Pathogenicity Prediction of Gene Fusion in Structural Variations: A Knowledge Graph-Infused Explainable Artificial Intelligence (XAI) Framework
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Knowledge Graph
2.2. Explainable AI (XAI)
2.3. Our Explainable Learning Model
2.3.1. Correct Answer Set: Dataset 1
- The target text describes approved drugs in Japan for $DISEASENAME.
- The target text describes FDA-approved drugs for $DISEASENAME.
- The target text is referenced by guidelines about $DISEASENAME.
- The target text describes highly statistically reliable clinical trials/meta-analyses and consensus among experts on $DISEASENAME.
- The target text describes FDA-approved drugs for other cancer types.
- The target text describes highly statistically reliable clinical trials/meta-analyses and consensus among experts regarding other cancer types.
- The target text describes small-scale clinical trials that have shown usefulness regardless of cancer type.
- The target text describes the usefulness shown in case reports regardless of cancer type.
- The usefulness of target text has been reported in preclinical studies (in vitro and in vivo).
2.3.2. Features
- The amount of literature regarding the same fusion genes in Mitelman;
- The number of entries about the same fusion genes in COSMIC;
- The sequence of domains registered with Pfam on each gene;
- The lengths of both UTRs;
- TargetScan-registered miRNAs that affect UTR;
- Families of miRNAs;
- Whether each domain is within the breakpoint;
2.4. Benchmark
2.4.1. Existing Methods
2.4.2. Learning Set: Dataset 2 and Dataset 3
2.4.3. Method of Comparing the Performance of the Models
- Cross Validation
- 2.
- Holdout Validation
2.4.4. Our Benchmark Learning Model
- Sequences of domains registered with Pfam for each gene;
- Lengths of both UTRs;
- TargetScan-registered miRNAs that affect UTR;
- Families of miRNAs;
- Whether each domain was within the breakpoint.
3. Results
3.1. Evaluation of Prediction Accuracy
3.1.1. Evaluation using Cross-Validation
3.1.2. Evaluation Using Holdout Validation
3.2. Evaluation of the Explanation of Prediction
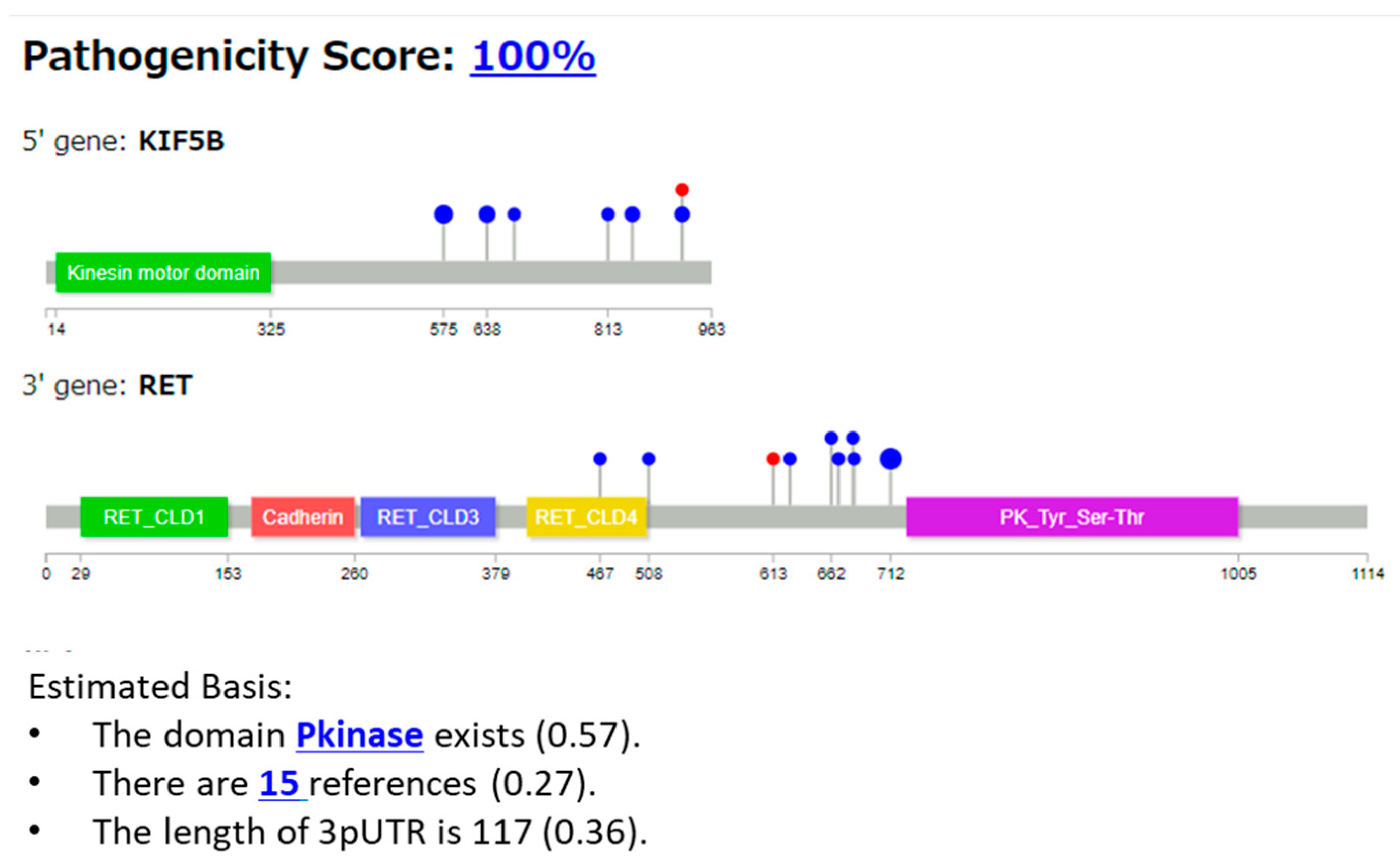
3.2.1. Case 1: KIF5B::RET Fusion
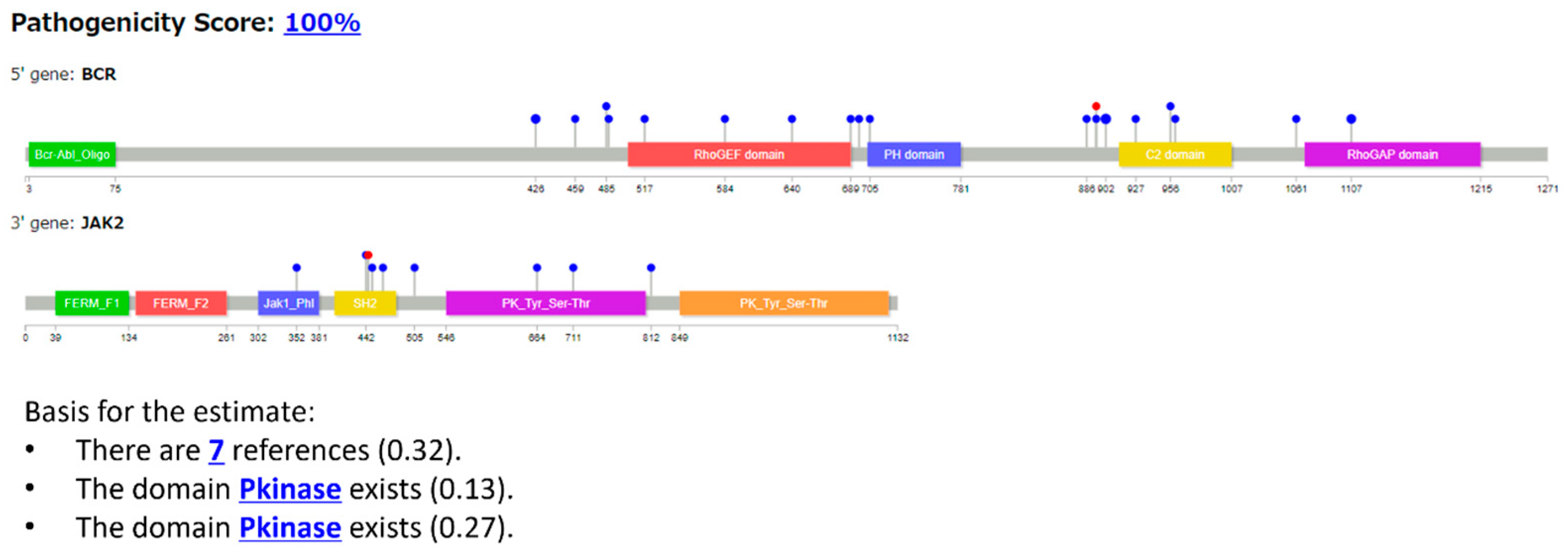
3.2.2. Case 2: BCR::JAK2 T Fusion
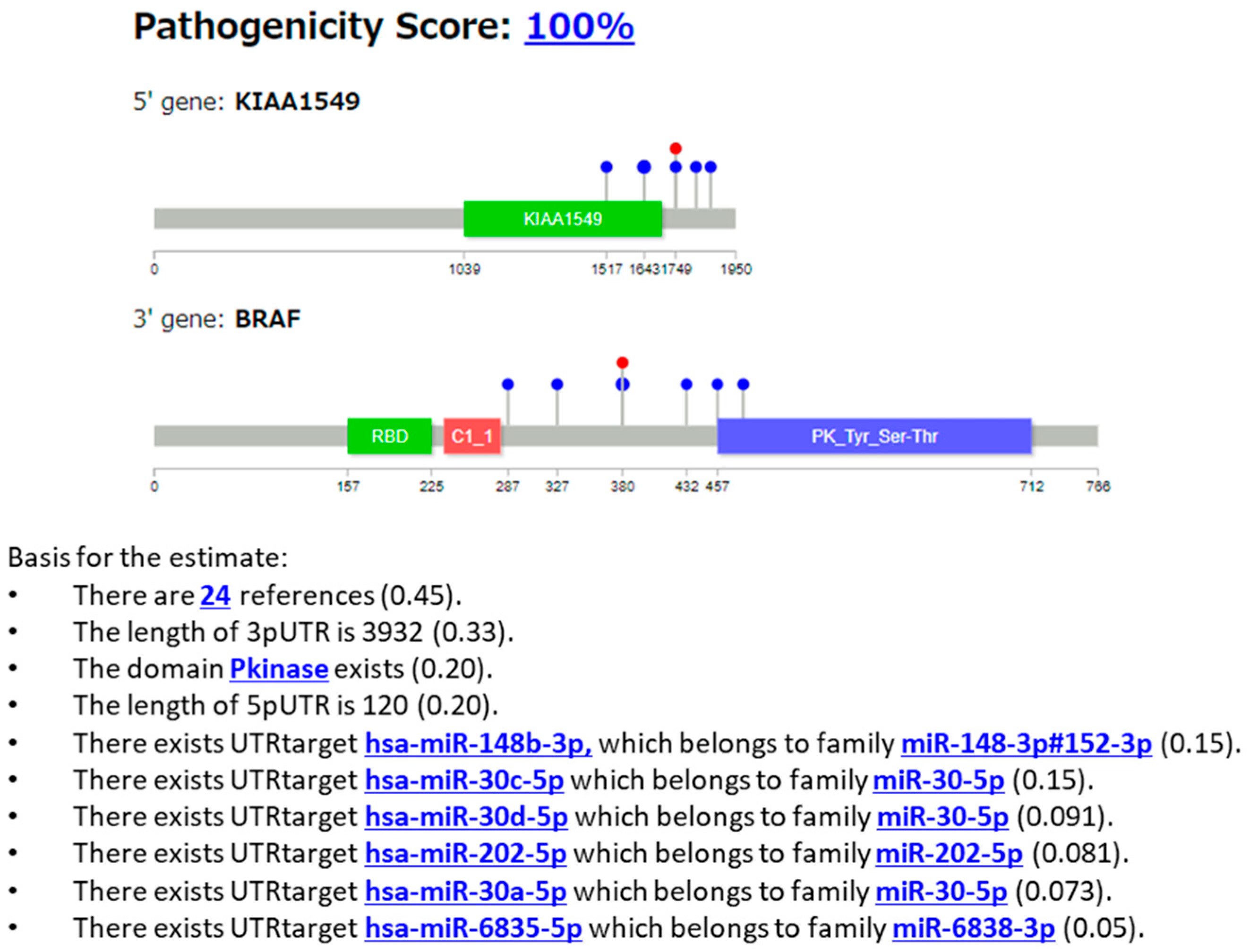
3.2.3. Case 3: KIAA1549::BRAF Fusion
3.2.4. Case 4: IKZF1::LRBA Fusion
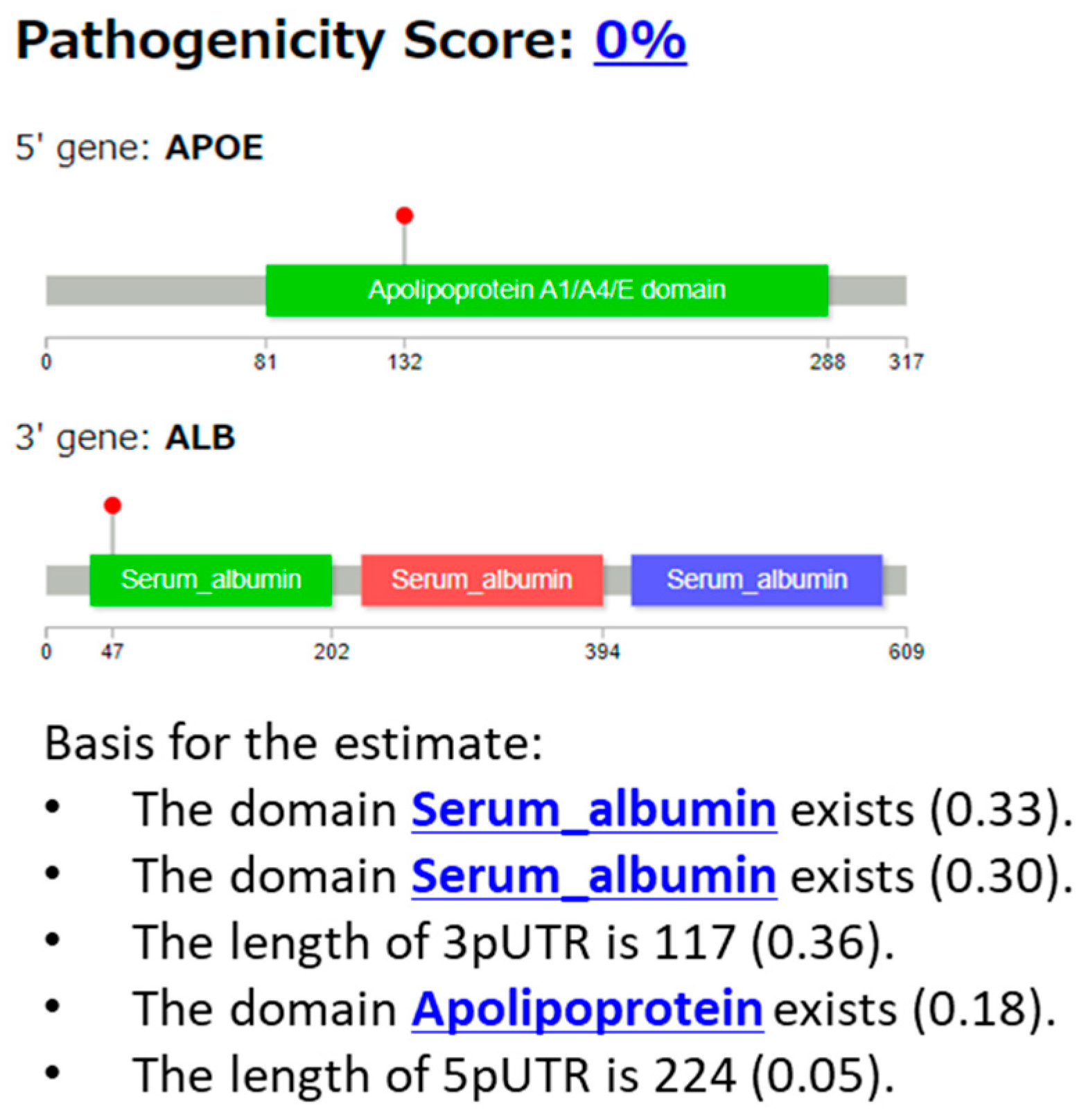
3.2.5. Case 5: APOE::ALB Fusion
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitelman, F.; Johansson, B.; Mertens, F. The Impact of Translocations and Gene Fusions on Cancer Causation. Nat. Rev. Cancer 2007, 7, 233–245. [Google Scholar] [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Lovino, M.; Montemurro, M.; Barrese, V.S.; Ficarra, E. Identifying the Oncogenic Potential of Gene Fusions Exploiting MiRNAs. J. Biomed. Inform. 2022, 129, 104057. [Google Scholar] [CrossRef] [PubMed]
- Lovino, M.; Ciaburri, M.S.; Urgese, G.; Di Cataldo, S.; Ficarra, E. DEEPrior: A Deep Learning Tool for the Prioritization of Gene Fusions. Bioinformatics 2020, 36, 3248–3250. [Google Scholar] [CrossRef] [PubMed]
- Shugay, M.; Ortiz de Mendíbil, I.; Vizmanos, J.L.; Novo, F.J. Oncofuse: A Computational Framework for the Prediction of the Oncogenic Potential of Gene Fusions. Bioinformatics 2013, 29, 2539–2546. [Google Scholar] [CrossRef]
- Sheu, R.-K.; Pardeshi, M.S. A Survey on Medical Explainable AI (XAI): Recent Progress, Explainability Approach, Human Interaction and Scoring System. Sensors 2022, 22, 8068. [Google Scholar] [CrossRef]
- Abe, S.; Tago, S.; Yokoyama, K.; Ogawa, M.; Takei, T.; Imoto, S.; Fuji, M. Explainable AI for Estimating Pathogenicity of Genetic Variants Using Large-Scale Knowledge Graphs. Cancers 2023, 15, 1118. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Resource Description Framework (RDF): Concepts and Abstract Syntax. Available online: https://www.w3.org/TR/rdf-concepts/ (accessed on 28 March 2024).
- Med2RDF. Available online: http://med2rdf.org/ (accessed on 28 March 2024).
- Auer, S.; Bizer, C.; Kobilarov, G.; Lehmann, J.; Cyganiak, R.; Ives, Z. DBpedia: A Nucleus for a Web of Open Data. In Proceedings of the The Semantic Web, Busan, Republic of Korea, 11–15 November 2007; Springer: Berlin/Heidelberg, Germany, 2007; pp. 722–735. [Google Scholar]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Raney, B.J.; Barber, G.P.; Benet-Pagès, A.; Casper, J.; Clawson, H.; Cline, M.S.; Diekhans, M.; Fischer, C.; Navarro Gonzalez, J.; Hickey, G.; et al. The UCSC Genome Browser Database: 2024 Update. Nucleic Acids Res. 2024, 52, D1082–D1088. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The Biochemical Basis of MicroRNA Targeting Efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef] [PubMed]
- Johansson, B.; Mertens, F.; Mitelman, F. Geographic Heterogeneity of Neoplasia-Associated Chromosome Aberrations. Genes Chromosomes Cancer 1991, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mitelman Database Chromosome Aberrations and Gene Fusions in Cancer. Available online: https://mitelmandatabase.isb-cgc.org/about (accessed on 25 March 2024).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology Knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, Q.; Tang, M.; Barthel, F.; Amin, S.; Yoshihara, K.; Lang, F.M.; Martinez-Ledesma, E.; Lee, S.H.; Zheng, S.; et al. TumorFusions: An Integrative Resource for Cancer-Associated Transcript Fusions. Nucleic Acids Res. 2018, 46, D1144–D1149. [Google Scholar] [CrossRef] [PubMed]
- Maruhashi, K.; Todoriki, M.; Ohwa, T.; Goto, K.; Hasegawa, Y.; Inakoshi, H.; Anai, H. Learning Multi-Way Relations via Tensor Decomposition with Neural Networks. AAAI 2018, 32, 3770–3777. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Demonstrations, San Diego, CA, USA, 12–17 June 2016. [Google Scholar] [CrossRef]
- Brown, T.B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models Are Few-Shot Learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Azure OpenAI Service. Available online: https://azure.microsoft.com/en-us/products/ai-services/openai-service/ (accessed on 25 March 2024).
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Liang, W.; Zhang, Y.; Cao, H.; Wang, B.; Ding, D.; Yang, X.; Vodrahalli, K.; He, S.; Smith, D.; Yin, Y.; et al. Can Large Language Models Provide Useful Feedback on Research Papers? A Large-Scale Empirical Analysis. arXiv 2023, arXiv:2310.01783. [Google Scholar]
- JSMO Guideline. Available online: https://www.jsmo.or.jp/about/doc/20200310.pdf (accessed on 22 March 2024).
- Abate, F.; Zairis, S.; Ficarra, E.; Acquaviva, A.; Wiggins, C.H.; Frattini, V.; Lasorella, A.; Iavarone, A.; Inghirami, G.; Rabadan, R. Pegasus: A Comprehensive Annotation and Prediction Tool for Detection of Driver Gene Fusions in Cancer. BMC Syst. Biol. 2014, 8, 97. [Google Scholar] [CrossRef]
- Babiceanu, M.; Qin, F.; Xie, Z.; Jia, Y.; Lopez, K.; Janus, N.; Facemire, L.; Kumar, S.; Pang, Y.; Qi, Y.; et al. Recurrent Chimeric Fusion RNAs in Non-Cancer Tissues and Cells. Nucleic Acids Res. 2016, 44, 2859–2872. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Ichikawa, H.; Totoki, Y.; Yasuda, K.; Hiramoto, M.; Nammo, T.; Sakamoto, H.; Tsuta, K.; Furuta, K.; Shimada, Y.; et al. KIF5B-RET Fusions in Lung Adenocarcinoma. Nat. Med. 2012, 18, 375–377. [Google Scholar] [CrossRef] [PubMed]
- Jay, J.J.; Brouwer, C. Lollipops in the Clinic: Information Dense Mutation Plots for Precision Medicine. PLoS ONE 2016, 11, e0160519. [Google Scholar] [CrossRef] [PubMed]
- Cirmena, G.; Aliano, S.; Fugazza, G.; Bruzzone, R.; Garuti, A.; Bocciardi, R.; Bacigalupo, A.; Ravazzolo, R.; Ballestrero, A.; Sessarego, M. A BCR-JAK2 Fusion Gene as the Result of a t(9;22)(P24;Q11) in a Patient with Acute Myeloid Leukemia. Cancer Genet. Cytogenet. 2008, 183, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Ryall, S.; Krishnatry, R.; Arnoldo, A.; Buczkowicz, P.; Mistry, M.; Siddaway, R.; Ling, C.; Pajovic, S.; Yu, M.; Rubin, J.B.; et al. Targeted Detection of Genetic Alterations Reveal the Prognostic Impact of H3K27M and MAPK Pathway Aberrations in Paediatric Thalamic Glioma. Acta Neuropathol. Commun. 2016, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Yokota, K.; Sasaki, H.; Okuda, K.; Shimizu, S.; Shitara, M.; Hikosaka, Y.; Moriyama, S.; Yano, M.; Fujii, Y. KIF5B/RET Fusion Gene in Surgically-Treated Adenocarcinoma of the Lung. Oncol. Rep. 2012, 28, 1187–1192. [Google Scholar] [CrossRef]
- Ju, Y.S.; Lee, W.-C.; Shin, J.-Y.; Lee, S.; Bleazard, T.; Won, J.-K.; Kim, Y.T.; Kim, J.-I.; Kang, J.-H.; Seo, J.-S. A Transforming KIF5B and RET Gene Fusion in Lung Adenocarcinoma Revealed from Whole-Genome and Transcriptome Sequencing. Genome Res. 2012, 22, 436–445. [Google Scholar] [CrossRef]
- Cuesta-Domínguez, Á.; Ortega, M.; Ormazábal, C.; Santos-Roncero, M.; Galán-Díez, M.; Steegmann, J.L.; Figuera, Á.; Arranz, E.; Vizmanos, J.L.; Bueren, J.A.; et al. Transforming and Tumorigenic Activity of JAK2 by Fusion to BCR: Molecular Mechanisms of Action of a Novel BCR-JAK2 Tyrosine-Kinase. PLoS ONE 2012, 7, e32451. [Google Scholar] [CrossRef]
- McWhirter, J.R.; Galasso, D.L.; Wang, J.Y. A Coiled-Coil Oligomerization Domain of Bcr Is Essential for the Transforming Function of Bcr-Abl Oncoproteins. Mol. Cell. Biol. 1993, 13, 7587–7595. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.-L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, M.; Badiali, M.; Moi, L.; Buttarelli, F.R.; Baldi, C.; Massimino, M.; Sanson, M.; Giangaspero, F. KIAA1549:BRAF Fusion Gene in Pediatric Brain Tumors of Various Histogenesis. Pediatr. Blood Cancer 2015, 62, 724–727. [Google Scholar] [CrossRef] [PubMed]
- Appay, R.; Fina, F.; Macagno, N.; Padovani, L.; Colin, C.; Barets, D.; Ordioni, J.; Scavarda, D.; Giangaspero, F.; Badiali, M.; et al. Duplications of KIAA1549 and BRAF Screening by Droplet Digital PCR from Formalin-Fixed Paraffin-Embedded DNA Is an Accurate Alternative for KIAA1549-BRAF Fusion Detection in Pilocytic Astrocytomas. Mod. Pathol. 2018, 31, 1490–1501. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, X.; Zeng, X.; Peng, X. The Role of Mir-148a in Cancer. J. Cancer 2016, 7, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, X.; Wang, Y.; Peng, R.; Lin, Z.; Wang, Y.; Hu, B.; Wang, J.; Shi, G. Low Expression of MicroRNA-30c Promotes Prostate Cancer Cells Invasion Involved in Downregulation of KRAS Protein. Oncol. Lett. 2017, 14, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.A.; Rajendran, P.; Scherthan, H. The MicroRNA-202 as a Diagnostic Biomarker and a Potential Tumor Suppressor. Int. J. Mol. Sci. 2022, 23, 5870. [Google Scholar] [CrossRef] [PubMed]
- Lind, K.T.; Chatwin, H.V.; DeSisto, J.; Coleman, P.; Sanford, B.; Donson, A.M.; Davies, K.D.; Willard, N.; Ewing, C.A.; Knox, A.J.; et al. Novel RAF Fusions in Pediatric Low-Grade Gliomas Demonstrate MAPK Pathway Activation. J. Neuropathol. Exp. Neurol. 2021, 80, 1099–1107. [Google Scholar] [CrossRef]
- Helgager, J.; Lidov, H.G.; Mahadevan, N.R.; Kieran, M.W.; Ligon, K.L.; Alexandrescu, S. A Novel GIT2-BRAF Fusion in Pilocytic Astrocytoma. Diagn. Pathol. 2017, 12, 82. [Google Scholar] [CrossRef]
- Yan, L.; Ping, N.; Zhu, M.; Sun, A.; Xue, Y.; Ruan, C.; Drexler, H.G.; Macleod, R.A.F.; Wu, D.; Chen, S. Clinical, Immunophenotypic, Cytogenetic, and Molecular Genetic Features in 117 Adult Patients with Mixed-Phenotype Acute Leukemia Defined by WHO-2008 Classification. Haematologica 2012, 97, 1708–1712. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Miller, C.B.; Radtke, I.; Phillips, L.A.; Dalton, J.; Ma, J.; White, D.; Hughes, T.P.; Le Beau, M.M.; Pui, C.-H.; et al. BCR-ABL1 Lymphoblastic Leukaemia Is Characterized by the Deletion of Ikaros. Nature 2008, 453, 110–114. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murakami, K.; Tago, S.-i.; Takishita, S.; Morikawa, H.; Kojima, R.; Yokoyama, K.; Ogawa, M.; Fukushima, H.; Takamori, H.; Nannya, Y.; et al. Pathogenicity Prediction of Gene Fusion in Structural Variations: A Knowledge Graph-Infused Explainable Artificial Intelligence (XAI) Framework. Cancers 2024, 16, 1915. https://doi.org/10.3390/cancers16101915
Murakami K, Tago S-i, Takishita S, Morikawa H, Kojima R, Yokoyama K, Ogawa M, Fukushima H, Takamori H, Nannya Y, et al. Pathogenicity Prediction of Gene Fusion in Structural Variations: A Knowledge Graph-Infused Explainable Artificial Intelligence (XAI) Framework. Cancers. 2024; 16(10):1915. https://doi.org/10.3390/cancers16101915
Chicago/Turabian StyleMurakami, Katsuhiko, Shin-ichiro Tago, Sho Takishita, Hiroaki Morikawa, Rikuhiro Kojima, Kazuaki Yokoyama, Miho Ogawa, Hidehito Fukushima, Hiroyuki Takamori, Yasuhito Nannya, and et al. 2024. "Pathogenicity Prediction of Gene Fusion in Structural Variations: A Knowledge Graph-Infused Explainable Artificial Intelligence (XAI) Framework" Cancers 16, no. 10: 1915. https://doi.org/10.3390/cancers16101915
APA StyleMurakami, K., Tago, S.-i., Takishita, S., Morikawa, H., Kojima, R., Yokoyama, K., Ogawa, M., Fukushima, H., Takamori, H., Nannya, Y., Imoto, S., & Fuji, M. (2024). Pathogenicity Prediction of Gene Fusion in Structural Variations: A Knowledge Graph-Infused Explainable Artificial Intelligence (XAI) Framework. Cancers, 16(10), 1915. https://doi.org/10.3390/cancers16101915






