Driver Mutations in Pancreatic Cancer and Opportunities for Targeted Therapy
Abstract
Simple Summary
Abstract
1. Introduction
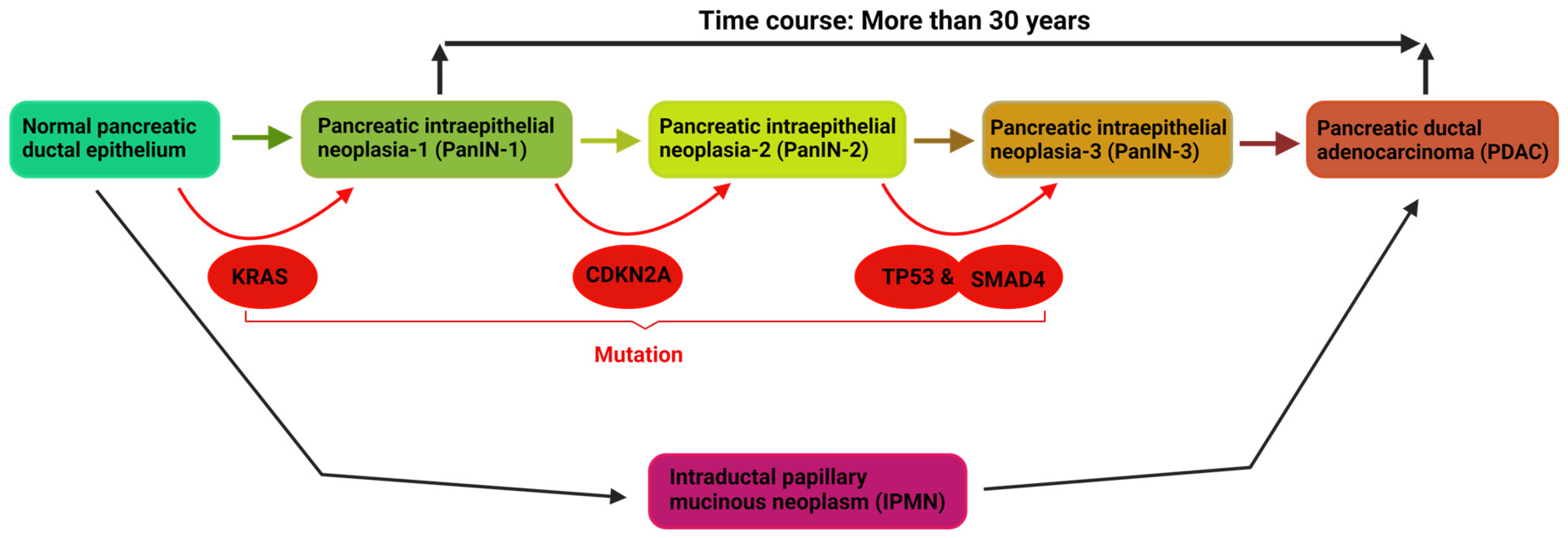
2. Driver Mutations, and Mechanisms of PDAC
3. Targeting Oncogenic Signaling Pathways for PDAC Treatment: KRAS
Targeting KRAS: Clinical and Therapeutic Investigations
4. Targeting Tumor Suppressor Mutations for PDAC Treatment
4.1. TP53
4.1.1. Targeting Negative Regulators of wt-p53
4.1.2. Targeting Mutant p53
4.2. SMAD4
4.3. CDKN2A
| Target | Agent(s) | Phase | Disease(s) | Mechanism | Reference(s) |
|---|---|---|---|---|---|
| MDM2 | RG7112 | I | WDLPS/DDLPS | Occupy p53 binding pocket of MDM2 for p53 stabilization/activation | [94,95,96,97,98] |
| MDM2 | Idasanutlin | I | Polycythemia Vera and Essential Thrombocythemia | Promote TP53 expression and prevent p53 degradation | [99] |
| MDM2 | Venetoclax-Idasanutlin | Ib | Relapsed/Refractory Acute Myeloid Leukemia | p53 activation to inactivate MCL-1 and BCL-xL | [100] |
| MDM2 | RO6839921 | I | Acute Myeloid Leukemia | Plasma esterase cleaves inactive prodrug to release active Idasanutlin to restore p53 activity | [101,102] |
| MDM2 and CDK4/6 | Siremadlin + Ribociclib | Ib | WDLPS/DDLPS | p53 pathway activation and inhibition of CDK enzymes | [103] |
| MDM2 | AMG-232 | I | P53WT Solid Tumors or Multiple Myeloma | Strong binding blocking MDM2-p53 interaction | [105,106,107] |
| MDM2 | KRT-232 | I | Solid Tumors or Multiple Myeloma and Acute Myeloid Leukemia | Binds to MDM2 and inhibits interaction with p53 for p53 activation | [104] |
| Mutant p53 | APR-246 + azacytidine | Ib/II | Myelodysplastic Syndrome | Induce apoptosis and reprogram TAMS to enhance immune checkpoint inhibitors | [113] |
| Mutant p53 | COTI-2 | I | Head and Neck Squamous Cell Carcinoma and Gynecologic Malignancies | Restore functionality to mutated p53 | [118] |
| SMAD4 | DTLL | Pre-Clinical | PDAC | Block ATR/mTOR pathway and restore SMAD4 mediated activation of Nf-кβ shunt | [129] |
| SMAD4 | Qianlongtong | Randomized Control | Benign Prostate Hyperplasia | Increase expression of SMAD4 | [131] |
| SMAD4 | Duvelisib | I | T-cell Lymphoma | PI3K-δ/γ inhibition in TGFβ-PI3/AKT Axis | [134] |
| SMAD4 | Perifosine and MK-2206 | II | Recurrent Glioblastoma and Breast Cancer | MSP/RON pathway in TGFβ-PI3/AKT Axis | [135,136] |
| CDK4/6 | Abemaciclib | Open label III | HR+, HER2-, node-positive, Breast Cancer | Inhibition of cell cycle progression through CDK4/6 inhibitor | [148] |
| CDK4/6 | Abemaciclib + Bevacizumab | I | Recurrent GBM with loss of CDKN2A | Inhibition of cell cycle progression through CDK4/6 inhibitor and anti-angiogenic therapy | NCT04074785 |
| CDK4/6 | Palbociclib + Fulvestrant | Placebo Controlled Randomized Trial | HR+, HER2-, Breast Cancer | Inhibition cell cycle progression through CDK4/6 inhibitor | NCT01942135 |
| CDK4/6 | Trilaciclib | Ib/II | Naïve Extensive Stage Small Cell Lung Cancer | Chemotherapy damage prevention by HSPC remaining in G1 arrest | [150] |
| CDNK2A | Ilorasertinib | II | Advanced Solid Tumors | Multikinase Inhibition to induce cell cycle arrest | NCT02478320 |
| CDK4/6 | Palbociclib | Non-randomized Open Label II | Metastatic Grade 1 and 2 Pancreatic Neuroendocrine Tumors | Inhibition of cell cycle progression in RB+ cells through CDK4/6 inhibitor | [153] |
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Panchal, K.; Sahoo, R.K.; Gupta, U.; Chaurasiya, A. Role of targeted immunotherapy for pancreatic ductal adenocarcinoma (PDAC) treatment: An overview. Int. Immunopharmacol. 2021, 95, 107508. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Olaoba, O.T.; Ligali, F.C.; Alabi, Z.O.; Akinyemi, A.O.; Ayinde, K.S. Of immune checkpoint maladies and remedies: The throwing of jabs in the oncogenic ring of PDAC. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2021, 1875, 188483. [Google Scholar] [CrossRef]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, W. Pancreatic Cancer: A Review of Risk Factors, Diagnosis, and Treatment. Technol. Cancer Res. Treat. 2020, 19, 1533033820962117. [Google Scholar] [CrossRef]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef]
- McMenamin, Ú.; McCain, S.; Kunzmann, A.T. Do smoking and alcohol behaviours influence GI cancer survival? Best Pract. Res. Clin. Gastroenterol. 2017, 31, 569–577. [Google Scholar] [CrossRef]
- Midha, S.; Chawla, S.; Garg, P.K. Modifiable and non-modifiable risk factors for pancreatic cancer: A review. Cancer Lett. 2016, 381, 269–277. [Google Scholar] [CrossRef]
- Yeo, T.P. Demographics, epidemiology, and inheritance of pancreatic ductal adenocarcinoma. Semin. Oncol. 2015, 42, 8–18. [Google Scholar] [CrossRef]
- Brown, D.G.; Rao, S.; Weir, T.L.; O’Malia, J.; Bazan, M.; Brown, R.J.; Ryan, E.P. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016, 4, 11. [Google Scholar] [CrossRef]
- Meng, C.; Bai, C.; Brown, T.D.; Hood, L.E.; Tian, Q. Human Gut Microbiota and Gastrointestinal Cancer. Genom. Proteom. Bioinform. 2018, 16, 33–49. [Google Scholar] [CrossRef]
- Downes, D.P.; Daurio, N.A.; McLaren, D.G.; Carrington, P.; Previs, S.F.; Williams, K.B. Impact of Extracellular Fatty Acids and Oxygen Tension on Lipid Synthesis and Assembly in Pancreatic Cancer Cells. ACS Chem. Biol. 2020, 15, 1892–1900. [Google Scholar] [CrossRef]
- Genkinger, J.M.; Spiegelman, D.; Anderson, K.E.; Bergkvist, L.; Bernstein, L.; Brandt, P.A.v.D.; English, D.R.; Freudenheim, J.L.; Fuchs, C.S.; Giles, G.G.; et al. Alcohol intake and pancreatic cancer risk: A pooled analysis of fourteen cohort studies. Cancer Epidemiol. Biomark. Prev. 2009, 18, 765–776. [Google Scholar] [CrossRef]
- Molina-Montes, E.; Van Hoogstraten, L.; Gomez-Rubio, P.; Löhr, M.; Sharp, L.; Molero, X.; Márquez, M.; Michalski, C.W.; Farré, A.; Perea, J.; et al. Pancreatic Cancer Risk in Relation to Lifetime Smoking Patterns, Tobacco Type, and Dose–Response Relationships. Cancer Epidemiol. Biomark. Prev. 2020, 29, 1009–1018. [Google Scholar] [CrossRef]
- Samokhvalov, A.V.; Rehm, J.; Roerecke, M. Alcohol Consumption as a Risk Factor for Acute and Chronic Pancreatitis: A Systematic Review and a Series of Meta-analyses. EBioMedicine 2015, 2, 1996–2002. [Google Scholar] [CrossRef]
- Kolbeinsson, H.M.; Chandana, S.; Wright, G.P.; Chung, M. Pancreatic Cancer: A Review of Current Treatment and Novel Therapies. J. Investig. Surg. 2023, 36, 2129884. [Google Scholar] [CrossRef]
- Ostroverkhova, D.; Przytycka, T.M.; Panchenko, A.R. Cancer driver mutations: Predictions and reality. Trends Mol. Med. 2023, 29, 554–566. [Google Scholar] [CrossRef]
- Hu, H.-F.; Ye, Z.; Qin, Y.; Xu, X.-W.; Yu, X.-J.; Zhuo, Q.-F.; Ji, S.-R. Mutations in key driver genes of pancreatic cancer: Molecularly targeted therapies and other clinical implications. Acta Pharmacol. Sin. 2021, 42, 1725–1741. [Google Scholar] [CrossRef]
- Vitellius, C.; Griveaux, O.; Morvant, B.; Pedrono, E.; Venara, A.; Ingster, O.; Baize, N.; Dincuff, E.; Rousselet, M.-C.; Guardiola, P.; et al. Impact of Driver Mutations on the Evolution of Isolated Metachronous Lung Metastasis of Pancreatic Ductal adenocarcinoma. Mol. Diagn. Ther. 2020, 24, 443–449. [Google Scholar] [CrossRef]
- Luo, J. KRAS mutation in pancreatic cancer. Semin. Oncol. 2021, 48, 10–18. [Google Scholar] [CrossRef]
- Peters, M.L.B.; Eckel, A.; Mueller, P.P.; Tramontano, A.C.; Weaver, D.T.; Lietz, A.; Hur, C.; Kong, C.Y.; Pandharipande, P.V. Progression to pancreatic ductal adenocarcinoma from pancreatic intraepithelial neoplasia: Results of a simulation model. Pancreatology 2018, 18, 928–934. [Google Scholar] [CrossRef]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef]
- Wood, K.; Hensing, T.; Malik, R.; Salgia, R. Prognostic and Predictive Value in KRAS in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol. 2016, 2, 805–812. [Google Scholar] [CrossRef]
- Dearden, S.; Stevens, J.; Wu, Y.-L.; Blowers, D. Mutation incidence and coincidence in non small-cell lung cancer: Meta-analyses by ethnicity and histology (mutMap). Ann. Oncol. 2013, 24, 2371–2376. [Google Scholar] [CrossRef]
- Shen, H.; Lundy, J.; Strickland, A.H.; Harris, M.; Swan, M.; Desmond, C.; Jenkins, B.J.; Croagh, D. KRAS G12D Mutation Subtype in Pancreatic Ductal Adenocarcinoma: Does It Influence Prognosis or Stage of Disease at Presentation? Cells 2022, 11, 3175. [Google Scholar] [CrossRef]
- Bryant, K.L.; Mancias, J.D.; Kimmelman, A.C.; Der, C.J. KRAS: Feeding pancreatic cancer proliferation. Trends Biochem. Sci. 2014, 39, 91–100. [Google Scholar] [CrossRef]
- Neumann, J.; Zeindl-Eberhart, E.; Kirchner, T.; Jung, A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol.-Res. Pract. 2009, 205, 858–862. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Saiki, A.Y.; Gaida, K.; Rex, K.; Achanta, P.; Miguel, T.S.; Koppada, N.; Bagal, D.; Lanman, B.A.; Foti, R.S.; McCarter, J.D.; et al. Abstract 4484: Discovery and in vitro characterization of AMG 510–a potent and selective covalent small-molecule inhibitor of KRASG12C. Cancer Res. 2019, 79 (Suppl. S13), 4484. [Google Scholar] [CrossRef]
- Sikdar, N.; Saha, G.; Dutta, A.; Ghosh, S.; Shreekhande, S.V.; Banerjee, S. Genetic Alterations of Periampullary and Pancreatic Ductal Adenocarcinoma: An Overview. Curr. Genom. 2018, 19, 444–463. [Google Scholar] [CrossRef]
- Mohamadkhani, A.; Naderi, E.; Sharafkhah, M.; Fazli, H.R.; Moradzadeh, M.; Pourshams, A. Detection of TP53 R249 Mutation in Iranian Patients with Pancreatic Cancer. J. Oncol. 2013, 2013, 738915. [Google Scholar] [CrossRef]
- Simanshu, D.K.; Nissley, D.V.; McCormick, F. RAS Proteins and Their Regulators in Human Disease. Cell 2017, 170, 17–33. [Google Scholar] [CrossRef]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef]
- Warne, P.H.; Vician, P.R.; Downward, J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature 1993, 364, 352–355. [Google Scholar] [CrossRef]
- Roux, P.P.; Blenis, J. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 2004, 68, 320–344. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Settleman, J.; Kyriakis, J.; Takeuchi-Suzuki, E.; Elledge, S.J.; Marshall, M.S.; Bruder, J.T.; Rapp, U.R.; Avruch, J. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature 1993, 364, 308–313. [Google Scholar] [CrossRef]
- Molina, J.R.; Adjei, A.A. The Ras/Raf/MAPK pathway. J. Thorac. Oncol. 2006, 1, 7–9. [Google Scholar] [CrossRef]
- Oyedele, A.-Q.K.; Ogunlana, A.T.; Boyenle, I.D.; Ibrahim, N.O.; Gbadebo, I.O.; Owolabi, N.A.; Ayoola, A.M.; Francis, A.C.; Eyinade, O.H.; Adelusi, T.I. Pharmacophoric analogs of sotorasib-entrapped KRAS G12C in its inactive GDP-bound conformation: Covalent docking and molecular dynamics investigations. Mol. Divers. 2022, 27, 1795–1807. [Google Scholar] [CrossRef]
- Scheffzek, K.; Ahmadian, M.R.; Kabsch, W.; Wiesmuller, L.; Lautwein, A.; Schmitz, F.; Wittinghofer, A. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 1997, 277, 333–338. [Google Scholar] [CrossRef]
- Haigis, K.M. KRAS Alleles: The Devil Is in the Detail. Trends Cancer 2017, 3, 686–697. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118 Pt 5, 843–846. [Google Scholar] [CrossRef]
- Astrain, G.B.; Nikolova, M.; Smith, M.J. Functional diversity in the RAS subfamily of small GTPases. Biochem. Soc. Trans. 2022, 50, 921–933. [Google Scholar] [CrossRef]
- Lu, Z.; Luo, R.Z.; Lu, Y.; Zhang, X.; Yu, Q.; Khare, S.; Kondo, S.; Kondo, Y.; Yu, Y.; Mills, G.B.; et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J. Clin. Investig. 2008, 118, 3917–3929. [Google Scholar] [CrossRef]
- Sutton, M.N.; Lu, Z.; Li, Y.-C.; Zhou, Y.; Huang, T.; Reger, A.S.; Hurwitz, A.M.; Palzkill, T.; Logsdon, C.; Liang, X.; et al. DIRAS3 (ARHI) Blocks RAS/MAPK Signaling by Binding Directly to RAS and Disrupting RAS Clusters. Cell Rep. 2019, 29, 3448–3459.e6. [Google Scholar] [CrossRef]
- Sutton, M.N.; Yang, H.; Huang, G.Y.; Fu, C.; Pontikos, M.; Wang, Y.; Mao, W.; Pang, L.; Yang, M.; Liu, J.; et al. RAS-related GTPases DIRAS1 and DIRAS2 induce autophagic cancer cell death and are required for autophagy in murine ovarian cancer cells. Autophagy 2018, 14, 637–653. [Google Scholar] [CrossRef]
- Castellano, E.; Downward, J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes. Cancer 2011, 2, 261–274. [Google Scholar] [CrossRef]
- Pawson, T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 2004, 116, 191–203. [Google Scholar] [CrossRef]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef]
- Castellano, E.; Downward, J. Role of RAS in the regulation of PI 3-kinase. Curr. Top. Microbiol. Immunol. 2010, 346, 143–169. [Google Scholar]
- He, Q.; Liu, Z.; Wang, J. Targeting KRAS in PDAC: A New Way to Cure It? Cancers 2022, 14, 4982. [Google Scholar] [CrossRef]
- Pantsar, T. The current understanding of KRAS protein structure and dynamics. Comput. Struct. Biotechnol. J. 2020, 18, 189–198. [Google Scholar] [CrossRef]
- Tang, D.; Kroemer, G.; Kang, R. Oncogenic KRAS blockade therapy: Renewed enthusiasm and persistent challenges. Mol. Cancer 2021, 20, 128. [Google Scholar] [CrossRef]
- Cruz-Migoni, A.; Canning, P.; Quevedo, C.E.; Bataille, C.J.R.; Bery, N.; Miller, A.; Russell, A.J.; Phillips, S.E.V.; Carr, S.B.; Rabbitts, T.H. Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc. Natl. Acad. Sci. USA 2019, 116, 2545–2550. [Google Scholar] [CrossRef]
- Kessler, D.; Gmachl, M.; Mantoulidis, A.; Martin, L.J.; Zoephel, A.; Mayer, M.; Gollner, A.; Covini, D.; Fischer, S.; Gerstberger, T.; et al. Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. USA 2019, 116, 15823–15829. [Google Scholar] [CrossRef]
- McCarthy, M.J.; Pagba, C.V.; Prakash, P.; Naji, A.K.; van der Hoeven, D.; Liang, H.; Gupta, A.K.; Zhou, Y.; Cho, K.-J.; Hancock, J.F.; et al. Discovery of High-Affinity Noncovalent Allosteric KRAS Inhibitors That Disrupt Effector Binding. ACS Omega 2019, 4, 2921–2930. [Google Scholar] [CrossRef]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503, 548–551. [Google Scholar] [CrossRef]
- Welsch, M.E.; Kaplan, A.; Chambers, J.M.; Stokes, M.E.; Bos, P.H.; Zask, A.; Zhang, Y.; Sanchez-Martin, M.; Badgley, M.A.; Huang, C.S.; et al. Multivalent Small-Molecule Pan-RAS Inhibitors. Cell 2017, 168, 878–889.e29. [Google Scholar] [CrossRef]
- Liu, J.; Kang, R.; Tang, D. The KRAS-G12C inhibitor: Activity and resistance. Cancer Gene Ther. 2022, 29, 875–878. [Google Scholar] [CrossRef]
- Lee, A. Sotorasib: A Review in KRAS G12C Mutation-Positive Non-small Cell Lung Cancer. Target. Oncol. 2022, 17, 727–733. [Google Scholar] [CrossRef]
- Kwan, A.K.; Piazza, G.A.; Keeton, A.B.; Leite, C.A. The path to the clinic: A comprehensive review on direct KRAS(G12C) inhibitors. J. Exp. Clin. Cancer Res. 2022, 41, 27. [Google Scholar] [CrossRef]
- Nagasaka, M.; Li, Y.; Sukari, A.; Ou, S.H.; Al-Hallak, M.N.; Azmi, A.S. KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer Treat. Rev. 2020, 84, 101974. [Google Scholar] [CrossRef]
- Fakih, M.; O’Neil, B.; Price, T.J.; Falchook, G.S.; Desai, J.; Kuo, J.; Govindan, R.; Rasmussen, E.; Morrow, P.K.; Ngang, J.; et al. Phase 1 Study Evaluating the Safety, Tolerability, Pharmacokinetics (PK), and Efficacy of AMG 510, a Novel Small Molecule KRASG12C Inhibitor, in Advanced Solid Tumors; American Society of Clinical Oncology: Alexandria, VA, USA, 2019. [Google Scholar]
- Papadopoulos, K.P.; Ou, S.-H.I.; Johnson, M.L.; Christensen, J.; Velastegui, K.; Potvin, D.; Faltaos, D.; Chao, R.C. A phase I/II multiple expansion cohort trial of MRTX849 in patients with advanced solid tumors with KRAS G12C mutation. J. Clin. Oncol. 2019, 37 (Suppl. S15), TPS3161. [Google Scholar] [CrossRef]
- Janes, M.R.; Zhang, J.; Li, L.-S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589.e17. [Google Scholar] [CrossRef]
- Gentile, D.R.; Rathinaswamy, M.K.; Jenkins, M.L.; Moss, S.M.; Siempelkamp, B.D.; Renslo, A.R.; Burke, J.E.; Shokat, K.M. Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States. Cell Chem. Biol. 2017, 24, 1455–1466.e14. [Google Scholar] [CrossRef]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Robert, C.; Grob, J.J.; Stroyakovskiy, D.; Karaszewska, B.; Hauschild, A.; Levchenko, E.; Chiarion Sileni, V.; Schachter, J.; Garbe, C.; Bondarenko, I.; et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N. Engl. J. Med. 2019, 381, 626–636. [Google Scholar] [CrossRef]
- Hallin, J.; Engstrom, L.D.; Hargis, L.; Calinisan, A.; Aranda, R.; Briere, D.M.; Sudhakar, N.; Bowcut, V.; Baer, B.R.; Ballard, J.A.; et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020, 10, 54–71. [Google Scholar] [CrossRef]
- Karp, J.E.; Flatten, K.; Feldman, E.J.; Greer, J.M.; Loegering, D.A.; Ricklis, R.M.; Morris, L.E.; Ritchie, E.; Smith, B.D.; Ironside, V.; et al. Active oral regimen for elderly adults with newly diagnosed acute myelogenous leukemia: A preclinical and phase 1 trial of the farnesyltransferase inhibitor tipifarnib (R115777, Zarnestra) combined with etoposide. Blood 2009, 113, 4841–4852. [Google Scholar] [CrossRef]
- Youssef, M.E.; Cavalu, S.; Hasan, A.M.; Yahya, G.; Abd-Eldayem, M.A.; Saber, S. Role of Ganetespib, an HSP90 Inhibitor, in Cancer Therapy: From Molecular Mechanisms to Clinical Practice. Int. J. Mol. Sci. 2023, 24, 5014. [Google Scholar] [CrossRef]
- Kim, M.J.; Lee, S.J.; Ryu, J.H.; Kim, S.H.; Kwon, I.C.; Roberts, T.M. Combination of KRAS gene silencing and PI3K inhibition for ovarian cancer treatment. J. Control. Release 2020, 318, 98–108. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Wu, Y.; Walker, J.R.; Westberg, M.; Ning, L.; Monje, M.; Kirkland, T.A.; Lin, M.Z.; Su, Y. Kinase-Modulated Bioluminescent Indicators Enable Noninvasive Imaging of Drug Activity in the Brain. ACS Central Sci. 2023, 9, 719–732. [Google Scholar] [CrossRef]
- Sakamoto, K.; Lin, B.; Nunomura, K.; Izawa, T.; Nakagawa, S. The K-Ras(G12D)-inhibitory peptide KS-58 suppresses growth of murine CT26 colorectal cancer cell-derived tumors. Sci. Rep. 2022, 12, 8121. [Google Scholar] [CrossRef]
- Wang, A.; Liu, J.; Li, X.; Zou, F.; Qi, Z.; Qi, S.; Liu, Q.; Wang, Z.; Cao, J.; Jiang, Z.; et al. Discovery of a highly potent pan-RAF inhibitor IHMT-RAF-128 for cancer treatment. Eur. J. Pharmacol. 2023, 952, 175752. [Google Scholar] [CrossRef]
- Ghosh, S.; Fan, F.; Powell, R.T.; Roszik, J.; Park, Y.S.; Stephan, C.; Sebastian, M.; Tan, L.; Sorokin, A.V.; Lorenzi, P.L.; et al. Vincristine Enhances the Efficacy of MEK Inhibitors in Preclinical Models of KRAS-mutant Colorectal Cancer. Mol. Cancer Ther. 2023, 22, 962–975. [Google Scholar] [CrossRef]
- Carbone, D.; De Franco, M.; Pecoraro, C.; Bassani, D.; Pavan, M.; Cascioferro, S.; Parrino, B.; Cirrincione, G.; Dall’acqua, S.; Moro, S.; et al. Discovery of the 3-Amino-1,2,4-triazine-Based Library as Selective PDK1 Inhibitors with Therapeutic Potential in Highly Aggressive Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2023, 24, 3679. [Google Scholar] [CrossRef]
- Hassin, O.; Oren, M. Drugging p53 in cancer: One protein, many targets. Nat. Rev. Drug Discov. 2023, 22, 127–144. [Google Scholar] [CrossRef]
- Hu, J.; Cao, J.; Topatana, W.; Juengpanich, S.; Li, S.; Zhang, B.; Shen, J.; Cai, L.; Cai, X.; Chen, M. Targeting mutant p53 for cancer therapy: Direct and indirect strategies. J. Hematol. Oncol. 2021, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Maddalena, M.; Mallel, G.; Nataraj, N.B.; Shreberk-Shaked, M.; Hassin, O.; Mukherjee, S.; Arandkar, S.; Rotkopf, R.; Kapsack, A.; Lambiase, G.; et al. TP53 missense mutations in PDAC are associated with enhanced fibrosis and an immunosuppressive microenvironment. Proc. Natl. Acad. Sci. USA 2021, 118, e2025631118. [Google Scholar] [CrossRef]
- McLornan, D.P.; List, A.; Mufti, G.J. Applying synthetic lethality for the selective targeting of cancer. N. Engl. J. Med. 2014, 371, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef] [PubMed]
- Wellenstein, M.D.; Coffelt, S.B.; Duits, D.E.M.; Van Miltenburg, M.H.; Slagter, M.; De Rink, I.; Henneman, L.; Kas, S.M.; Prekovic, S.; Hau, C.-S.; et al. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature 2019, 572, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.P.; Lozano, G. Mutant p53 partners in crime. Cell Death Differ. 2018, 25, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Pilley, S.; Rodriguez, T.A.; Vousden, K.H. Mutant p53 in cell-cell interactions. Genes Dev. 2021, 35, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Sabapathy, K.; Lane, D.P. Therapeutic targeting of p53: All mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 2018, 15, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Stein, Y.; Rotter, V.; Aloni-Grinstein, R. Gain-of-function mutant p53: All the roads lead to tumorigenesis. Int. J. Mol. Sci. 2019, 20, 6197. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Xu, D.; Zhang, T.; Hu, W.; Feng, Z. Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell Biol. 2020, 12, 674–687. [Google Scholar] [CrossRef]
- Zhu, G.; Pan, C.; Bei, J.-X.; Li, B.; Liang, C.; Xu, Y.; Fu, X. Mutant p53 in cancer progression and targeted therapies. Front. Oncol. 2020, 10, 595187. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, A.; Tolcher, A.; Beeram, M.; Nemunaitis, J.; Weiss, G.J.; Bhalla, K.; Agrawal, M.; Nichols, G.; Middleton, S.; Beryozkina, A.; et al. Clinical pharmacology characterization of RG7112, an MDM2 antagonist, in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2015, 76, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Andreeff, M.; Kelly, K.R.; Yee, K.; Assouline, S.; Strair, R.; Popplewell, L.; Bowen, D.; Martinelli, G.; Drummond, M.W.; Vyas, P.; et al. Results of the Phase I Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin. Cancer Res. 2016, 22, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Chen, G.; Jukofsky, L.; Geho, D.; Han, S.W.; Birzele, F.; Bader, S.; Himmelein, L.; Cai, J.; Albertyn, Z.; et al. MDM2 antagonist clinical response association with a gene expression signature in acute myeloid leukaemia. Br. J. Haematol. 2015, 171, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Ray-Coquard, I.; Blay, J.-Y.; Italiano, A.; Le Cesne, A.; Penel, N.; Zhi, J.; Heil, F.; Rueger, R.; Graves, B.; Ding, M.; et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol. 2012, 13, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.; Wovkulich, P.; Pizzolato, G.; Lovey, A.; Ding, Q.; Jiang, N.; Liu, J.-J.; Zhao, C.; Glenn, K.; Wen, Y.; et al. Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development. ACS Med. Chem. Lett. 2013, 4, 466–469. [Google Scholar] [CrossRef]
- Mascarenhas, J.; Lu, M.; Kosiorek, H.; Virtgaym, E.; Xia, L.; Sandy, L.; Mesa, R.; Petersen, B.; Farnoud, N.; Najfeld, V.; et al. Oral idasanutlin in patients with polycythemia vera. Blood 2019, 134, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.G.; Dail, M.; Garcia, J.S.; Jonas, B.A.; Yee, K.W.L.; Kelly, K.R.; Vey, N.; Assouline, S.; Roboz, G.J.; Paolini, S.; et al. Venetoclax and idasanutlin in relapsed/refractory AML: A nonrandomized, open-label phase 1b trial. Blood 2023, 141, 1265–1276. [Google Scholar] [CrossRef]
- Uy, G.L.; Assouline, S.; Young, A.-M.; Blotner, S.; Higgins, B.; Chen, L.-C.; Yee, K. Phase 1 study of the MDM2 antagonist RO6839921 in patients with acute myeloid leukemia. Investig. New Drugs 2020, 38, 1430–1441. [Google Scholar] [CrossRef]
- Razak, A.R.A.; Miller, W.H.; Uy, G.L.; Blotner, S.; Young, A.-M.; Higgins, B.; Chen, L.-C.; Gore, L. A phase 1 study of the MDM2 antagonist RO6839921, a pegylated prodrug of idasanutlin, in patients with advanced solid tumors. Investig. New Drugs 2020, 38, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Abdul Razak, A.R.; Bauer, S.; Suarez, C.; Lin, C.C.; Quek, R.; Hütter-Krönke, M.L.; Cubedo, R.; Ferretti, S.; Guerreiro, N.; Jullion, A.; et al. Co-Targeting of MDM2 and CDK4/6 with Siremadlin and Ribociclib for the Treatment of Patients with Well-Differentiated or Dedifferentiated Liposarcoma: Results from a Proof-of-Concept, Phase Ib Study. Clin. Cancer Res. 2022, 28, 1087–1097. [Google Scholar] [CrossRef]
- Taylor, A.; Lee, D.; Allard, M.; Poland, B.; Greg Slatter, J. Phase 1 Concentration-QTc and Cardiac Safety Analysis of the MDM2 Antagonist KRT-232 in Patients with Advanced Solid Tumors, Multiple Myeloma, or Acute Myeloid Leukemia. Clin. Pharmacol. Drug Dev. 2021, 10, 918–926. [Google Scholar] [CrossRef]
- Gluck, W.L.; Gounder, M.M.; Frank, R.; Eskens, F.; Blay, J.Y.; Cassier, P.A.; Soria, J.-C.; Chawla, S.; De Weger, V.; Wagner, A.J.; et al. Phase 1 study of the MDM2 inhibitor AMG 232 in patients with advanced P53 wild-type solid tumors or multiple myeloma. Investig. New Drugs 2020, 38, 831–843. [Google Scholar] [CrossRef]
- Erba, H.P.; Becker, P.S.; Shami, P.J.; Grunwald, M.R.; Flesher, D.L.; Zhu, M.; Rasmussen, E.; Henary, H.A.; Anderson, A.A.; Wang, E.S. Phase 1b study of the MDM2 inhibitor AMG 232 with or without trametinib in relapsed/refractory acute myeloid leukemia. Blood Adv. 2019, 3, 1939–1949. [Google Scholar] [CrossRef] [PubMed]
- Jeay, S.; Ferretti, S.; Holzer, P.; Fuchs, J.; Chapeau, E.A.; Wartmann, M.; Sterker, D.; Romanet, V.; Murakami, M.; Kerr, G.; et al. Dose and Schedule Determine Distinct Molecular Mechanisms Underlying the Efficacy of the p53–MDM2 Inhibitor HDM201. Cancer Res. 2018, 78, 6257–6267. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Fujiwara, Y.; Nakano, K.; Shimizu, T.; Tomomatsu, J.; Koyama, T.; Ogura, M.; Tachibana, M.; Kakurai, Y.; Yamashita, T.; et al. Safety and pharmacokinetics of milademetan, a MDM2 inhibitor, in Japanese patients with solid tumors: A phase I study. Cancer Sci. 2021, 112, 2361–2370. [Google Scholar] [CrossRef] [PubMed]
- De Weger, V.A.; de Jonge, M.; Langenberg, M.H.; Schellens, J.H.; Lolkema, M.; Varga, A.; Demers, B.; Thomas, K.; Hsu, K.; Tuffal, G.; et al. A phase I study of the HDM2 antagonist SAR405838 combined with the MEK inhibitor pimasertib in patients with advanced solid tumours. Br. J. Cancer 2019, 120, 286–293. [Google Scholar] [CrossRef]
- Bidou, L.; Bugaud, O.; Belakhov, V.; Baasov, T.; Namy, O. Characterization of new-generation aminoglycoside promoting premature termination codon readthrough in cancer cells. RNA Biol. 2017, 14, 378–388. [Google Scholar] [CrossRef]
- Xie, X.; Fan, C.; Luo, B.; Zhang, J.; Jensen, L.D.; Burman, J.; Jönsson, C.; Ljusberg, A.; Larsson, P.; Zhao, Z.; et al. APR-246 enhances colorectal cancer sensitivity to radiotherapy. Mol. Cancer Ther. 2023, 22, 947–961. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Abrams, S.L.; Steelman, L.S.; Cocco, L.; Ratti, S.; Martelli, A.M.; Lombardi, P.; Gizak, A.; Duda, P. APR-246—The Mutant TP53 Reactivator—Increases the Effectiveness of Berberine and Modified Berberines to Inhibit the Proliferation of Pancreatic Cancer Cells. Biomolecules 2022, 12, 276. [Google Scholar] [CrossRef]
- Sallman, D.A.; DeZern, A.E.; Garcia-Manero, G.; Steensma, D.P.; Roboz, G.J.; Sekeres, M.A.; Cluzeau, T.; Sweet, K.L.; McLemore, A.; McGraw, K.L.; et al. Eprenetapopt (APR-246) and Azacitidine in TP53-Mutant Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1584–1594. [Google Scholar] [CrossRef] [PubMed]
- Zatloukalová, P.; Galoczová, M.; Vojtěšek, B. Prima-1 and APR-246 in Cancer Therapy. Klin. Onkol. 2018, 31 (Suppl. S2), 71–76. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Michels, J.; Mezzadra, R.; Venkatesh, D.; Dong, L.; Gomez, R.; Samaan, F.; Ho, Y.-J.; Campesato, L.F.; Mangarin, L.; et al. Increased p53 expression induced by APR-246 reprograms tumor-associated macrophages to augment immune checkpoint blockade. J. Clin. Investig. 2022, 132. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, A.; Patel, A.A.; Silver, N.L.; Tang, L.; Liu, Z.; Wang, L.; Tanaka, N.; Rao, X.; Takahashi, H.; Maduka, N.K.; et al. COTI-2, A Novel Thiosemicarbazone Derivative, Exhibits Antitumor Activity in HNSCC through p53-dependent and -independent Mechanisms. Clin. Cancer Res. 2019, 25, 5650–5662. [Google Scholar] [CrossRef] [PubMed]
- Salim, K.Y.; Vareki, S.M.; Danter, W.R.; San-Marina, S.; Koropatnick, J. COTI-2, a novel small molecule that is active against multiple human cancer cell lines in vitro and in vivo. Oncotarget 2016, 7, 41363–41379. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.H.B.; Hager, S.; Mathuber, M.; Pósa, V.; Roller, A.; Enyedy, É.A.; Stefanelli, A.; Berger, W.; Keppler, B.K.; Heffeter, P.; et al. Cancer Cell Resistance Against the Clinically Investigated Thiosemicarbazone COTI-2 Is Based on Formation of Intracellular Copper Complex Glutathione Adducts and ABCC1-Mediated Efflux. J. Med. Chem. 2020, 63, 13719–13732. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.A.; Schutte, M.; Hoque, A.T.M.S.; Moskaluk, C.A.; da Costa, L.T.; Rozenblum, E.; Weinstein, C.L.; Fischer, A.; Yeo, C.J.; Hruban, R.H.; et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996, 271, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Sunamura, M.; Horii, A. Molecular mechanisms of pancreatic carcinogenesis. Cancer Sci. 2006, 97, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef]
- Xia, X.; Wu, W.; Huang, C.; Cen, G.; Jiang, T.; Cao, J.; Huang, K.; Qiu, Z. SMAD4 and its role in pancreatic cancer. Tumor Biol. 2015, 36, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Saiki, Y.; Horii, A. Molecular pathology of pancreatic cancer. Pathol. Int. 2014, 64, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.H. Non-Smad TGF-beta signals. J. Cell Sci. 2005, 118 Pt 16, 3573–3584. [Google Scholar] [CrossRef] [PubMed]
- Alhopuro, P.; Alazzouzi, H.; Sammalkorpi, H.; Dávalos, V.; Salovaara, R.; Hemminki, A.; Järvinen, H.; Mecklin, J.-P.; Schwartz, S.; Aaltonen, L.A.; et al. SMAD4 levels and response to 5-fluorouracil in colorectal cancer. Clin. Cancer Res. 2005, 11, 6311–6316. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M.M.; von Eyben, R.; Pai, J.; Vossler, S.R.; Limaye, M.; Jayachandran, P.; Anderson, E.M.; Shaffer, J.L.; Longacre, T.; Pai, R.K.; et al. Smad4 inactivation predicts for worse prognosis and response to fluorouracil-based treatment in colorectal cancer. J. Clin. Pathol. 2015, 68, 341–345. [Google Scholar] [CrossRef]
- Cheng, H.; Fertig, E.J.; Ozawa, H.; Hatakeyama, H.; Howard, J.D.; Perez, J.; Considine, M.; Thakar, M.; Ranaweera, R.; Krigsfeld, G.; et al. Decreased SMAD4 expression is associated with induction of epithelial-to-mesenchymal transition and cetuximab resistance in head and neck squamous cell carcinoma. Cancer Biol. Ther. 2015, 16, 1252–1258. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, K.; Hablesreiter, R.; Inoue, Y.; Yoshida, K.; Briest, F.; Christen, F.; Kakiuchi, N.; Yoshizato, T.; Shiozawa, Y.; Shiraishi, Y.; et al. A genetically defined signature of responsiveness to erlotinib in early-stage pancreatic cancer patients: Results from the CONKO-005 trial. EBioMedicine 2021, 66, 103327. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Song, W.; Cao, R.; Ye, C.; Zhang, L.; Chen, H.; Wang, J.; Shi, Y.; Li, R.; Li, Y.; et al. An EGFR/HER2-targeted conjugate sensitizes gemcitabine-sensitive and resistant pancreatic cancer through different SMAD4-mediated mechanisms. Nat. Commun. 2022, 13, 5506. [Google Scholar] [CrossRef] [PubMed]
- Hufford, D.J. Folk medicine and health culture in contemporary society. Prim. Care Clin. Off. Pract. 1997, 24, 723–741. [Google Scholar] [CrossRef]
- Ling, Z.; Liu, H.; Yang, J.; He, J.-Q.; Yang, S.; Chen, Q.-H. Qianlongtong capsule elevates the Smad4 gene expression in prostate stromal cells. Zhonghua Nan Ke Xue Natl. J. Androl. 2014, 20, 730–733. [Google Scholar]
- Cappuccio, G.; Caiazza, M.; Roca, A.; Melis, D.; Iuliano, A.; Matyas, G.; Rubino, M.; Limongelli, G.; Brunetti-Pierri, N. A pilot clinical trial with losartan in Myhre syndrome. Am. J. Med. Genet. Part A 2021, 185, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.-M.; Liu, X.; Wheler, J.; Naing, A.; Hong, D.; Coleman, R.L.; Tsimberidou, A.; Janku, F.; Zinner, R.; Lu, K.; et al. Targeted PI3K/AKT/mTOR therapy for metastatic carcinomas of the cervix: A phase I clinical experience. Oncotarget 2014, 5, 11168–11179. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, S.M.; Koch, R.; Porcu, P.; Oki, Y.; Moskowitz, A.; Perez, M.; Myskowski, P.; Officer, A.; Jaffe, J.D.; Morrow, S.N.; et al. Activity of the PI3K-δ,γ inhibitor duvelisib in a phase 1 trial and preclinical models of T-cell lymphoma. Blood 2018, 131, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Kaley, T.J.; Panageas, K.S.; Mellinghoff, I.K.; Nolan, C.; Gavrilovic, I.T.; DeAngelis, L.M.; Abrey, L.E.; Holland, E.C.; Lassman, A.B. Phase II trial of an AKT inhibitor (perifosine) for recurrent glioblastoma. J. Neuro-Oncol. 2019, 144, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Lin, N.U.; Maurer, M.A.; Chen, H.; Mahvash, A.; Sahin, A.; Akcakanat, A.; Li, Y.; Abramson, V.; Litton, J.; et al. Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res. 2019, 21, 78. [Google Scholar] [CrossRef]
- LoRusso, P.M.; Gounder, M.; Jalal, S.I.; André, V.; Kambhampati, S.R.P.; Loizos, N.; Hall, J.; Holzer, T.R.; Nasir, A.; Cosaert, J.; et al. Phase 1 study of narnatumab, an anti-RON receptor monoclonal antibody, in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 442–450. [Google Scholar] [CrossRef]
- Kreuger, I.Z.; Slieker, R.C.; van Groningen, T.; van Doorn, R. Therapeutic Strategies for Targeting CDKN2A Loss in Melanoma. J. Investig. Dermatol. 2023, 143, 18–25.e1. [Google Scholar] [CrossRef]
- Liggett, W.H.; Sidransky, D. Role of the p16 tumor suppressor gene in cancer. J. Clin. Oncol. 1998, 16, 1197–1206. [Google Scholar] [CrossRef]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D.; Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef]
- Stott, F.J.; Bates, S.; James, M.C.; McConnell, B.B.; Starborg, M.; Brookes, S.; Palmero, I.; Ryan, K.; Hara, E.; Vousden, K.H.; et al. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998, 17, 5001–5014. [Google Scholar] [CrossRef]
- Xing, X.; Cai, W.; Shi, H.; Wang, Y.; Li, M.; Jiao, J.; Chen, M. The prognostic value of CDKN2A hypermethylation in colorectal cancer: A meta-analysis. Br. J. Cancer 2013, 108, 2542–2548. [Google Scholar] [CrossRef]
- Liu, M.T.; Liu, J.Y.; Su, J. CDKN2A gene in melanoma. Zhonghua Bing Li Xue Za Zhi 2019, 48, 909–912. [Google Scholar]
- Tsuriel, S.; Hannes, V.; Hasona, A.; Raz, M.; Hershkovitz, D. Digital PCR-Based Method for Detecting CDKN2A Loss in Brain Tumours. Mol. Diagn. Ther. 2022, 26, 689–698. [Google Scholar] [CrossRef]
- Ladanyi, M. Implications of P16/CDKN2A deletion in pleural mesotheliomas. Lung Cancer 2005, 49 (Suppl. S1), S95–S98. [Google Scholar] [CrossRef]
- Wang, J.; Patil, V.; Liu, J.; Dogan, H.; Tabatabai, G.S.; Yefet, L.; Behling, F.; Hoffman, E.; Bunda, S.; Yakubov, R.; et al. Increased mRNA expression of CDKN2A is a transcriptomic marker of clinically aggressive meningiomas. Acta Neuropathol. 2023, 146, 145–162. [Google Scholar] [CrossRef]
- Kimura, H.; Klein, A.P.M.; Hruban, R.H.; Roberts, N.J.V. The Role of Inherited Pathogenic CDKN2A Variants in Susceptibility to Pancreatic Cancer. Pancreas 2021, 50, 1123–1130. [Google Scholar] [CrossRef]
- Johnston, S.R.D.; Harbeck, N.; Hegg, R.; Toi, M.; Martin, M.; Shao, Z.M.; Zhang, Q.Y.; Rodriguez, J.L.M.; Campone, M.; Hamilton, E.; et al. Abemaciclib Combined With Endocrine Therapy for the Adjuvant Treatment of HR+, HER2−, Node-Positive, High-Risk, Early Breast Cancer (monarchE). J. Clin. Oncol. 2020, 38, 3987–3998. [Google Scholar] [CrossRef]
- Turner, N.C.; Slamon, D.J.; Ro, J.; Bondarenko, I.; Im, S.-A.; Masuda, N.; Colleoni, M.; DeMichele, A.; Loi, S.; Verma, S.; et al. Overall Survival with Palbociclib and Fulvestrant in Advanced Breast Cancer. N. Engl. J. Med. 2018, 379, 1926–1936. [Google Scholar] [CrossRef]
- Weiss, J.M.; Csoszi, T.; Maglakelidze, M.; Hoyer, R.J.; Beck, J.T.; Gomez, M.D.; Lowczak, A.; Aljumaily, R.; Lima, C.R.; Boccia, R.V.; et al. Myelopreservation with the CDK4/6 inhibitor trilaciclib in patients with small-cell lung cancer receiving first-line chemotherapy: A phase Ib/randomized phase II trial. Ann. Oncol. 2019, 30, 1613–1621. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Tibes, R.; Kadia, T.; Kantarjian, H.; Arellano, M.; Knight, E.A.; Xiong, H.; Qin, Q.; Munasinghe, W.; Roberts-Rapp, L.; et al. Phase 1 dose escalation trial of ilorasertib, a dual Aurora/VEGF receptor kinase inhibitor, in patients with hematologic malignancies. Investig. New Drugs 2015, 33, 870–880. [Google Scholar] [CrossRef]
- Maitland, M.L.; Piha-Paul, S.; Falchook, G.; Kurzrock, R.; Nguyen, L.; Janisch, L.; Karovic, S.; McKee, M.; Hoening, E.; Wong, S.; et al. Clinical pharmacodynamic/exposure characterisation of the multikinase inhibitor ilorasertib (ABT-348) in a phase 1 dose-escalation trial. Br. J. Cancer 2018, 118, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Grande, E.; Teulé, A.; Alonso-Gordoa, T.; Jiménez-Fonseca, P.; Benavent, M.; Capdevila, J.; Custodio, A.; Vera, R.; Munarriz, J.; La Casta, A.; et al. The PALBONET Trial: A Phase II Study of Palbociclib in Metastatic Grade 1 and 2 Pancreatic Neuroendocrine Tumors (GETNE-1407). Oncology 2020, 25, 745-e1265. [Google Scholar] [CrossRef] [PubMed]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; Koay, E.J.; Chari, S.T.; Maitra, A. Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology 2019, 156, 2024–2040. [Google Scholar] [CrossRef]

| S/No | Gene | Frequency of Mutation in PDACs | Types of Mutation | Amino Acid Residues | References |
|---|---|---|---|---|---|
| 1 | KRAS | 90% | Missense point | G12C, G12D, and G12V | [28,29,30,31] |
| 2 | CDNK2A | 30–40% | Point mutation, deletion and loss of heterozygousity, insertion and frame shift mutation, promoter methylation, splice site mutation | P16INK4A (p16-Leu148) and P14ARF mutations | [32,33] |
| 3 | DPC4/SMAD4 | 50% | Nonsense mutation, missense mutation, frameshift mutation, splice site mutation, deletion and insertion, point mutation, promoter methylation, large rearrangement, silent mutation | MH1 domain: R361C, R361H, R361S, and R361G MH2 domain: R100C, D351N, L384P, P529L. Linker region: E249K, G253V C-terminal region: Q408P and G437E | [19,32] |
| 4 | TP53 | ≥50% | Missense, nonsense, frameshift, splice site, deletion and insertion, promoter methylation, hotspot, wild-type p53. | R17H, R28Q, R273H, R282W, and Y220C | [19,34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olaoba, O.T.; Adelusi, T.I.; Yang, M.; Maidens, T.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Li, G. Driver Mutations in Pancreatic Cancer and Opportunities for Targeted Therapy. Cancers 2024, 16, 1808. https://doi.org/10.3390/cancers16101808
Olaoba OT, Adelusi TI, Yang M, Maidens T, Kimchi ET, Staveley-O’Carroll KF, Li G. Driver Mutations in Pancreatic Cancer and Opportunities for Targeted Therapy. Cancers. 2024; 16(10):1808. https://doi.org/10.3390/cancers16101808
Chicago/Turabian StyleOlaoba, Olamide T., Temitope I. Adelusi, Ming Yang, Tessa Maidens, Eric T. Kimchi, Kevin F. Staveley-O’Carroll, and Guangfu Li. 2024. "Driver Mutations in Pancreatic Cancer and Opportunities for Targeted Therapy" Cancers 16, no. 10: 1808. https://doi.org/10.3390/cancers16101808
APA StyleOlaoba, O. T., Adelusi, T. I., Yang, M., Maidens, T., Kimchi, E. T., Staveley-O’Carroll, K. F., & Li, G. (2024). Driver Mutations in Pancreatic Cancer and Opportunities for Targeted Therapy. Cancers, 16(10), 1808. https://doi.org/10.3390/cancers16101808






