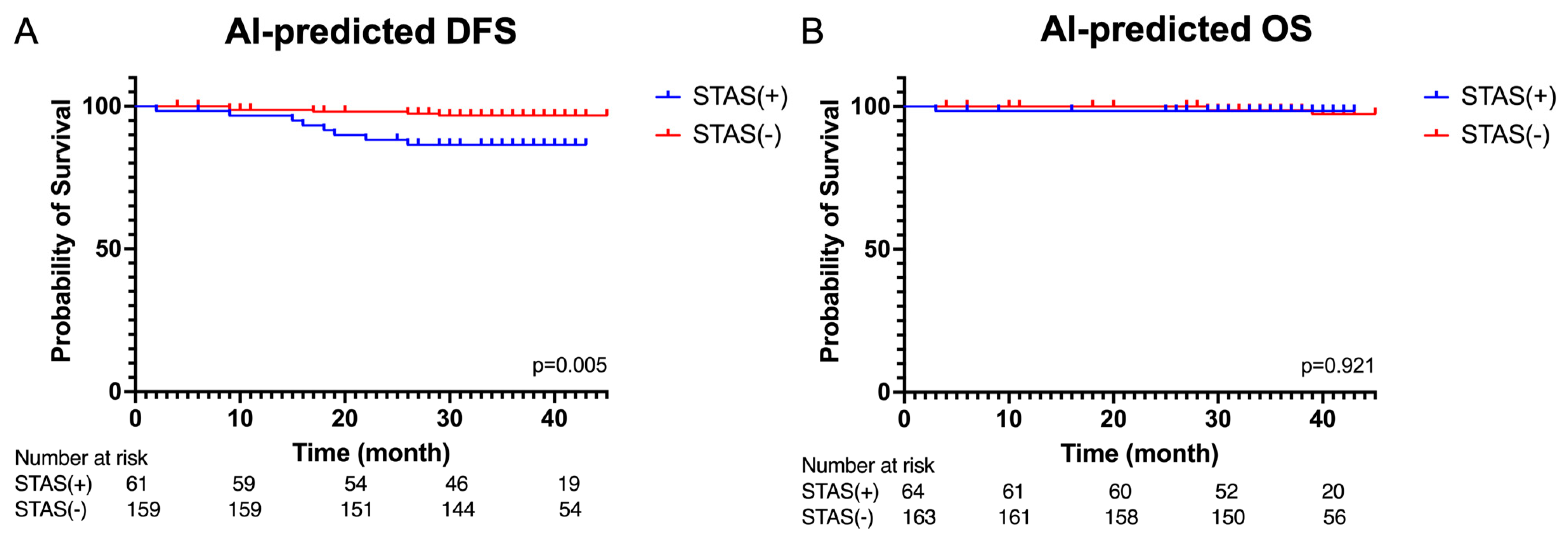
3.4. Survival Analysis Based on Artificail Intelligence (AI) Pathological Feature Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, Y.B.; Kim, H.; Mino-Kenudson, M.; Cho, S.; Kwon, H.J.; Lee, K.R.; Kwon, S.; Lee, J.; Kim, K.; Jheon, S.; et al. Tumor spread through air spaces (STAS): Prognostic significance of grading in non-small cell lung cancer. Mod. Pathol. 2021, 34, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Bassi, M.; Russomando, A.; Vannucci, J.; Ciardiello, A.; Dolciami, M.; Ricci, P.; Pernazza, A.; D’Amati, G.; Mancini Terracciano, C.; Faccini, R.; et al. Role of radiomics in predicting lung cancer spread through air spaces in a heterogeneous dataset. Transl. Lung Cancer Res. 2022, 11, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, T.; Kameda, K.; Lu, S.; Bott, M.J.; Tan, K.S.; Montecalvo, J.; Chang, J.C.; Rekhtman, N.; Jones, D.R.; Travis, W.D.; et al. Lobectomy is associated with better outcomes than sublobar resection in spread through air spaces (STAS)-positive T1 lung adenocarcinoma: A propensity score-matched analysis. J. Thorac. Oncol. 2019, 14, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Kadota, K.; Kushida, Y.; Kagawa, S.; Ishikawa, R.; Ibuki, E.; Inoue, K.; Go, T.; Yokomise, H.; Ishii, T.; Kadowaki, N.; et al. Limited resection is associated with a higher risk of locoregional recurrence than lobectomy in Stage I lung adenocarcinoma with tumor spread through air spaces. Am. J. Surg. Pathol. 2019, 43, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, A.; Tsukamoto, T.; Kiriyama, Y.; Fujita, H. Automated classification of lung cancer types from cytological images using deep convolutional neural networks. BioMed. Res. Int. 2017, 2017, 4067832. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Yang, L.; Wang, S.; Guo, L.; Huang, C.; Xie, Y.; Xiao, G. Microvessel prediction in H&E Stained Pathology Images using fully convolutional neural networks. BMC Bioinform. 2018, 19, 64. [Google Scholar] [CrossRef] [PubMed]
- Sha, L.; Osinski, B.L.; Ho, I.Y.; Tan, T.L.; Willis, C.; Weiss, H.; Beaubier, N.; Mahon, B.M.; Taxter, T.J.; Yip, S.S.F. Multi-field-of-view deep learning model predicts nonsmall cell lung cancer programmed death-ligand 1 status from whole-slide hematoxylin and eosin images. J. Pathol. Inform. 2019, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- de Haan, K.; Zhang, Y.; Zuckerman, J.E.; Liu, T.; Sisk, A.E.; Diaz, M.F.P.; Jen, K.Y.; Nobori, A.; Liou, S.; Zhang, S.; et al. Deep learning-based transformation of H&E stained tissues into special stains. Nat. Commun. 2021, 12, 4884. [Google Scholar] [CrossRef]
- Kosaraju, S.; Park, J.; Lee, H.; Yang, J.W.; Kang, M. Deep learning-based framework for slide-based histopathological image analysis. Sci. Rep. 2022, 12, 19075. [Google Scholar] [CrossRef]
- Shafi, S.; Parwani, A.V. Artificial intelligence in diagnostic pathology. Diagn. Pathol. 2023, 18, 109. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.Y.; Chao, H.S.; Chen, Y.M. Application of Artificial Intelligence in Lung Cancer. Cancers 2022, 14, 1370. [Google Scholar] [CrossRef] [PubMed]
- Ladbury, C.; Amini, A.; Govindarajan, A.; Mambetsariev, I.; Raz, D.J.; Massarelli, E.; Williams, T.; Rodin, A.; Salgia, R. Integration of artificial intelligence in lung cancer: Rise of the machine. Cell Rep. Med. 2023, 4, 100933. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Fu, C.; Tie, M.; Sham, C.W.; Ma, H. RGSB-UNet: Hybrid Deep Learning Framework for Tumour Segmentation in Digital Pathology Images. Bioengineering 2023, 10, 957. [Google Scholar] [CrossRef]
- Zhang, S.; Yuan, Z.; Zhou, X.; Wang, H.; Chen, B.; Wang, Y. VENet: Variational energy network for gland segmentation of pathological images and early gastric cancer diagnosis of whole slide images. Comput. Methods Programs Biomed. 2024, 250, 108178. [Google Scholar] [CrossRef] [PubMed]
- Salido, J.; Vallez, N.; González-López, L.; Deniz, O.; Bueno, G. Comparison of deep learning models for digital H&E staining from unpaired label-free multispectral microscopy images. Comput. Methods Programs Biomed. 2023, 235, 107528. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Fawi, M.; Brychcy, A.; Abouzid, M.; Witt, M.; Kaczmarek, E. Development and Validation of a Deep Learning Model for Histopathological Slide Analysis in Lung Cancer Diagnosis. Cancers 2024, 16, 1506. [Google Scholar] [CrossRef] [PubMed]
- Abdeltawab, H.A.; Khalifa, F.A.; Ghazal, M.A.; Cheng, L.; El-Baz, A.S.; Gondim, D.D. A deep learning framework for automated classification of histopathological kidney whole-slide images. J. Pathol. Inform. 2022, 13, 100093. [Google Scholar] [CrossRef]
- Kanavati, F.; Ichihara, S.; Tsuneki, M. A deep learning model for breast ductal carcinoma in situ classification in whole slide images. Virchows Arch. 2022, 480, 1009–1022. [Google Scholar] [CrossRef]
- Lu, W.; Toss, M.; Dawood, M.; Rakha, E.; Rajpoot, N.; Minhas, F. SlideGraph(+): Whole slide image level graphs to predict HER2 status in breast cancer. Med. Image Anal. 2022, 80, 102486. [Google Scholar] [CrossRef]
- Tsuneki, M.; Kanavati, F. Weakly supervised learning for multi-organ adenocarcinoma classification in whole slide images. PLoS ONE 2022, 17, e0275378. [Google Scholar] [CrossRef]
- Wang, C.W.; Huang, S.C.; Lee, Y.C.; Shen, Y.J.; Meng, S.I.; Gaol, J.L. Deep learning for bone marrow cell detection and classification on whole-slide images. Med. Image Anal. 2022, 75, 102270. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, Y.; Tang, D.; Ni, M.; Zheng, J.; Xu, G.; Peng, C.; Shen, S.; Zhan, Q.; Wang, X.; et al. A deep learning-based segmentation system for rapid onsite cytologic pathology evaluation of pancreatic masses: A retrospective, multicenter, diagnostic study. eBioMedicine 2022, 80, 104022. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Zhou, M.; Wang, H.; Gevaert, O.; Metaxas, D.; Zhang, S. A Large-scale Synthetic Pathological Dataset for Deep Learning-enabled Segmentation of Breast Cancer. Sci. Data 2023, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Su, F.; Zhang, W.; Liu, Y.; Chen, S.; Lin, M.; Feng, M.; Yin, J.; Tan, L.; Shen, Y. The development and validation of pathological sections based U-Net deep learning segmentation model for the detection of esophageal mucosa and squamous cell neoplasm. J. Gastrointest. Oncol. 2023, 14, 1982–1992. [Google Scholar] [CrossRef] [PubMed]
- Elazab, N.; Gab-Allah, W.A.; Elmogy, M. A multi-class brain tumor grading system based on histopathological images using a hybrid YOLO and RESNET networks. Sci. Rep. 2024, 14, 4584. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Re, C.; Rubin, D.L.; Snyder, M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016, 7, 12474. [Google Scholar] [CrossRef]
- Shim, W.S.; Yim, K.; Kim, T.J.; Sung, Y.E.; Lee, G.; Hong, J.H.; Chun, S.H.; Kim, S.; An, H.J.; Na, S.J.; et al. DeepRePath: Identifying the Prognostic Features of Early-Stage Lung Adenocarcinoma Using Multi-Scale Pathology Images and Deep Convolutional Neural Networks. Cancers 2021, 13, 3308. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cao, Y.; Li, W.; Liu, Z.; Liu, P.; Tian, X.; Sun, C.; Wang, W.; Gao, H.; Kang, S.; et al. The pathological risk score: A new deep learning-based signature for predicting survival in cervical cancer. Cancer Med. 2023, 12, 1051–1063. [Google Scholar] [CrossRef]
- Kim, P.J.; Hwang, H.S.; Choi, G.; Sung, H.J.; Ahn, B.; Uh, J.S.; Yoon, S.; Kim, D.; Chun, S.M.; Jang, S.J.; et al. A new model using deep learning to predict recurrence after surgical resection of lung adenocarcinoma. Sci. Rep. 2024, 14, 6366. [Google Scholar] [CrossRef]
- Ivanova, M.; Pescia, C.; Trapani, D.; Venetis, K.; Frascarelli, C.; Mane, E.; Cursano, G.; Sajjadi, E.; Scatena, C.; Cerbelli, B.; et al. Early Breast Cancer Risk Assessment: Integrating Histopathology with Artificial Intelligence. Cancers 2024, 16, 1981. [Google Scholar] [CrossRef]
- Lin, M.W.; Chen, L.W.; Yang, S.M.; Hsieh, M.S.; Ou, D.X.; Lee, Y.H.; Chen, J.S.; Chang, Y.C.; Chen, C.M. CT-based deep-learning model for spread-through-air-spaces prediction in ground glass-predominant lung adenocarcinoma. Ann. Surg. Oncol. 2023, 31, 1536–1545. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Yin, R.; Sun, L.; Gao, N.; Wang, X.; Zhang, L.; Zhang, Z. CT-based radiomics model to predict spread through air space in resectable lung cancer. Cancer Med. 2023, 12, 18755–18766. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, C.; Gong, J.; Wu, X.; Luo, Y.; Sun, G. A CT-based logistic regression model to predict spread through air space in lung adenocarcinoma. Quant. Imaging Med. Surg. 2020, 10, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Y.; Feng, M.; Yang, S.; Zhou, L.; Ge, D.; Lu, S.; Liu, L.; Shan, F.; Zhang, Z. Radiomics nomograms of tumors and peritumoral regions for the preoperative prediction of spread through air spaces in lung adenocarcinoma. Transl. Oncol. 2020, 13, 100820. [Google Scholar] [CrossRef] [PubMed]
- Onozato, Y.; Nakajima, T.; Yokota, H.; Morimoto, J.; Nishiyama, A.; Toyoda, T.; Inage, T.; Tanaka, K.; Sakairi, Y.; Suzuki, H.; et al. Radiomics is feasible for prediction of spread through air spaces in patients with nonsmall cell lung cancer. Sci. Rep. 2021, 11, 13526. [Google Scholar] [CrossRef]
- Liao, G.; Huang, L.; Wu, S.; Zhang, P.; Xie, D.; Yao, L.; Zhang, Z.; Yao, S.; Shanshan, L.; Wang, S.; et al. Preoperative CT-based peritumoral and tumoral radiomic features prediction for tumor spread through air spaces in clinical stage I lung adenocarcinoma. Lung Cancer 2022, 163, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Takehana, K.; Sakamoto, R.; Fujimoto, K.; Matsuo, Y.; Nakajima, N.; Yoshizawa, A.; Menju, T.; Nakamura, M.; Yamada, R.; Mizowaki, T.; et al. Peritumoral radiomics features on preoperative thin-slice CT images can predict the spread through air spaces of lung adenocarcinoma. Sci. Rep. 2022, 12, 10323. [Google Scholar] [CrossRef]
- Suh, Y.J.; Han, K.; Kwon, Y.; Kim, H.; Lee, S.; Hwang, S.H.; Kim, M.H.; Shin, H.J.; Lee, C.Y.; Shim, H.S. Computed Tomography Radiomics for Preoperative Prediction of Spread Through Air Spaces in the Early Stage of Surgically Resected Lung Adenocarcinomas. Yonsei Med. J. 2024, 65, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lyu, D.; Hu, L.; Wu, J.; Duan, S.; Zhou, T.; Tu, W.; Xiao, Y.; Fan, L.; Liu, S. CT-Based Intratumoral and Peritumoral Radiomics Nomograms for the Preoperative Prediction of Spread Through Air Spaces in Clinical Stage IA Non-small Cell Lung Cancer. J. Imaging Inform. Med. 2024, 37, 520–535. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.; Jiang, C.; Kang, W.; Pan, Y.; Tang, X.; Luo, Y.; Gong, J. CT-Based Super-Resolution Deep Learning Models with Attention Mechanisms for Predicting Spread Through Air Spaces of Solid or Part-Solid Lung Adenocarcinoma. Acad. Radiol. 2024; in press. [Google Scholar] [CrossRef]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO classification of lung tumors: Impact of advances since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef]
- Moreira, A.L.; Ocampo, P.S.S.; Xia, Y.; Zhong, H.; Russell, P.A.; Minami, Y.; Cooper, W.A.; Yoshida, A.; Bubendorf, L.; Papotti, M.; et al. A grading system for invasive pulmonary adenocarcinoma: A proposal from the International Association for the Study of Lung Cancer pathology committee. J. Thorac. Oncol. 2020, 15, 1599–1610. [Google Scholar] [CrossRef]
- Redmon, J. Darknet: Open-Source Neural Networks in C. Available online: https://pjreddie.com/darknet/ (accessed on 22 October 2022).
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Jocher, G. Yolov5, GitHub. Available online: https://github.com/ultralytics/yolov5 (accessed on 10 November 2022).
- Sharma, A. Training the YOLOv5 Object Detector on a Custom Dataset. Available online: https://pyimg.co/fq0a3 (accessed on 22 October 2022).
- Jocher, G.; Chaurasia, A.; Stoken, A.; Borovec, J.; Kwon, Y.; Michael, K.; Xie, T.; Fang, J.; Imyhxy; Lorna; et al. ultralytics/yolov5: v7.0—YOLOv5 SOTA Realtime Instance Segmentation (v7.0). Zenodo. Available online: https://ui.adsabs.harvard.edu/abs/2022zndo...7347926J/abstract (accessed on 22 November 2022).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2016, arXiv:1512.03385. [Google Scholar]
- Jiang, C.; Luo, Y.; Yuan, J.; You, S.; Chen, Z.; Wu, M.; Wang, G.; Gong, J. CT-based radiomics and machine learning to predict spread through air space in lung adenocarcinoma. Eur. Radiol. 2020, 30, 4050–4057. [Google Scholar] [CrossRef]
- Jin, W.; Shen, L.; Tian, Y.; Zhu, H.; Zou, N.; Zhang, M.; Chen, Q.; Dong, C.; Yang, Q.; Jiang, L.; et al. Improving the prediction of spreading through air spaces (STAS) in primary lung cancer with a dynamic dual-delta hybrid machine learning model: A multicenter cohort study. Biomark. Res. 2023, 11, 102. [Google Scholar] [CrossRef]
- Dai, C.; Xie, H.; Su, H.; She, Y.; Zhu, E.; Fan, Z.; Zhou, F.; Ren, Y.; Xie, D.; Zheng, H.; et al. Tumor Spread through Air Spaces Affects the Recurrence and Overall Survival in Patients with Lung adenocarcinoma >2 to 3 cm. J. Thorac. Oncol. 2017, 12, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Onozato, M.L.; Kovach, A.E.; Yeap, B.Y.; Morales-Oyarvide, V.; Klepeis, V.E.; Tammireddy, S.; Heist, R.S.; Mark, E.J.; Dias-Santagata, D.; Iafrate, A.J.; et al. Tumor islands in resected early-stage lung adenocarcinomas are associated with unique clinicopathologic and molecular characteristics and worse prognosis. Am. J. Surg. Pathol. 2013, 37, 287–294. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Warth, A.; Muley, T.; Kossakowski, C.A.; Goeppert, B.; Schirmacher, P.; Dienemann, H.; Weichert, W. Prognostic impact of intra-alveolar tumor spread in pulmonary adenocarcinoma. Am. J. Surg. Pathol. 2015, 39, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Kadota, K.; Nitadori, J.I.; Sima, C.S.; Ujiie, H.; Rizk, N.P.; Jones, D.R.; Adusumilli, P.S.; Travis, W.D. Tumor spread through air spaces is an important pattern of invasion and impacts the frequency AND location of recurrences after limited resection for small Stage I lung adenocarcinomas. J. Thorac. Oncol. 2015, 10, 806–814. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, D.; She, Y.; Wang, T.; Xie, H.; Li, J.; Jiang, G.; Chen, Y.; Zhang, L.; Xie, D.; Chen, C. Radiomics-based prediction for tumour spread through air spaces in stage i lung adenocarcinoma using machine learning. Eur. J. Cardiothorac. Surg. 2020, 58, 51–58. [Google Scholar] [CrossRef]
- Chen, L.W.; Lin, M.W.; Hsieh, M.S.; Yang, S.M.; Wang, H.J.; Chen, Y.C.; Chen, H.Y.; Hu, Y.H.; Lee, C.E.; Chen, J.S.; et al. Radiomic values from high-grade subtypes to predict spread through air spaces in lung adenocarcinoma. Ann. Thorac. Surg. 2022, 114, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Liang, C.; Yin, K.; Fang, J.; Chen, B.; Wang, Z.; Lan, X.; Zhang, J. 3D convolutional neural network model from contrast-enhanced CT to predict spread through air spaces in non-small cell lung cancer. Diagn. Interv. Imaging 2022, 103, 535–544. [Google Scholar] [CrossRef]

| N = 227 | |
|---|---|
| Age (year) | 61.1 ± 10.5 |
| Gender | |
| Female | 147 (64.8%) |
| Male | 80 (35.2%) |
| Smoking history | |
| Smoker | 38 (16.7%) |
| Non-smoker | 189 (83.3%) |
| Tumor size (cm) | 1.7 ± 1.0 |
| T stage | |
| T1a | 141(62.1%) |
| T1b | 31 (13.7%) |
| T2a | 47 (20.7%) |
| Location | |
| RUL | 82 (36.1%) |
| RML | 19 (8.4%) |
| RLL | 34 (15.0%) |
| LUL | 58 (25.6%) |
| LLL | 32 (14.1%) |
| Surgical procedure | |
| Lobectomy | 82 (36.1%) |
| Sublobar resection | 145 (63.9%) |
| Post-operative hospital stay (days) | 4.1 ± 4.9 |
| Complication | |
| Chylothorax | 2 (0.9%) |
| Air leakage | 1 (0.4%) |
| Atrial fibrillation | 1 (0.4%) |
| STAS | |
| Present | 63 (27.7%) |
| Absent | 164 (72.3%) |
| Histological grading | |
| 1 | 60 (26.4%) |
| 2 | 126 (55.5%) |
| 3 | 32 (14.1%) |
| Methods | Accuracy (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | AUC (%) |
|---|---|---|---|---|---|---|
| Proposed model | 72 (163/227) | 81 (52/64) | 68 (111/163) | 50 (52/104) | 90 (111/123) | 83 |
| Histological Grades † | Number of Detection Candidates | Number of Strong-Confidence Candidates * (%) | Number of Low-Confidence Candidates * (%) | p-Value ** |
|---|---|---|---|---|
| Grade 1 (60/227) | 9498 | 21 (1962/9498) | 79 (7536/9498) | <0.001 |
| Grade 2 (126/227) | 35,296 | 36 (12,842/35,296) | 64 (22,454/35,296) | <0.001 |
| Grade 3 (32/227) | 11,698 | 51 (5934/11,698) | 49 (5764/11,698) | reference |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ou, D.-X.; Lu, C.-W.; Chen, L.-W.; Lee, W.-Y.; Hu, H.-W.; Chuang, J.-H.; Lin, M.-W.; Chen, K.-Y.; Chiu, L.-Y.; Chen, J.-S.; et al. Deep Learning Analysis for Predicting Tumor Spread through Air Space in Early-Stage Lung Adenocarcinoma Pathology Images. Cancers 2024, 16, 2132. https://doi.org/10.3390/cancers16112132
Ou D-X, Lu C-W, Chen L-W, Lee W-Y, Hu H-W, Chuang J-H, Lin M-W, Chen K-Y, Chiu L-Y, Chen J-S, et al. Deep Learning Analysis for Predicting Tumor Spread through Air Space in Early-Stage Lung Adenocarcinoma Pathology Images. Cancers. 2024; 16(11):2132. https://doi.org/10.3390/cancers16112132
Chicago/Turabian StyleOu, De-Xiang, Chao-Wen Lu, Li-Wei Chen, Wen-Yao Lee, Hsiang-Wei Hu, Jen-Hao Chuang, Mong-Wei Lin, Kuan-Yu Chen, Ling-Ying Chiu, Jin-Shing Chen, and et al. 2024. "Deep Learning Analysis for Predicting Tumor Spread through Air Space in Early-Stage Lung Adenocarcinoma Pathology Images" Cancers 16, no. 11: 2132. https://doi.org/10.3390/cancers16112132
APA StyleOu, D.-X., Lu, C.-W., Chen, L.-W., Lee, W.-Y., Hu, H.-W., Chuang, J.-H., Lin, M.-W., Chen, K.-Y., Chiu, L.-Y., Chen, J.-S., Chen, C.-M., & Hsieh, M.-S. (2024). Deep Learning Analysis for Predicting Tumor Spread through Air Space in Early-Stage Lung Adenocarcinoma Pathology Images. Cancers, 16(11), 2132. https://doi.org/10.3390/cancers16112132






