GAS-Luc2 Reporter Cell Lines for Immune Checkpoint Drug Screening in Solid Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Surface Protein Profiling via Flow Cytometry
2.3. Plasmid Transduction and Single Cell Cloning
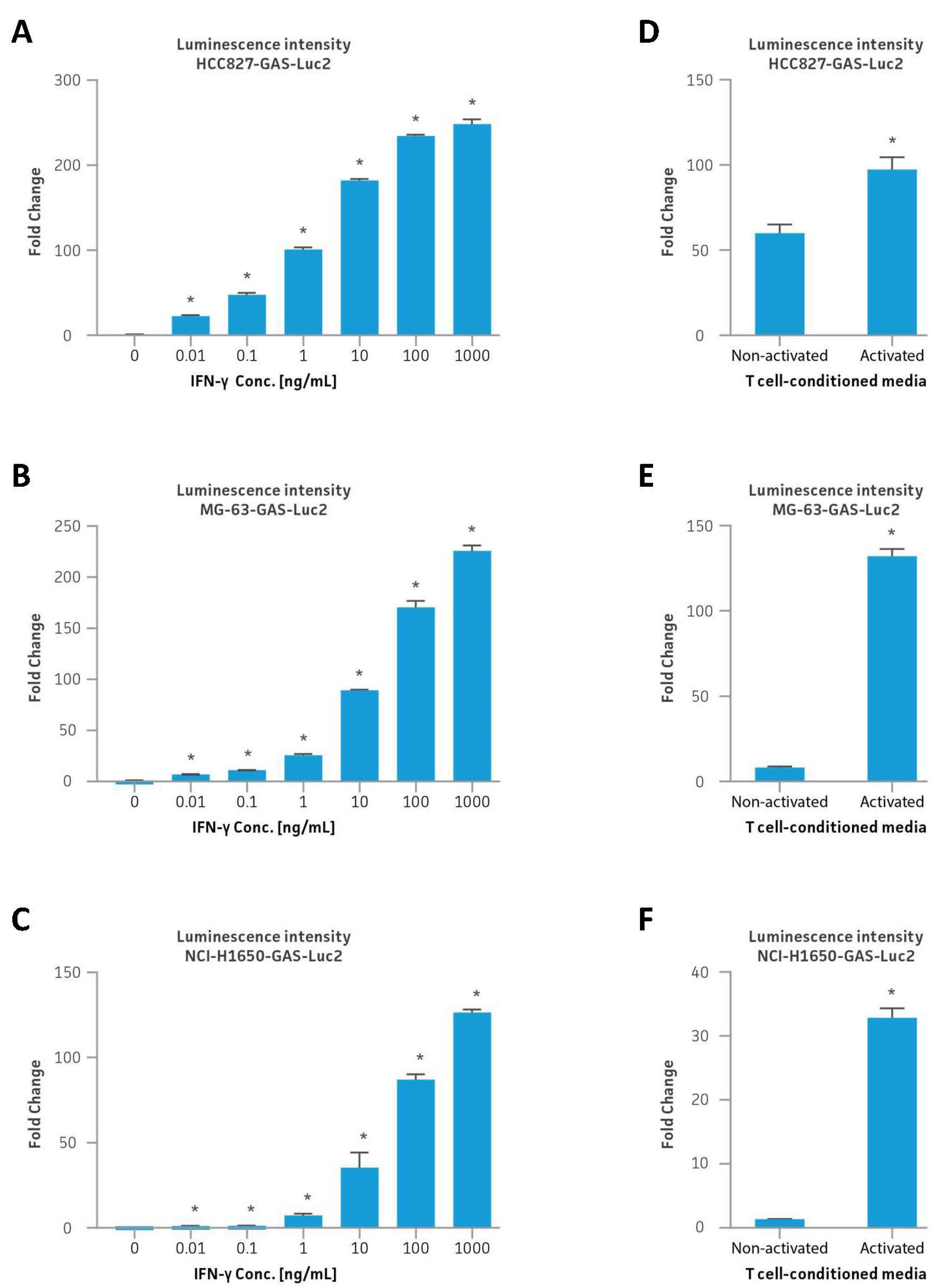
2.4. Interferon Gamma Stimulation
2.5. T Cell-Conditioned Media Stimulation
2.6. Cancer Cell and Immune Cell Co-Culture
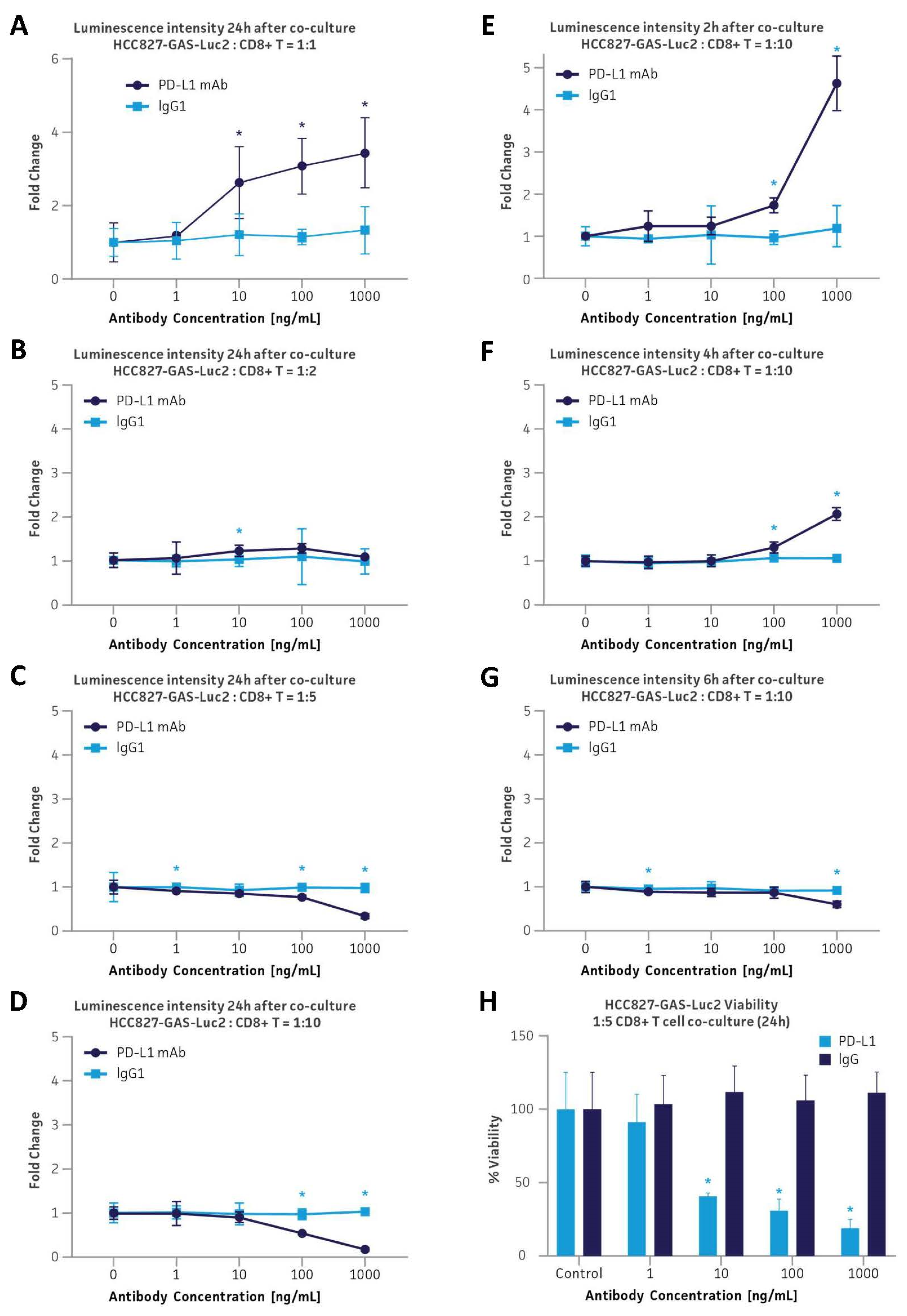
2.7. CD8+ Cytotoxic T Cell Co-Culture on Cancer Cell Viability
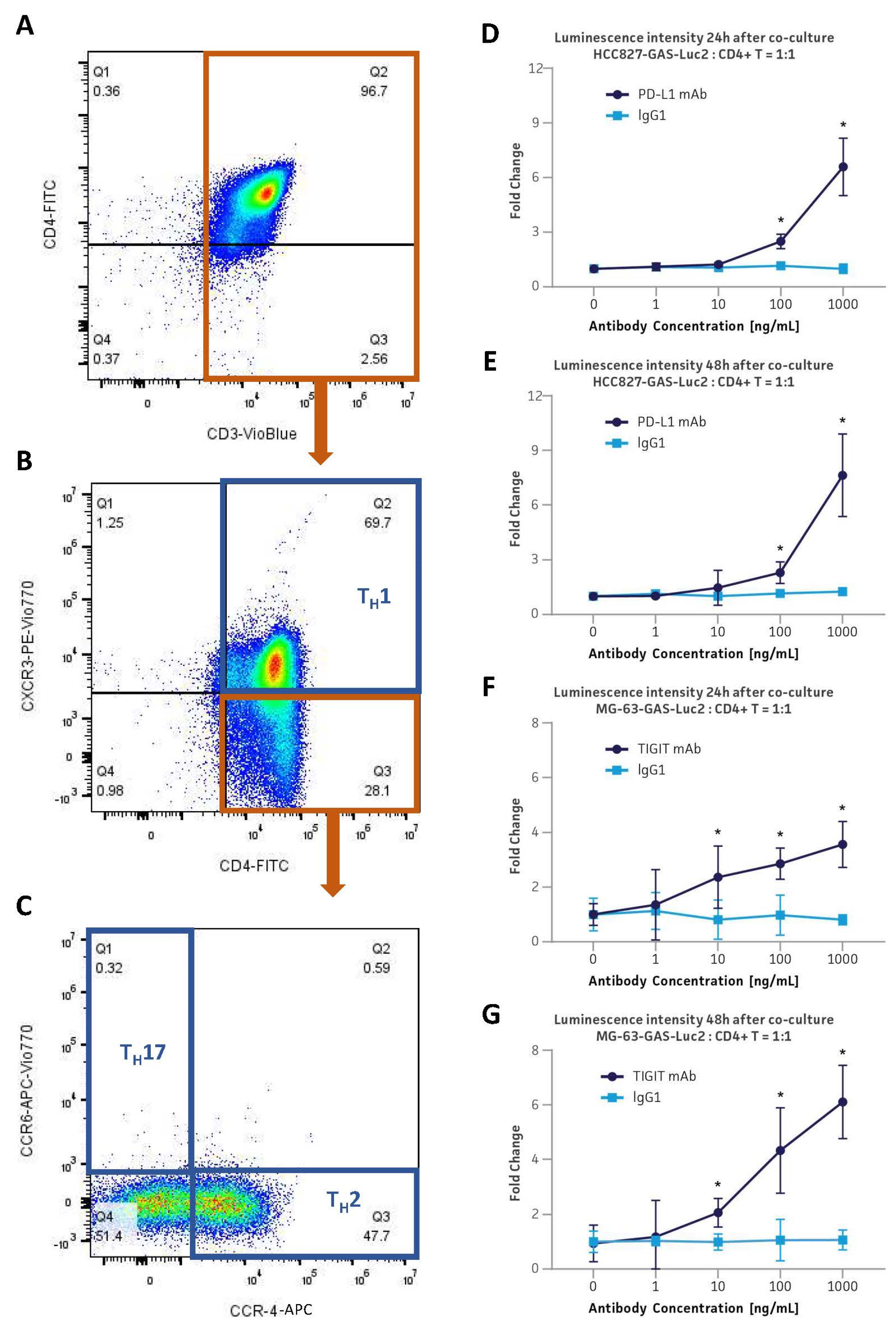
2.8. CD4+ Helper T Cell Subset Phenotyping
2.9. Generation of Artificial Antigen-Presenting Cell Line
2.10. Quantification and Statistical Analysis
3. Results
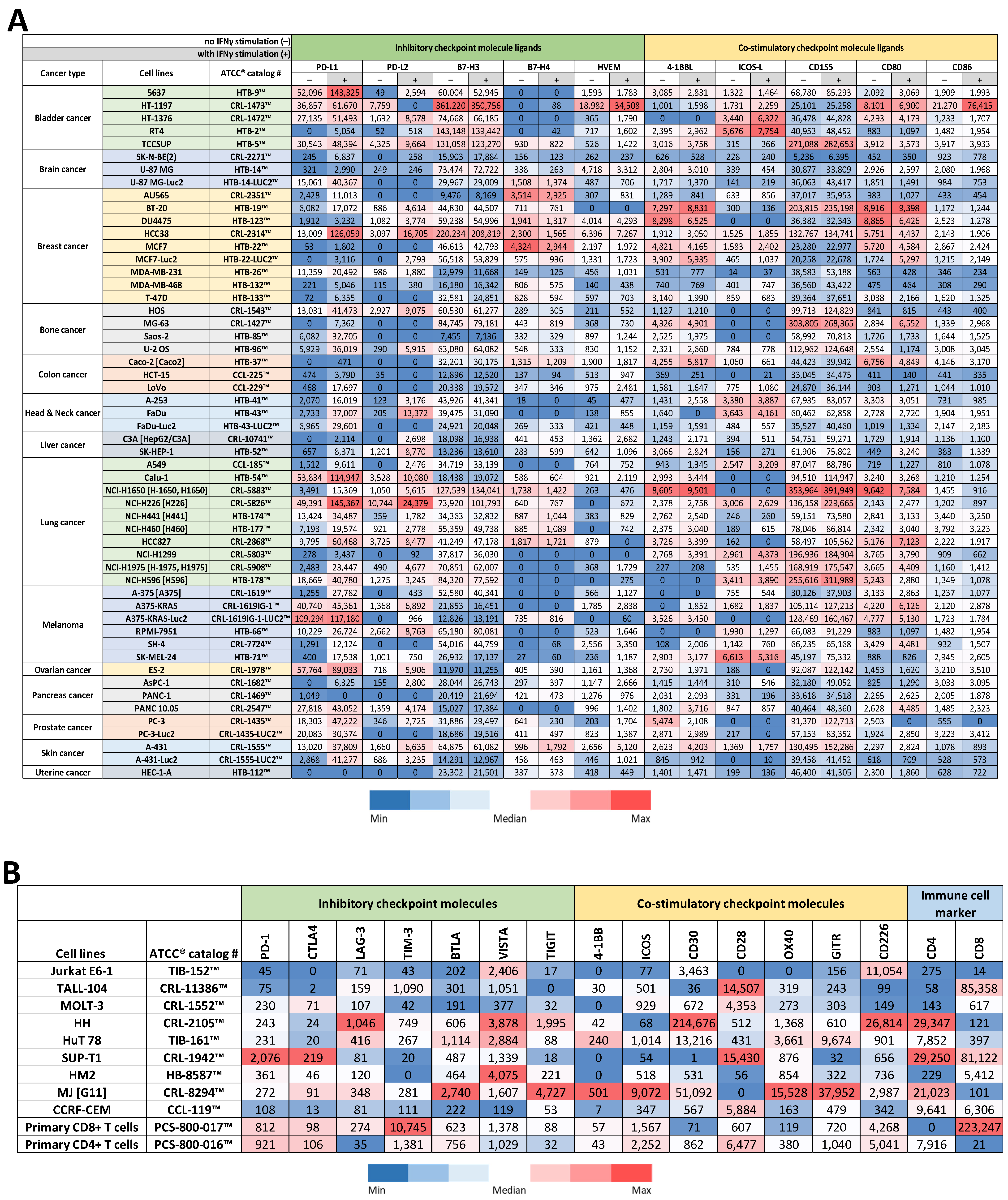
3.1. Protein Profiling of Cancer Cell Lines for Immune Checkpoint Molecule Ligand Expression
3.2. Protein Profiling of T Cell Lines & Primary T Cells for Immune Checkpoint Molecule Expression
3.3. Selection of Candidate Reporter Cell Lines
3.4. Comparisons of Physical Properties of Reporter Cell Lines to Parental Cell Lines
3.5. IFNγ and T Cell-Conditioned Media Stimulation of Reporter Cell Lines
3.6. Application in the PD-L1 Immune Checkpoint Inhibitor Platform via Co-Culture with Primary Human CD8+ Cytotoxic T Cells
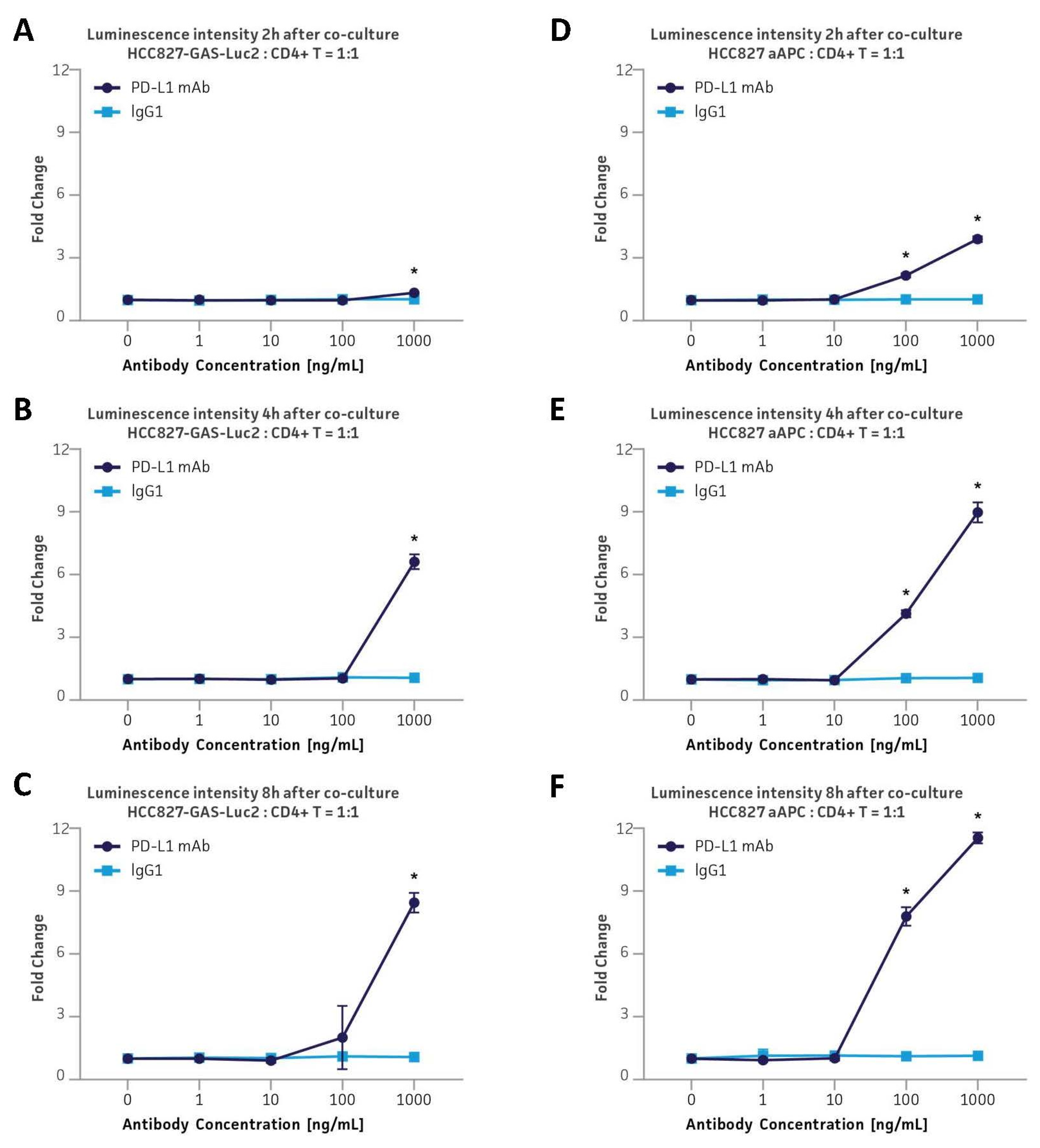
3.7. Application in the PD-L1 & TIGIT Immune Checkpoint Inhibitor Platforms via Co-Culture with Primary Human CD4+ Helper T Cells
3.8. Application in the PD-L1 Immune Checkpoint Inhibitor Platform via Co-Culture with Primary Human CD4+ Helper T Cells by Employing an Anti-CD3/CD28-Expressing aAPC
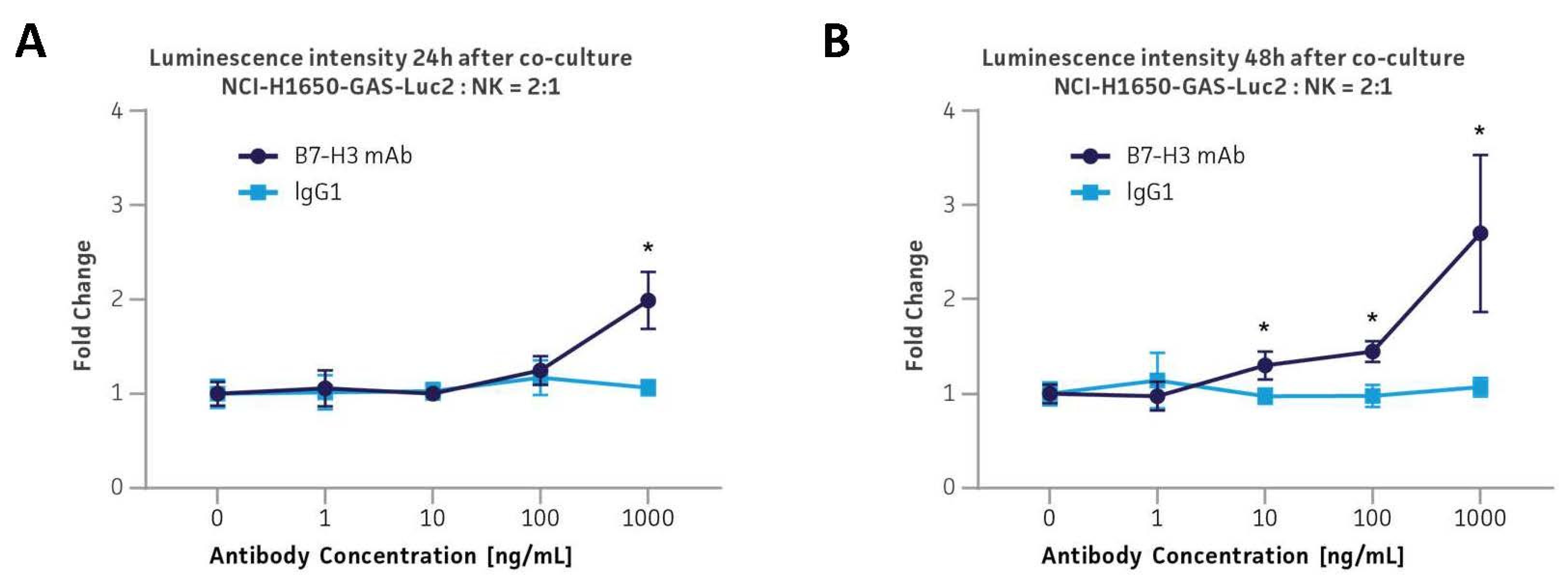
3.9. Application in the B7-H3 Immune Checkpoint ADCC Platform via Co-Culture with Primary Human CD56+ NK Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Korman, A.J.; Garrett-Thomson, S.C.; Lonberg, N. The foundations of immune checkpoint blockade and the ipilimumab approval decennial. Nat. Rev. Drug Discov. 2022, 21, 509–528. [Google Scholar] [CrossRef]
- Kubli, S.P.; Berger, T.; Araujo, D.V.; Siu, L.L.; Mak, T.W. Beyond immune checkpoint blockade: Emerging immunological strategies. Nat. Rev. Drug Discov. 2021, 20, 899–919. [Google Scholar] [CrossRef]
- Olson, B.; Li, Y.; Lin, Y.; Liu, E.T.; Patnaik, A. Mouse Models for Cancer Immunotherapy Research. Cancer Discov. 2018, 8, 1358–1365. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988 e1916. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598 e1512. [Google Scholar] [CrossRef]
- Versteven, M.; Van den Bergh, J.M.J.; Broos, K.; Fujiki, F.; Campillo-Davo, D.; De Reu, H.; Morimoto, S.; Lecocq, Q.; Keyaerts, M.; Berneman, Z.; et al. A versatile T cell-based assay to assess therapeutic antigen-specific PD-1-targeted approaches. Oncotarget 2018, 9, 27797–27808. [Google Scholar] [CrossRef]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef]
- Yang, S.; Wei, W.; Zhao, Q. B7-H3, a checkpoint molecule, as a target for cancer immunotherapy. Int. J. Biol. Sci. 2020, 16, 1767–1773. [Google Scholar] [CrossRef]
- Zhou, W.-T.; Jin, W.-L. B7-H3/CD276: An Emerging Cancer Immunotherapy. Front. Immunol. 2021, 12, 701006. [Google Scholar] [CrossRef]
- Kučan Brlić, P.; Lenac Roviš, T.; Cinamon, G.; Tsukerman, P.; Mandelboim, O.; Jonjić, S. Targeting PVR (CD155) and its receptors in anti-tumor therapy. Cell. Mol. Immunol. 2019, 16, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.; Aguilera, A.R.; Sundarrajan, A.; Corvino, D.; Stannard, K.; Krumeich, S.; Das, I.; Lima, L.G.; Meza Guzman, L.G.; Li, K.; et al. CD155 on Tumor Cells Drives Resistance to Immunotherapy by Inducing the Degradation of the Activating Receptor CD226 in CD8(+) T Cells. Immunity 2020, 53, 805–823.e815. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.G.; Marabelle, A.; Caramella, C.; Garcia, C.; Aldea, M.; Chaput, N.; Robert, C.; Besse, B. Tumour burden and efficacy of immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2022, 19, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Larson, R.C.; Kann, M.C.; Bailey, S.R.; Haradhvala, N.J.; Llopis, P.M.; Bouffard, A.A.; Scarfo, I.; Leick, M.B.; Grauwet, K.; Berger, T.R.; et al. CAR T cell killing requires the IFNgammaR pathway in solid but not liquid tumours. Nature 2022, 604, 563–570. [Google Scholar] [CrossRef]
- Chen, D.; Varanasi, S.K.; Hara, T.; Traina, K.; Sun, M.; McDonald, B.; Farsakoglu, Y.; Clanton, J.; Xu, S.; Garcia-Rivera, L.; et al. CTLA-4 blockade induces CD4(+) T cell IFNgamma-driven microglial phagocytosis and anti-tumor function in glioblastoma. Immunity 2023, 56, 2086–2104.e8. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Brasier, A.R.; Tate, J.E.; Habener, J.F. Optimized use of the firefly luciferase assay as a reporter gene in mammalian cell lines. Biotechniques 1989, 7, 1116–1122. [Google Scholar] [PubMed]
- Grewal, T.; Priceputu, E.; Davignon, J.; Bernier, L. Identification of a gamma-interferon-responsive element in the promoter of the human macrophage scavenger receptor A gene. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 825–831. [Google Scholar] [CrossRef]
- Schmidts, A.; Marsh, L.C.; Srivastava, A.A.; Bouffard, A.A.; Boroughs, A.C.; Scarfo, I.; Larson, R.C.; Bedoya, F.; Choi, B.D.; Frigault, M.J.; et al. Cell-based artificial APC resistant to lentiviral transduction for efficient generation of CAR-T cells from various cell sources. J. Immunother. Cancer 2020, 8, e000990. [Google Scholar] [CrossRef]
- Shrestha, B.; Zhang, Y.; Yu, B.; Li, G.; Boucher, J.C.; Beatty, N.J.; Tsai, H.C.; Wang, X.; Mishra, A.; Sweet, K.; et al. Generation of Antitumor T Cells For Adoptive Cell Therapy With Artificial Antigen Presenting Cells. J. Immunother. 2020, 43, 79–88. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R., 3rd; Kalocsay, M.; Jane-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402 e316. [Google Scholar] [CrossRef]
- Upadhaya, S.; Hubbard-Lucey, V.M.; Yu, J.X. Immuno-oncology drug development forges on despite COVID-19. Nat. Rev. Drug Discov. 2020, 19, 751–752. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Cancer Cell Line Encyclopedia, C.; Genomics of Drug Sensitivity in Cancer, C. Pharmacogenomic agreement between two cancer cell line data sets. Nature 2015, 528, 84–87. [Google Scholar] [CrossRef]
- Li, H.; Ning, S.; Ghandi, M.; Kryukov, G.V.; Gopal, S.; Deik, A.; Souza, A.; Pierce, K.; Keskula, P.; Hernandez, D.; et al. The landscape of cancer cell line metabolism. Nat. Med. 2019, 25, 850–860. [Google Scholar] [CrossRef]
- Jaffe, J.D.; Wang, Y.; Chan, H.M.; Zhang, J.; Huether, R.; Kryukov, G.V.; Bhang, H.E.; Taylor, J.E.; Hu, M.; Englund, N.P.; et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 1386–1391. [Google Scholar] [CrossRef]
- Guo, Y.; Xiao, P.; Lei, S.; Deng, F.; Xiao, G.G.; Liu, Y.; Chen, X.; Li, L.; Wu, S.; Chen, Y.; et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin. 2008, 40, 426–436. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef]
- Liu, B.; Hu, X.; Feng, K.; Gao, R.; Xue, Z.; Zhang, S.; Zhang, Y.; Corse, E.; Hu, Y.; Han, W.; et al. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat. Cancer 2022, 3, 108–121. [Google Scholar] [CrossRef]
- Melenhorst, J.J.; Chen, G.M.; Wang, M.; Porter, D.L.; Chen, C.; Collins, M.A.; Gao, P.; Bandyopadhyay, S.; Sun, H.; Zhao, Z.; et al. Decade-long leukaemia remissions with persistence of CD4(+) CAR T cells. Nature 2022, 602, 503–509. [Google Scholar] [CrossRef]
- Xu, Y.; Wei, Z.; Feng, M.; Zhu, D.; Mei, S.; Wu, Z.; Feng, Q.; Chang, W.; Ji, M.; Liu, C.; et al. Tumor-infiltrated activated B cells suppress liver metastasis of colorectal cancers. Cell Rep. 2022, 40, 111295. [Google Scholar] [CrossRef]
- Borriello, L.; Coste, A.; Traub, B.; Sharma, V.P.; Karagiannis, G.S.; Lin, Y.; Wang, Y.; Ye, X.; Duran, C.L.; Chen, X.; et al. Primary tumor associated macrophages activate programs of invasion and dormancy in disseminating tumor cells. Nat. Commun. 2022, 13, 626. [Google Scholar] [CrossRef]
- Buechler, M.B.; Pradhan, R.N.; Krishnamurty, A.T.; Cox, C.; Calviello, A.K.; Wang, A.W.; Yang, Y.A.; Tam, L.; Caothien, R.; Roose-Girma, M.; et al. Cross-tissue organization of the fibroblast lineage. Nature 2021, 593, 575–579. [Google Scholar] [CrossRef]
- Xie, Y.; He, L.; Lugano, R.; Zhang, Y.; Cao, H.; He, Q.; Chao, M.; Liu, B.; Cao, Q.; Wang, J.; et al. Key molecular alterations in endothelial cells in human glioblastoma uncovered through single-cell RNA sequencing. JCI Insight 2021, 6, 15. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Gibson, J.; Ou, K.; Lopez, L.S.; Ng, R.H.; Leggett, N.; Jonsson, V.D.; Zarif, J.C.; Lee, P.P.; Wang, X.; et al. PD-L1 blockade restores CAR T cell activity through IFN-gamma-regulation of CD163+ M2 macrophages. J. Immunother. Cancer 2022, 10, e004400. [Google Scholar] [CrossRef]
- Eggermont, L.J.; Paulis, L.E.; Tel, J.; Figdor, C.G. Towards efficient cancer immunotherapy: Advances in developing artificial antigen-presenting cells. Trends Biotechnol. 2014, 32, 456–465. [Google Scholar] [CrossRef]
- Sunshine, J.C.; Green, J.J. Nanoengineering approaches to the design of artificial antigen-presenting cells. Nanomedicine 2013, 8, 1173–1189. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.; Foulke, J.G.; Chen, L.; Tian, F.; Gu, Z. GAS-Luc2 Reporter Cell Lines for Immune Checkpoint Drug Screening in Solid Tumors. Cancers 2024, 16, 1965. https://doi.org/10.3390/cancers16111965
Chang H, Foulke JG, Chen L, Tian F, Gu Z. GAS-Luc2 Reporter Cell Lines for Immune Checkpoint Drug Screening in Solid Tumors. Cancers. 2024; 16(11):1965. https://doi.org/10.3390/cancers16111965
Chicago/Turabian StyleChang, Hyeyoun, John G. Foulke, Luping Chen, Fang Tian, and Zhizhan Gu. 2024. "GAS-Luc2 Reporter Cell Lines for Immune Checkpoint Drug Screening in Solid Tumors" Cancers 16, no. 11: 1965. https://doi.org/10.3390/cancers16111965
APA StyleChang, H., Foulke, J. G., Chen, L., Tian, F., & Gu, Z. (2024). GAS-Luc2 Reporter Cell Lines for Immune Checkpoint Drug Screening in Solid Tumors. Cancers, 16(11), 1965. https://doi.org/10.3390/cancers16111965






