Cyclooxygenase-2 Blockade Is Crucial to Restore Natural Killer Cell Activity before Anti-CTLA-4 Therapy against High-Grade Serous Ovarian Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
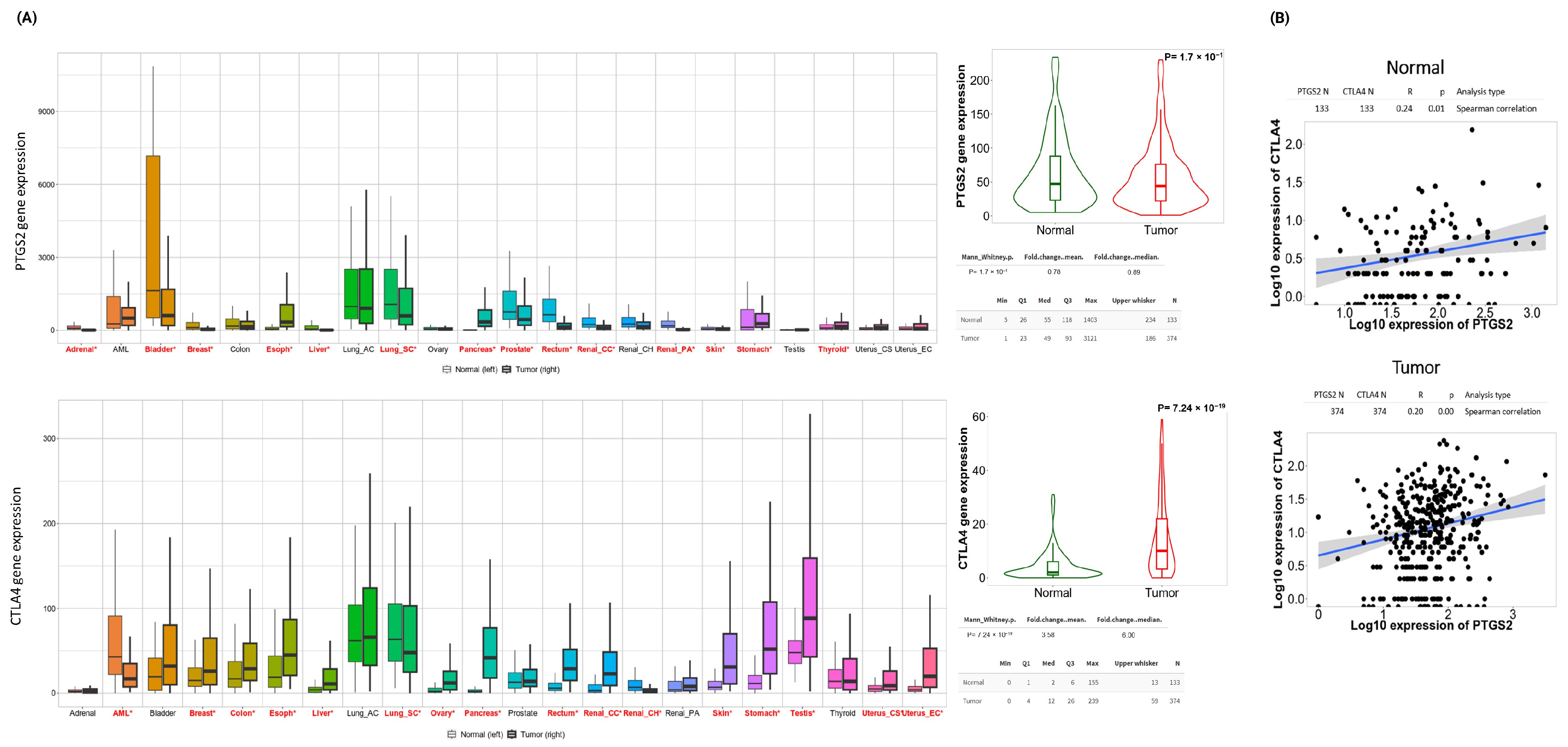
2.2. Genetic Assessment of COX-2, CTLA-4, and Ovarian Tumor Immune Microenvironment
2.3. Patients
2.4. NK Cells Isolation and Cell Cultures
2.5. Drugs Assessment
2.6. Treatment Protocol
2.7. Immunophenotyping of NK Cells
2.8. Cytotoxicity Assay in NK Cells and HGSOC Cell Lines Co-Culture
2.9. Statistical Analyses
3. Results
3.1. The PTGS2/CTLA4 Ratio Defines Survival in Multiple HGSOC Cohorts
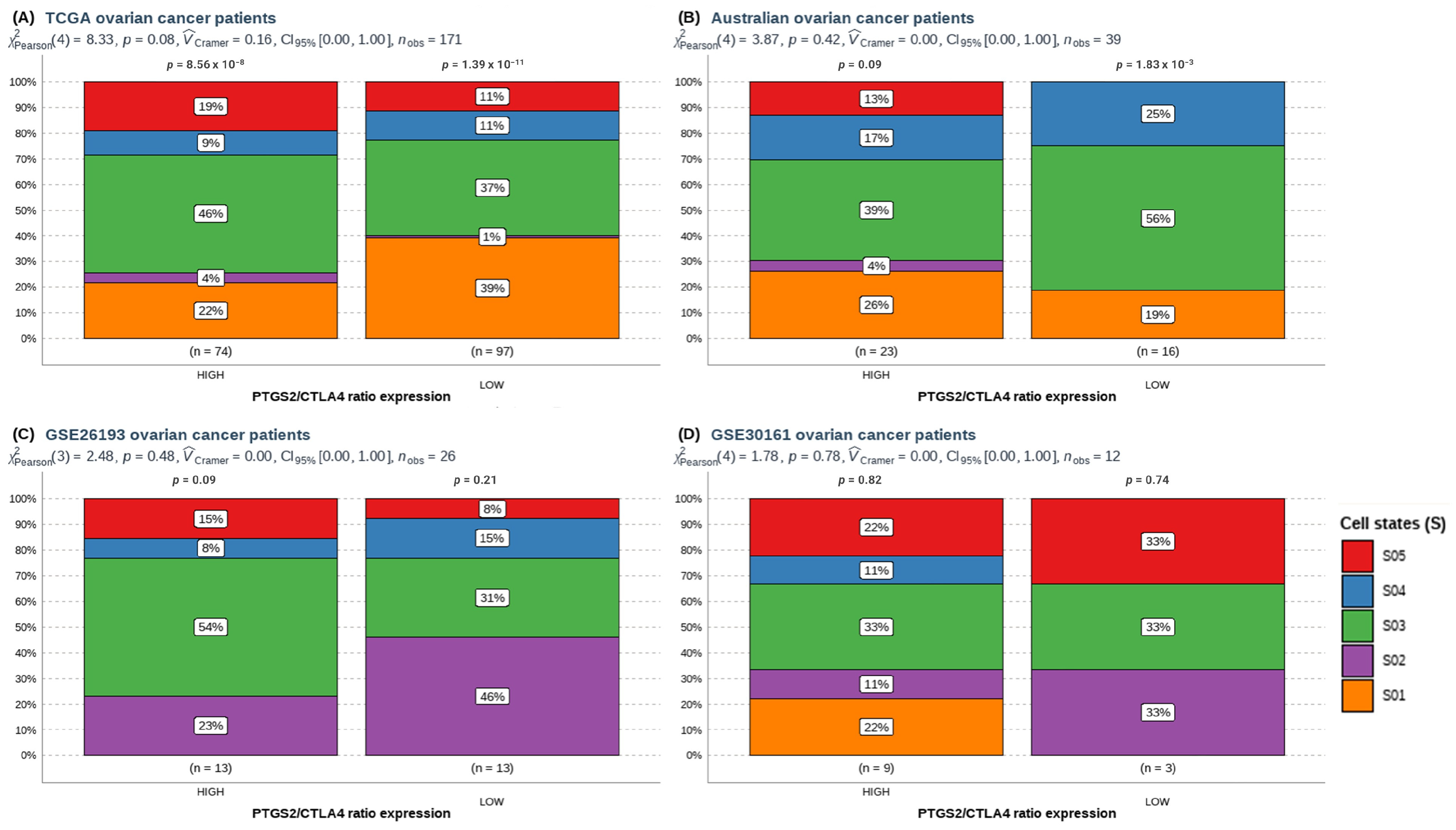
3.2. A High PTGS2/CTLA4 Ratio Determines the Abundance and Cell States of Immune Cells in HGSOC
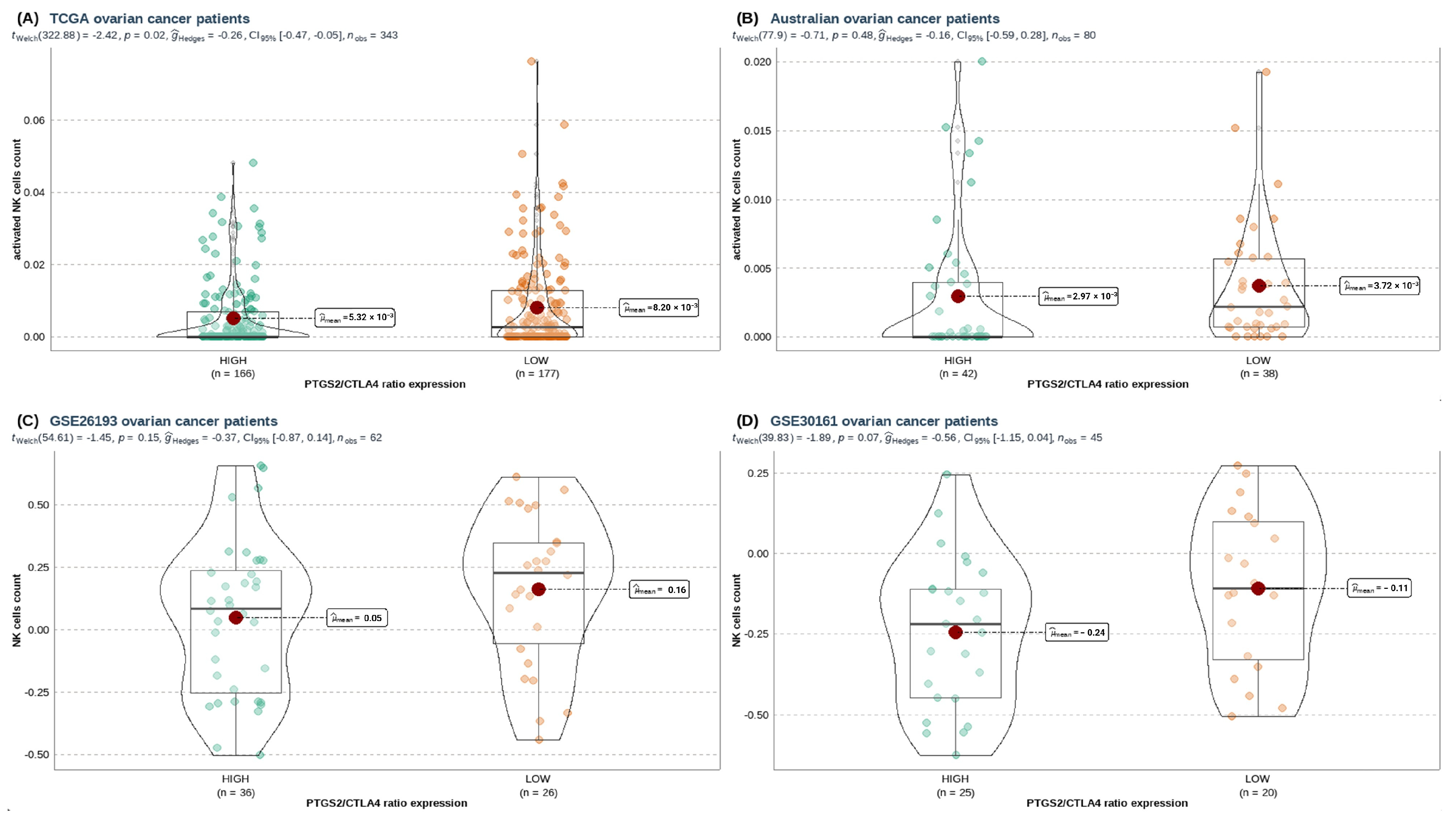
3.3. A High PTGS2/CTLA4 Ratio Hinders NK Gene Score Associated with NK Infiltration, Cytotoxicity, and Survival in HGSOC
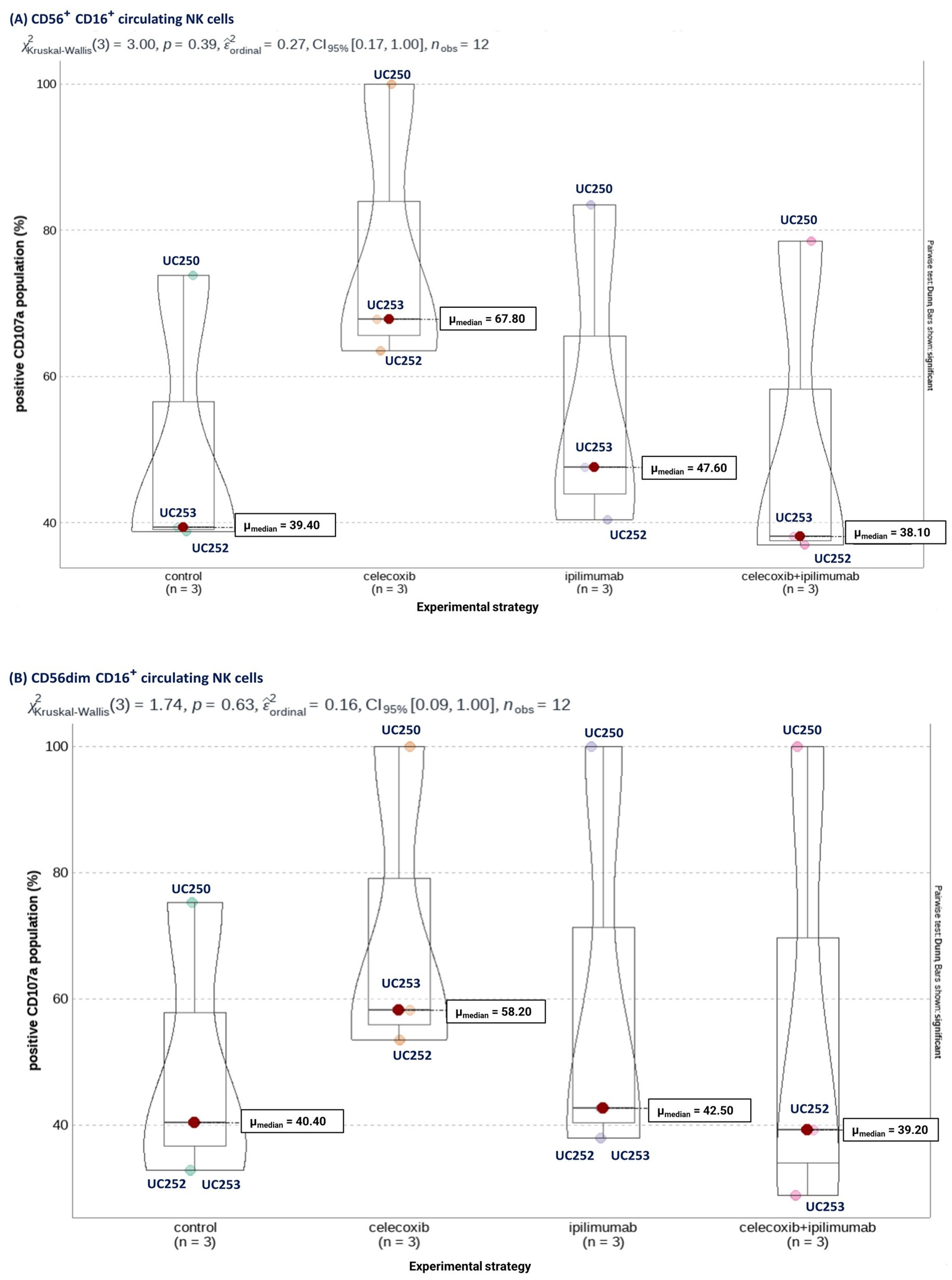
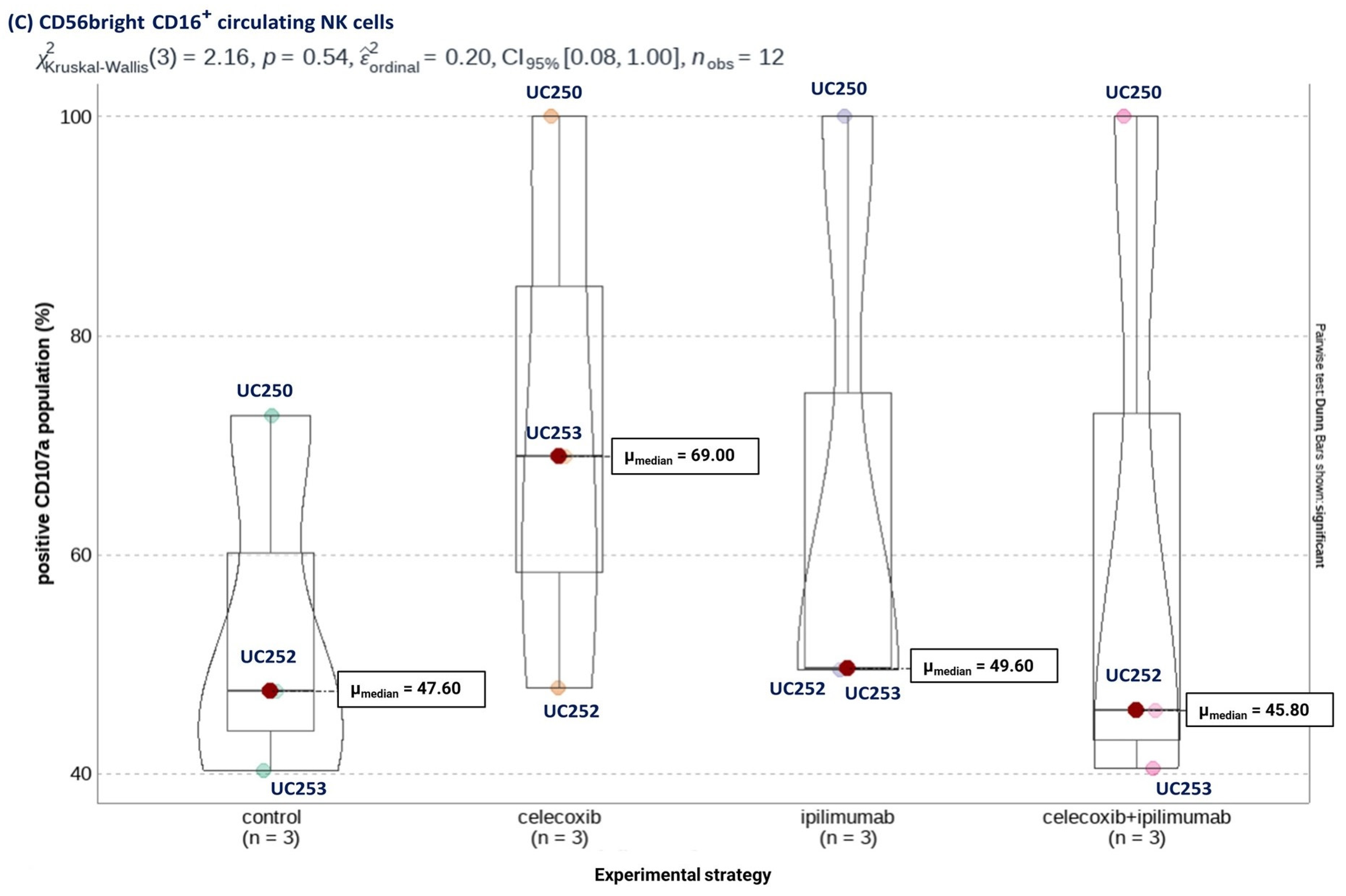
3.4. Selective COX-2 Blockade through Celecoxib Improves NK Cells’ Cytotoxic Capacity Independent of CTLA-4 Signaling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, Z.; Mirza, H.; Ennis, D.P.; Smith, P.; Morrill Gavarró, L.; Sokota, C.; Giannone, G.; Goranova, T.; Bradley, T.; Piskorz, A.; et al. The Genomic Landscape of Early-Stage Ovarian High-Grade Serous Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2022, 28, 2911–2922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, D.; Saw, P.E.; Song, E. Turning cold tumors hot: From molecular mechanisms to clinical applications. Trends Immunol. 2022, 43, 523–545. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.P.; Balmaceda, C.; Bravo, M.L.; Kato, S.; Villarroel, A.; Owen, G.I.; Roa, J.C.; Cuello, M.A.; Ibañez, C. Patient inflammatory status and CD4+/CD8+ intraepithelial tumor lymphocyte infiltration are predictors of outcomes in high-grade serous ovarian cancer. Gynecol. Oncol. 2018, 151, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Knutson, K.L.; Maurer, M.J.; Preston, C.C.; Moysich, K.B.; Goergen, K.; Hawthorne, K.M.; Cunningham, J.M.; Odunsi, K.; Hartmann, L.C.; Kalli, K.R.; et al. Regulatory T cells, inherited variation, and clinical outcome in epithelial ovarian cancer. Cancer Immunol. Immunother. CII 2015, 64, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Worzfeld, T.; Pogge von Strandmann, E.; Huber, M.; Adhikary, T.; Wagner, U.; Reinartz, S.; Müller, R. The Unique Molecular and Cellular Microenvironment of Ovarian Cancer. Front. Oncol. 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Huppert, L.A.; Green, M.D.; Kim, L.; Chow, C.; Leyfman, Y.; Daud, A.I.; Lee, J.C. Tissue-specific tregs in cancer metastasis: Opportunities for precision immunotherapy. Cell. Mol. Immunol. 2021, 19, 33–45. [Google Scholar] [CrossRef]
- Landskron, J.; Helland, Ø.; Torgersen, K.M.; Aandahl, E.M.; Gjertsen, B.T.; Bjørge, L.; Taskén, K. Activated regulatory and memory T-cells accumulate in malignant ascites from ovarian carcinoma patients. Cancer Immunol. Immunother. CII 2015, 64, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Chardin, L.; Leary, A. Immunotherapy in Ovarian Cancer: Thinking Beyond PD-1/PD-L1. Front. Oncol. 2021, 11, 795547. [Google Scholar] [CrossRef]
- James, N.E.; Miller, K.; LaFranzo, N.; Lips, E.; Woodman, M.; Ou, J.; Ribeiro, J.R. Immune Modeling Analysis Reveals Immunologic Signatures Associated with Improved Outcomes in High Grade Serous Ovarian Cancer. Front. Oncol. 2021, 11, 622182. [Google Scholar] [CrossRef]
- Lisi, L.; Lacal, P.M.; Martire, M.; Navarra, P.; Graziani, G. Clinical experience with CTLA-4 blockade for cancer immunotherapy: From the monospecific monoclonal antibody ipilimumab to probodies and bispecific molecules targeting the tumor microenvironment. Pharmacol. Res. 2022, 175, 105997. [Google Scholar] [CrossRef]
- Sakaguchi, S.; Wing, K.; Miyara, M. Regulatory T cells—A brief history and perspective. Eur. J. Immunol. 2007, 37, S116–S123. [Google Scholar] [CrossRef] [PubMed]
- Friese, C.; Harbst, K.; Borch, T.H.; Westergaard, M.C.W.; Pedersen, M.; Kverneland, A.; Jönsson, G.; Donia, M.; Svane, I.M.; Met, Ö. CTLA-4 blockade boosts the expansion of tumor-reactive CD8(+) tumor-infiltrating lymphocytes in ovarian cancer. Sci. Rep. 2020, 10, 3914. [Google Scholar] [CrossRef]
- Khan, M.; Arooj, S.; Wang, H. NK Cell-Based Immune Checkpoint Inhibition. Front. Immunol. 2020, 11, 167. [Google Scholar] [CrossRef] [PubMed]
- Keilson, J.M.; Knochelmann, H.M.; Paulos, C.M.; Kudchadkar, R.R.; Lowe, M.C. The evolving landscape of immunotherapy in solid tumors. J. Surg. Oncol. 2021, 123, 798–806. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, J.; Song, Q.; Sun, X.; Xue, M.; Yang, Z.; Shang, J. Comprehensive analysis of CTLA-4 in the tumor immune microenvironment of 33 cancer types. Int. Immunopharmacol. 2020, 85, 106633. [Google Scholar] [CrossRef] [PubMed]
- Maiorano, B.A.; Lorusso, D.; Maiorano, M.F.P.; Ciardiello, D.; Parrella, P.; Petracca, A.; Cormio, G.; Maiello, E. The Interplay between PARP Inhibitors and Immunotherapy in Ovarian Cancer: The Rationale behind a New Combination Therapy. Int. J. Mol. Sci. 2022, 23, 3871. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.M.; Kleczko, E.K.; Nemenoff, R.A. Eicosanoids in Cancer: New Roles in Immunoregulation. Front. Pharmacol. 2020, 11, 595498. [Google Scholar] [CrossRef]
- Smith, P.G.; Roque, D.; Ching, M.M.; Fulton, A.; Rao, G.; Reader, J.C. The Role of Eicosanoids in Gynecological Malignancies. Front. Pharmacol. 2020, 11, 1233. [Google Scholar] [CrossRef]
- Li, Y.; Wei, Y.; Zheng, F.; Guan, Y.; Zhang, X. Prostaglandin E2 in the Regulation of Water Transport in Renal Collecting Ducts. Int. J. Mol. Sci. 2017, 18, 2539. [Google Scholar] [CrossRef]
- Jin, K.; Qian, C.; Lin, J.; Liu, B. Cyclooxygenase-2-Prostaglandin E2 pathway: A key player in tumor-associated immune cells. Front. Oncol. 2023, 13, 1099811. [Google Scholar] [CrossRef]
- Chen, C.; Guan, J.; Gu, X.; Chu, Q.; Zhu, H. Prostaglandin E2 and Receptors: Insight into Tumorigenesis, Tumor Progression, and Treatment of Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2022, 10, 834859. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Huang, W.; Wang, B.; Liang, N.; Long, S.; Li, W.; Zhou, Q. Interplay between cyclooxygenase-2 and microRNAs in cancer (Review). Mol. Med. Rep. 2021, 23, 347. [Google Scholar] [CrossRef] [PubMed]
- Garrido, M.P.; Hurtado, I.; Valenzuela-Valderrama, M.; Salvatierra, R.; Hernández, A.; Vega, M.; Selman, A.; Quest, A.F.G.; Romero, C. NGF-enhanced vasculogenic properties of epithelial ovarian cancer cells is reduced by inhibition of the cox-2/pge2 signaling axis. Cancers 2019, 11, 1970. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, K.; Deng, L.; Liang, J.; Liang, H.; Feng, D.; Ling, B. Cyclooxygenase 2 Promotes Proliferation and Invasion in Ovarian Cancer Cells via the PGE2/NF-κB Pathway. Cell Transplant. 2019, 28, 1S–13S. [Google Scholar] [CrossRef] [PubMed]
- Bonavita, E.; Bromley, C.P.; Jonsson, G.; Pelly, V.S.; Sahoo, S.; Walwyn-Brown, K.; Mensurado, S.; Moeini, A.; Flanagan, E.; Bell, C.R.; et al. Antagonistic Inflammatory Phenotypes Dictate Tumor Fate and Response to Immune Checkpoint Blockade. Immunity 2020, 53, 1215–1229.e8. [Google Scholar] [CrossRef] [PubMed]
- Bonavita, E.; Bromley, C.P.; Jonsson, G.; Pelly, V.S.; Sahoo, S.; Walwyn-Brown, K.; Mensurado, S.; Moeini, A.; Flanagan, E.; Bell, C.R.; et al. Anti-Inflammatory Drugs Remodel the Tumor Immune Environment to Enhance Immune Checkpoint Blockade Efficacy. Cancer Discov. 2021, 11, 2602–2619. [Google Scholar] [CrossRef]
- Bell, C.R.; Pelly, V.S.; Moeini, A.; Chiang, S.C.; Flanagan, E.; Bromley, C.P.; Clark, C.; Earnshaw, C.H.; Koufaki, M.A.; Bonavita, E.; et al. Chemotherapy-induced COX-2 upregulation by cancer cells defines their inflammatory properties and limits the efficacy of chemoimmunotherapy combinations. Nat. Commun. 2022, 13, 2063. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Lánczky, A.; Györffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Fernández, E.A.; Mahmoud, Y.D.; Veigas, F.; Rocha, D.; Miranda, M.; Merlo, J.; Balzarini, M.; Lujan, H.D.; Rabinovich, G.A.; Girotti, M.R. Unveiling the immune infiltrate modulation in cancer and response to immunotherapy by MIXTURE-an enhanced deconvolution method. Brief. Bioinform. 2021, 22, bbaa317. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Alizadeh, A.A. High-throughput genomic profiling of tumor-infiltrating leukocytes. Curr. Opin. Immunol. 2016, 41, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Nava, A.; Alves da Quinta, D.; Prato, L.; Girotti, M.R.; Moron, G.; Llera, A.S.; Fernández, E.A. Novel evaluation approach for molecular signature-based deconvolution methods. J. Biomed. Inform. 2023, 142, 104387. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Nieto, P.; Elosua-Bayes, M.; Trincado, J.L.; Marchese, D.; Massoni-Badosa, R.; Salvany, M.; Henriques, A.; Nieto, J.; Aguilar-Fernández, S.; Mereu, E.; et al. A single-cell tumor immune atlas for precision oncology. Genome Res. 2021, 31, 1913–1926. [Google Scholar] [CrossRef]
- Luca, B.A.; Steen, C.B.; Matusiak, M.; Azizi, A.; Varma, S.; Zhu, C.; Przybyl, J.; Espín-Pérez, A.; Diehn, M.; Alizadeh, A.A.; et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 2021, 184, 5482–5496.e28. [Google Scholar] [CrossRef]
- Patil, I. Visualizations with statistical details: The’ggstatsplot’approach. J. Open Source Softw. 2021, 6, 3167. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Cursons, J.; Souza-Fonseca-Guimaraes, F.; Foroutan, M.; Anderson, A.; Hollande, F.; Hediyeh-Zadeh, S.; Behren, A.; Huntington, N.D.; Davis, M.J. A Gene Signature Predicting Natural Killer Cell Infiltration and Improved Survival in Melanoma Patients. Cancer Immunol. Res. 2019, 7, 1162–1174. [Google Scholar] [CrossRef]
- Martinet, L.; Jean, C.; Dietrich, G.; Fournié, J.J.; Poupot, R. PGE2 inhibits natural killer and gamma delta T cell cytotoxicity triggered by NKR and TCR through a cAMP-mediated PKA type I-dependent signaling. Biochem. Pharmacol. 2010, 80, 838–845. [Google Scholar] [CrossRef]
- Holt, D.; Ma, X.; Kundu, N.; Fulton, A. Prostaglandin E(2) (PGE (2)) suppresses natural killer cell function primarily through the PGE(2) receptor EP4. Cancer Immunol. Immunother. CII 2011, 60, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, M.; Rosenberg, D.W. Multifaceted roles of PGE2 in inflammation and cancer. Semin. Immunopathol. 2013, 35, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Finetti, F.; Travelli, C.; Ercoli, J.; Colombo, G.; Buoso, E.; Trabalzini, L. Prostaglandin E2 and Cancer: Insight into Tumor Progression and Immunity. Biology 2020, 9, 434. [Google Scholar] [CrossRef] [PubMed]
- Rowshanravan, B.; Halliday, N.; Sansom, D.M. CTLA-4: A moving target in immunotherapy. Blood 2018, 131, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Bjoern, J.; Lyngaa, R.; Andersen, R.; Hölmich, L.R.; Hadrup, S.R.; Donia, M.; Svane, I.M. Influence of ipilimumab on expanded tumour derived T cells from patients with metastatic melanoma. Oncotarget 2017, 8, 27062–27074. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, Y.; Cheng, H.; Zhong, M.; Hu, Y.; Li, Q.; Gao, X.; Liu, S. The efficacy and safety of combined ipilimumab and nivolumab versus ipilimumab in patients with Stage III/IV unresectable melanoma: A systematic review and meta-analysis. J. Cancer Res. Ther. 2021, 17, 1679–1688. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.M.Y.; Maleki Vareki, S. Addressing the Elephant in the Immunotherapy Room: Effector T-Cell Priming Versus Depletion of Regulatory T-Cells by Anti-CTLA-4 Therapy. Cancers 2022, 14, 1580. [Google Scholar] [CrossRef] [PubMed]
- Mittica, G.; Genta, S.; Aglietta, M.; Valabrega, G. Immune checkpoint inhibitors: A new opportunity in the treatment of ovarian cancer? Int. J. Mol. Sci. 2016, 17, 1169. [Google Scholar] [CrossRef]
- Kuske, M.; Haist, M.; Jung, T.; Grabbe, S.; Bros, M. Immunomodulatory Properties of Immune Checkpoint Inhibitors-More than Boosting T-Cell Responses? Cancers 2022, 14, 1710. [Google Scholar] [CrossRef]
- Giannini, A.; Di Dio, C.; Di Donato, V.; D’oria, O.; Salerno, M.G.; Capalbo, G.; Cuccu, I.; Perniola, G.; Muzii, L.; Bogani, G. PARP Inhibitors in Newly Diagnosed and Recurrent Ovarian Cancer. Am. J. Clin. Oncol. 2023, 46, 414–419. [Google Scholar] [CrossRef]
- Esen, F.; Deniz, G.; Aktas, E.C. PD-1, CTLA-4, LAG-3, and TIGIT: The roles of immune checkpoint receptors on the regulation of human NK cell phenotype and functions. Immunol. Lett. 2021, 240, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Davis-Marcisak, E.F.; Fitzgerald, A.A.; Kessler, M.D.; Danilova, L.; Jaffee, E.M.; Zaidi, N.; Weiner, L.M.; Fertig, E.J. Transfer learning between preclinical models and human tumors identifies a conserved NK cell activation signature in anti-CTLA-4 responsive tumors. Genome Med. 2021, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Buckle, I.; Guillerey, C. Inhibitory Receptors and Immune Checkpoints Regulating Natural Killer Cell Responses to Cancer. Cancers 2021, 24, 4263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Qin, C.; Lin, Y. Development and Validation of a Prognostic Risk Model Based on Nature Killer Cells for Serous Ovarian Cancer. J. Pers. Med. 2023, 13, 403. [Google Scholar] [CrossRef] [PubMed]
- Golia D’Augè, T.; Giannini, A.; Bogani, G.; Di Dio, C.; Laganà, A.S.; Di Donato, V.; Salerno, M.G.; Caserta, D.; Chiantera, V.; Vizza, E.; et al. Prevention, Screening, Treatment and Follow-Up of Gynecological Cancers: State of Art and Future Perspectives. Clin. Exp. Obstet. Gynecol. 2023, 50, 160. [Google Scholar] [CrossRef]
- Zheng, L.; Qin, S.; Si, W.; Wang, A.; Xing, B.; Gao, R.; Ren, X.; Wang, L.; Wu, X.; Zhang, J.; et al. The pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 2021, 374, abe6474. [Google Scholar] [CrossRef]
- Jiang, F.; Jiao, Y.; Yang, K.; Mao, M.; Yu, M.; Cao, D.; Xiang, Y. Single-Cell Profiling of the Immune Atlas of Tumor-Infiltrating Lymphocytes in Endometrial Carcinoma. Cancers 2022, 14, 4311. [Google Scholar] [CrossRef]
- Vidal, M.; Fraga, M.; Llerena, F.; Vera, A.; Hernández, M.; Koch, E.; Reyes-López, F.; Vallejos-Vidal, E.; Cabrera-Vives, G.; Nova-Lamperti, E. Analysis of Tumor-Infiltrating T-Cell Transcriptomes Reveal a Unique Genetic Signature across Different Types of Cancer. Int. J. Mol. Sci. 2022, 23, 11065. [Google Scholar] [CrossRef]





| Ecotype Category | TCGA | AOCS | GSE26193 | GSE0161 | ||||
|---|---|---|---|---|---|---|---|---|
| HIGH | LOW | HIGH | LOW | HIGH | LOW | HIGH | LOW | |
| CE01 + CE02 | 45 | 52 | 15 | 9 | 12 | 6 | 9 | 2 |
| CE03 | 4 | 4 | - | 1 | 1 | 1 | - | 1 |
| CE04 + CE05 | 11 | 10 | - | 3 | 1 | - | 1 | 4 |
| CE06 | 11 | 5 | 1 | 1 | 4 | 1 | 2 | 2 |
| CE07 + CE08 | 38 | 27 | 11 | 8 | 7 | 11 | 8 | 6 |
| CE09 + CE10 | 14 | 52 | 5 | 6 | 3 | 3 | 1 | 3 |
| Total | 123 | 150 | 32 | 28 | 28 | 22 | 21 | 18 |
| NK Cell State | TCGA | AOCS | GSE26193 | GSE0161 | ||||
|---|---|---|---|---|---|---|---|---|
| HIGH | LOW | HIGH | LOW | HIGH | LOW | HIGH | LOW | |
| S01 | 16 | 38 | 6 | 3 | 3 | 6 | 2 | - |
| S02 | 3 | 1 | 1 | - | - | - | 1 | 1 |
| S03 | 34 | 36 | 9 | 9 | 7 | 4 | 3 | 1 |
| S04 | 7 | 11 | 4 | 4 | 1 | 2 | 1 | - |
| S05 | 14 | 11 | 3 | - | 2 | 1 | 2 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Valenzuela, F.; Wichmann, I.; Suárez, F.; Kato, S.; Ossandón, E.; Hermoso, M.; Fernández, E.A.; Cuello, M.A. Cyclooxygenase-2 Blockade Is Crucial to Restore Natural Killer Cell Activity before Anti-CTLA-4 Therapy against High-Grade Serous Ovarian Cancer. Cancers 2024, 16, 80. https://doi.org/10.3390/cancers16010080
Gómez-Valenzuela F, Wichmann I, Suárez F, Kato S, Ossandón E, Hermoso M, Fernández EA, Cuello MA. Cyclooxygenase-2 Blockade Is Crucial to Restore Natural Killer Cell Activity before Anti-CTLA-4 Therapy against High-Grade Serous Ovarian Cancer. Cancers. 2024; 16(1):80. https://doi.org/10.3390/cancers16010080
Chicago/Turabian StyleGómez-Valenzuela, Fernán, Ignacio Wichmann, Felipe Suárez, Sumie Kato, Enrique Ossandón, Marcela Hermoso, Elmer A. Fernández, and Mauricio A. Cuello. 2024. "Cyclooxygenase-2 Blockade Is Crucial to Restore Natural Killer Cell Activity before Anti-CTLA-4 Therapy against High-Grade Serous Ovarian Cancer" Cancers 16, no. 1: 80. https://doi.org/10.3390/cancers16010080
APA StyleGómez-Valenzuela, F., Wichmann, I., Suárez, F., Kato, S., Ossandón, E., Hermoso, M., Fernández, E. A., & Cuello, M. A. (2024). Cyclooxygenase-2 Blockade Is Crucial to Restore Natural Killer Cell Activity before Anti-CTLA-4 Therapy against High-Grade Serous Ovarian Cancer. Cancers, 16(1), 80. https://doi.org/10.3390/cancers16010080








