Transforming a Large-Scale Prostate Cancer Outcomes Dataset to the OMOP Common Data Model—Experiences from a Scientific Data Holder’s Perspective
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
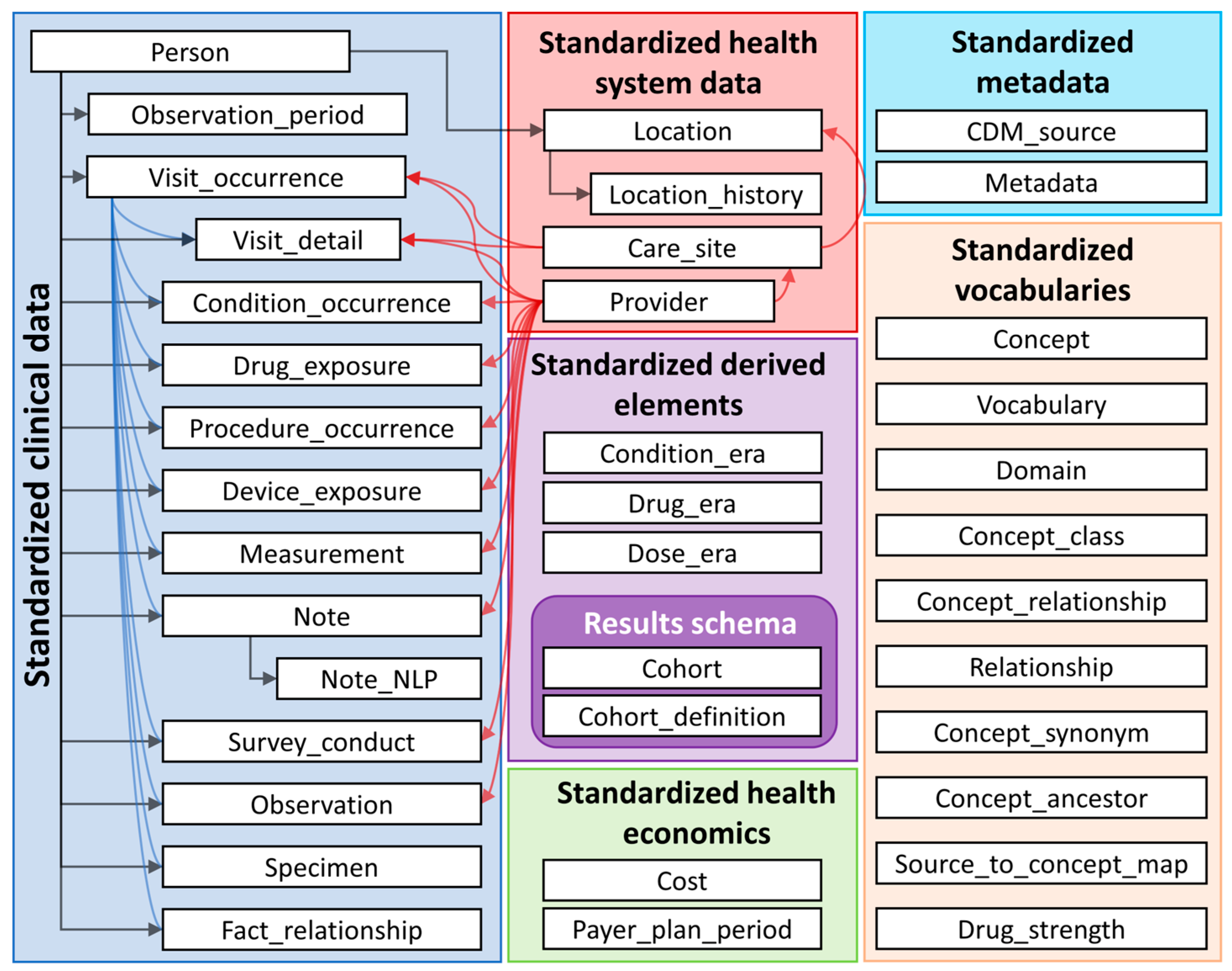
2.1. The OMOP CDM (Version 5.4)
- A common, person-centric structured database framework based on a relational database approach with dedicated data tables (i.e., person table, measurement table, condition table, provider table, cf. Figure 1) to harmonise the data structure.
- A huge variety of standardised vocabularies (i.e., SNOMED, LOINC, etc.) which serve as a reference for all unique concepts that are mandatory to use within the OMOP CDM. This results in a comprehensive repository of concept IDs which are unique within the OMOP CDM and enable one to distinctly link information from the source data to an OMOP CDM concept. Concept IDs are specific to OMOP CDM, and codes from the integrated vocabularies are mapped to OMOP CDM concept IDs to ensure the uniqueness of the identifier. The online repository https://athena.ohdsi.org (accessed on 23 May 2024) offers an extensive overview of all concept IDs and vocabularies used for OMOP CDM.
- A technologically independent data model that is applicable to any relational database solution (i.e., SQL, Oracle, MariaDB).
2.2. The PCO Study Dataset
- Health care provider’s information: unique identifier, localisation (country, federal state);
- Patient’s sociodemographic information: unique identifier, age, citizenship (self-reported and documented by health care provider), insurance status (self-reported), highest school degree (self-reported), gender, date of death (if applicable);
- Patient’s case anamnesis: comorbidities (according to ICHOM standard dataset), previous cancer diagnosis;
- General prostate cancer case information: risk classification according to German S3-guideline [25], diagnosis date, cTNM, pTNM, Gleason scores at diagnosis and after surgery, PSA value at diagnosis and after surgery, ICD10 code, cores used for diagnosis (amount, percentages involved);
- Prostate cancer case treatment information: type of treatment (active surveillance, watchful waiting, radical prostatectomy, radiation, androgen deprivation treatment, other local and systemically treatment options), surgical approach (nerve-sparing, open/robotic/laparoscopic, lymphadenectomy incl. amount of taken nodes and amount of involved nodes), surgical revision procedures, date of treatment procedures, complications (Clavien–Dindo, CTCAE), margins, fraction means and amount of radiation;
- Prostate cancer case procedures’ information: tumour board dates, psycho-oncological counselling, social counselling;
- Prostate cancer case follow-up information: biochemical recurrence, number and years of follow-up, vital status, local recurrence and other malignancies after prostate cancer;
- Patient-reported outcomes: EPIC-26 scores and single items before and one year after diagnosis [18].
2.3. German Cancer Society’s Data Transformation Approach
- Conceptual vocabulary mapping of each applicable source data field to an OMOP CDM concept and data table(s) using OMOP vocabularies;
- Developing and testing an efficient ETL (extract-transform-load) process based on a mock dataset within a VM (virtual machine) environment (ETL development and testing);
- Setting up of a production VM that hosts the final PostgreSQL database with data using the OMOP CDM format.
2.3.1. Vocabulary Mapping
2.3.2. ETL Development and Testing
2.3.3. Production VM with Final PostgreSQL Database
3. Results
3.1. Source Dataset Specific Characteristics
3.2. ETL Process
- -
- A new and unique ID for the visit is generated (visit_occurence_id);
- -
- The kind of visit is set to “inpatient visit” (conceptID: 9201, visit_concept_ID);
- -
- The visit_start_date and visit_end_date are both set on the same date (SurgeryDate), since no end date of the surgery is available in the PCO dataset and surgery is most likely finished on the same day as it started;
- -
- Provenance of visit entry is set to “EHR” (conceptID 32187, visit_type_concept_id);
- -
- The “care_site_id” is filled with the identifier of the care_site associated with the value of the source field “RegNr” (pseudonym of health care providers within the source dataset) and the fieldname “SurgeryDate” is copied to visit_source_value.
3.3. Quality Assurance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rosenbloom, S.T.; Carroll, R.J.; Warner, J.L.; Matheny, M.E.; Denny, J.C. Representing Knowledge Consistently Across Health Systems. Yearb. Med. Inform 2017, 26, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Belenkaya, R.; Gurley, M.J.; Golozar, A.; Dymshyts, D.; Miller, R.T.; Williams, A.E.; Ratwani, S.; Siapos, A.; Korsik, V.; Warner, J.; et al. Extending the OMOP Common Data Model and Standardized Vocabularies to Support Observational Cancer Research. JCO Clin. Cancer Inform. 2021, 5, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Schlomer, B.J.; Copp, H.L. Secondary Data Analysis of Large Data Sets in Urology: Successes and Errors to Avoid. J. Urol. 2014, 191, 587–596. [Google Scholar] [CrossRef] [PubMed]
- ICHOM International Consortium for Health Outcomes Measurement. Available online: https://www.ichom.org/ (accessed on 19 November 2022).
- Kirkham, J.J.; Gorst, S.; Altman, D.G.; Blazeby, J.M.; Clarke, M.; Tunis, S.; Williamson, P.R.; COS-STAP Group. Core Outcome Set-STAndardised Protocol Items: The COS-STAP Statement. Trials 2019, 20, 116. [Google Scholar] [CrossRef] [PubMed]
- Kirkham, J.J.; Gorst, S.; Altman, D.G.; Blazeby, J.M.; Clarke, M.; Devane, D.; Gargon, E.; Moher, D.; Schmitt, J.; Tugwell, P.; et al. Core Outcome Set–STAndards for Reporting: The COS-STAR Statement. PLOS Med. 2016, 13, e1002148. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, S.; Williamson, P.R. The Need for Core Outcome Sets in Urological Cancer Research. Transl. Androl. Urol. 2021, 10, 2832–2835. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, I.; Eckert, M.; Hutchinson, A.D.; Marker, J.; Corsini, N. Core Outcome Sets in Cancer and Their Approaches to Identifying and Selecting Patient-Reported Outcome Measures: A Systematic Review. J. Patient Rep. Outcomes 2020, 4, 77. [Google Scholar] [CrossRef] [PubMed]
- Stang, P.E.; Ryan, P.B.; Racoosin, J.A.; Overhage, J.M.; Hartzema, A.G.; Reich, C.; Welebob, E.; Scarnecchia, T.; Woodcock, J. Advancing the Science for Active Surveillance: Rationale and Design for the Observational Medical Outcomes Partnership. Ann. Intern Med. 2010, 153, 600–606. [Google Scholar] [CrossRef] [PubMed]
- The Book of OHDSI; Observational Health Data Sciences and Informatics Informatics , Ed.; 2021. [Google Scholar]
- Unberath, P.; Prokosch, H.U.; Gründner, J.; Erpenbeck, M.; Maier, C.; Christoph, J. EHR-Independent Predictive Decision Support Architecture Based on OMOP. Appl. Clin. Inform. 2020, 11, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Wood, W.A.; Marks, P.; Plovnick, R.M.; Hewitt, K.; Neuberg, D.S.; Walters, S.; Dolan, B.K.; Tucker, E.A.; Abrams, C.S.; Thompson, A.A.; et al. ASH Research Collaborative: A Real-World Data Infrastructure to Support Real-World Evidence Development and Learning Healthcare Systems in Hematology. Blood Adv. 2021, 5, 5429–5438. [Google Scholar] [CrossRef] [PubMed]
- H, J.; Sc, Y.; Sy, K.; Si, S.; Jl, W.; R, B.; Rw, P. Characterizing the Anticancer Treatment Trajectory and Pattern in Patients Receiving Chemotherapy for Cancer Using Harmonized Observational Databases: Retrospective Study. JMIR Med. Inform. 2021, 9, e25035. [Google Scholar] [CrossRef] [PubMed]
- Voss, E.A.; Blacketer, C.; van Sandijk, S.; Moinat, M.; Kallfelz, M.; van Speybroeck, M.; Prieto-Alhambra, D.; Schuemie, M.J.; Rijnbeek, P.R. European Health Data & Evidence Network-Learnings from Building out a Standardized International Health Data Network. J. Am. Med. Inform. Assoc. 2023, 31, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Heads of Medicines Agency; European Medicines Agency. HMA-EMA Joint Big Data Taskforce Phase II Report: Evolving Data-Driven Regulation; European Medicines Agency: Amsterdam, The Netherlands, 2024.
- Omar, M.I.; Roobol, M.J.; Ribal, M.J.; Abbott, T.; Agapow, P.-M.; Araujo, S.; Asiimwe, A.; Auffray, C.; Balaur, I.; Beyer, K.; et al. Introducing PIONEER: A Project to Harness Big Data in Prostate Cancer Research. Nat. Rev. Urol. 2020, 17, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Gandaglia, G.; Pellegrino, F.; Golozar, A.; De Meulder, B.; Abbott, T.; Achtman, A.; Imran Omar, M.; Alshammari, T.; Areia, C.; Asiimwe, A.; et al. Clinical Characterization of Patients Diagnosed with Prostate Cancer and Undergoing Conservative Management: A PIONEER Analysis Based on Big Data. Eur. Urol. 2024, 85, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, K.M.; Wei, J.T.; Dunn, R.L.; Sanda, M.G. Development and Validation of an Abbreviated Version of the Expanded Prostate Cancer Index Composite Instrument for Measuring Health-Related Quality of Life among Prostate Cancer Survivors. Urology 2010, 76, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.M.; Millar, J.L.; Moore, C.M.; Lewis, J.D.; Huland, H.; Sampurno, F.; Connor, S.E.; Villanti, P.; Litwin, M.S. Cohort Profile: The TrueNTH Global Registry—An International Registry to Monitor and Improve Localised Prostate Cancer Health Outcomes. BMJ Open 2017, 7, e017006. [Google Scholar] [CrossRef] [PubMed]
- Sibert, N.T.; Pfaff, H.; Breidenbach, C.; Wesselmann, S.; Roth, R.; Feick, G.; Carl, G.; Dieng, S.; Gaber, A.A.; Blana, A.; et al. Variation across Operating Sites in Urinary and Sexual Outcomes after Radical Prostatectomy in Localized and Locally Advanced Prostate Cancer. World J. Urol. 2022, 40, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, C.; Roth, R.; Carl, G.; Feick, G.; Oesterle, A.; Hinkel, A.; Steiner, T.; Brock, M.; Kaftan, B.; Borowitz, R.; et al. A Multicenter Paper-Based and Web-Based System for Collecting Patient-Reported Outcome Measures in Patients Undergoing Local Treatment for Prostate Cancer: First Experiences. J. Patient Rep. Outcomes 2020, 4, 56. [Google Scholar] [CrossRef] [PubMed]
- ICHOM. International Consortium for Health Outcomes Measurement Localized Prostate Cancer Data Collection Reference Guide 2015; ICHOM: Boston, MA, USA, 2015. [Google Scholar]
- Overhage, J.M.; Ryan, P.B.; Reich, C.G.; Hartzema, A.G.; Stang, P.E. Validation of a Common Data Model for Active Safety Surveillance Research. J. Am. Med. Inform. Assoc. 2012, 19, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Hripcsak, G.; Duke, J.D.; Shah, N.H.; Reich, C.G.; Huser, V.; Schuemie, M.J.; Suchard, M.A.; Park, R.W.; Wong, I.C.K.; Rijnbeek, P.R.; et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud. Health Technol. Inform. 2015, 216, 574–578. [Google Scholar] [PubMed]
- Leitlinienprogramm Onkologie (Deutsche Krebsgesellschaft, Deutsche Krebshilfe, AWMF) S3-Leitlinie Prostatakarzinom. Langversion 6.2; AWMF-Registernummer: 043/022OL, 2021; AWMF: Frankfurt, Germany, 2021.
- Observational Health Data Sciences and Informatics (OHDSI). OHDSI/WhiteRabbit 2024; Columbia University: New York, NY, USA, 2024. [Google Scholar]
- GitHub. OHDSI/Usagi 2024; GitHub: San Francisco, NY, USA, 2024. [Google Scholar]
- GitHub. EHDEN/CdmInspection 2024; GitHub: San Francisco, NY, USA, 2024. [Google Scholar]
- Observational Health Data Sciences and Informatics (OHDSI). OHDSI/Atlas 2024; Columbia University: New York, NY, USA, 2024. [Google Scholar]
- GitHub. OHDSI/Hades 2024; GitHub: San Francisco, NY, USA, 2024. [Google Scholar]
- Sibert, N.T.; Dieng, S.; Oesterle, A.; Feick, G.; Carl, G.; Steiner, T.; Minner, J.; Roghmann, F.; Kaftan, B.; Zengerling, F.; et al. Psychometric Validation of the German Version of the EPIC-26 Questionnaire for Patients with Localized and Locally Advanced Prostate Cancer. World J. Urol. 2021, 39, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, P.; Ong, R.; Davydov, A.; Orlova, A.; Solovyev, P.; Sun, H.; Wetherill, G.; Brand, M.; Didden, E.-M. Standardizing Registry Data to the OMOP Common Data Model: Experience from Three Pulmonary Hypertension Databases. BMC Med. Res. Methodol. 2021, 21, 238. [Google Scholar] [CrossRef] [PubMed]
- Carus, J.; Nürnberg, S.; Ückert, F.; Schlüter, C.; Bartels, S. Mapping Cancer Registry Data to the Episode Domain of the Observational Medical Outcomes Partnership Model (OMOP). Appl. Sci. 2022, 12, 4010. [Google Scholar] [CrossRef]
- Kim, H.; Choi, J.; Jang, I.; Quach, J.; Ohno-Machado, L. Feasibility of Representing Data from Published Nursing Research Using the OMOP Common Data Model. AMIA Annu. Symp. Proc. 2017, 2016, 715–723. [Google Scholar] [PubMed]
- Di Maio, M.; Basch, E.; Denis, F.; Fallowfield, L.J.; Ganz, P.A.; Howell, D.; Kowalski, C.; Perrone, F.; Stover, A.M.; Sundaresan, P.; et al. The Role of Patient-Reported Outcome Measures in the Continuum of Cancer Clinical Care: ESMO Clinical Practice Guideline. Ann. Oncol. 2022, 33, 878–892. [Google Scholar] [CrossRef] [PubMed]
- Donovan, J.L.; Hamdy, F.C.; Lane, J.A.; Mason, M.; Metcalfe, C.; Walsh, E.; Blazeby, J.M.; Peters, T.J.; Holding, P.; Bonnington, S.; et al. Patient-Reported Outcomes after Monitoring, Surgery, or Radiotherapy for Prostate Cancer. N. Engl. J. Med. 2016, 375, 1425–1437. [Google Scholar] [CrossRef] [PubMed]
- Al Hussein Al Awamlh, B.; Wallis, C.J.D.; Penson, D.F.; Huang, L.-C.; Zhao, Z.; Conwill, R.; Talwar, R.; Morgans, A.K.; Goodman, M.; Hamilton, A.S.; et al. Functional Outcomes After Localized Prostate Cancer Treatment. JAMA 2024, 331, 302. [Google Scholar] [CrossRef] [PubMed]
- Rajwa, P.; Borkowetz, A.; Abbott, T.; Alberti, A.; Bjartell, A.; Brash, J.T.; Campi, R.; Chilelli, A.; Conover, M.; Constantinovici, N.; et al. Research Protocol for an Observational Health Data Analysis on the Adverse Events of Systemic Treatment in Patients with Metastatic Hormone-Sensitive Prostate Cancer: Big Data Analytics Using the PIONEER Platform. Eur. Urol. Open Sci. 2024, 63, 81–88. [Google Scholar] [CrossRef]
- Kroes, J.A.; Bansal, A.T.; Berret, E.; Christian, N.; Kremer, A.; Alloni, A.; Gabetta, M.; Marshall, C.; Wagers, S.; Djukanovic, R.; et al. Blueprint for Harmonising Unstandardised Disease Registries to Allow Federated Data Analysis: Prepare for the Future. ERJ Open Res. 2022, 8, 00168–2022. [Google Scholar] [CrossRef]
- Raab, R.; Küderle, A.; Zakreuskaya, A.; Stern, A.D.; Klucken, J.; Kaissis, G.; Rueckert, D.; Boll, S.; Eils, R.; Wagener, H.; et al. Federated Electronic Health Records for the European Health Data Space. Lancet Digit. Health 2023, 5, e840–e847. [Google Scholar] [CrossRef]
| Tablename from OMOP CDM | Count | Number of Persons |
|---|---|---|
| observation | 1,507,707 | 49,300 |
| condition_occurrence | 862,669 | 49,286 |
| measurement | 801,580 | 49,300 |
| procedure_occurrence | 227,540 | 49,300 |
| observation_period | 175,675 | 49,300 |
| visit_occurrence | 65,098 | 48,358 |
| person | 49,300 | 49,300 |
| death | 721 | 721 |
| care_site | 148 | NA |
| location | 38 | NA |
| provider | 0 | NA |
| specimen | 0 | 0 |
| dose_era | 0 | 0 |
| device_exposure | 0 | 0 |
| visit_detail | 0 | 0 |
| drug_era | 0 | 0 |
| condition_era | 0 | 0 |
| cost | 0 | NA |
| note | 0 | 0 |
| drug_exposure | 0 | 0 |
| payer_plan_period | 0 | 0 |
| Topic | Definition |
|---|---|
| period | Diagnosis, pre-treatment PCO survey, treatment period |
| mapping condition | SurgeryDate is not null or empty |
| visit_occurrence_id | Generate a unique visit_occurrence_id |
| person_id | Find the person_id in the person table where person_source_value = PatientID |
| visit_concept_id | Use the concept_id 9201 “Inpatient Visit” |
| visit_start_date | Copy paste the date SurgeryDate |
| visit_start_datetime | |
| visit_end_date | Copy paste the date SurgeryDate |
| visit_end_datetime | |
| visit_type_concept_id | Use the concept_id 32817 “EHR” |
| provider_id | |
| care_site_id | Find the care_site_id in the care_site table where case_site_source_value is the same as the current value of RegNr |
| visit_source_value | Copy paste the source fieldname “SurgeryDate” |
| visit_source_concept_id | |
| admitted_from_concept_id | |
| admitted_from_value | |
| discharge_to_concept_id | |
| discharge_to_source_value | |
| preceding_visit_occurence_id |
| Topic | Definition |
|---|---|
| period | Diagnosis, pre-treatment PCO survey, treatment period |
| mapping condition | PreUrinaryFunction is not null or empty and PreDate is not null or empty |
| observation_id | Generate a unique observation_id |
| person_id | Find the person_id in the person table where person_source_value = PatientID |
| observation_concept_id | Use concept_id 36684305 “Assessment score” |
| observation_date | Copy paste the date PreDate |
| observation_datetime | |
| observation_type_concept_id | Use 32883 “Survey” |
| value_as_number | Copy paste the value of PreUrinaryFunction |
| value_as_string | |
| value_as_concept_id | |
| qualifier_concept_id | Use the concept_id associated to the source fieldname “PreUrinaryFunction” |
| unit_concept_id | |
| provider_id | |
| visit_occurence_id | |
| visit_detail_id | |
| observation_source_value | Copy paste the fieldname PreUrinaryFunction |
| observation_source_concept_id | |
| unit_source_value | |
| qualifier_source_value | |
| value_source_value | Copy paste the fieldname PreUrinaryFunction |
| observation_event_id | |
| obs_event_field_concept_id |
| Characteristic | Result Source Dataset (with Multiple Cases Excluded) | Result OMOPed Dataset | Reason for Discrepancy |
|---|---|---|---|
| number of patients with cM0 | 49,300 | 49,300 | - |
| number of patients with pM0 | 42,925 | 42,875 | Additional mapping rule: SurgeryDate must be unequal 0 |
| number of patients with Clavien–Dindo Grade I | 1931 | 1931 | - |
| number of patients with lymphadenectomy | 38,036 | 38,036 | - |
| minimum of year of birth | na | 1932 | Birth year is not a data field of the source dataset, but is calculated for the OMOPed dataset by subtracting the age at diagnosis from year of diagnosis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sibert, N.T.; Soff, J.; La Ferla, S.; Quaranta, M.; Kremer, A.; Kowalski, C. Transforming a Large-Scale Prostate Cancer Outcomes Dataset to the OMOP Common Data Model—Experiences from a Scientific Data Holder’s Perspective. Cancers 2024, 16, 2069. https://doi.org/10.3390/cancers16112069
Sibert NT, Soff J, La Ferla S, Quaranta M, Kremer A, Kowalski C. Transforming a Large-Scale Prostate Cancer Outcomes Dataset to the OMOP Common Data Model—Experiences from a Scientific Data Holder’s Perspective. Cancers. 2024; 16(11):2069. https://doi.org/10.3390/cancers16112069
Chicago/Turabian StyleSibert, Nora Tabea, Johannes Soff, Sebastiano La Ferla, Maria Quaranta, Andreas Kremer, and Christoph Kowalski. 2024. "Transforming a Large-Scale Prostate Cancer Outcomes Dataset to the OMOP Common Data Model—Experiences from a Scientific Data Holder’s Perspective" Cancers 16, no. 11: 2069. https://doi.org/10.3390/cancers16112069
APA StyleSibert, N. T., Soff, J., La Ferla, S., Quaranta, M., Kremer, A., & Kowalski, C. (2024). Transforming a Large-Scale Prostate Cancer Outcomes Dataset to the OMOP Common Data Model—Experiences from a Scientific Data Holder’s Perspective. Cancers, 16(11), 2069. https://doi.org/10.3390/cancers16112069





