Clinical Significance of microRNAs in Hematologic Malignancies and Hematopoietic Stem Cell Transplantation
Abstract
Simple Summary
Abstract
1. Introduction
2. MicroRNA in Hematopoiesis and Hematologic Malignancies
2.1. MiRNA Biology
2.2. The Role of microRNAs in Normal Hematopoiesis
2.3. MicroRNA Dysregulation in Malignant Hematopoiesis
2.4. Aberrant microRNA Expressions in Diagnosis, Prognosis, and Monitoring of Therapy
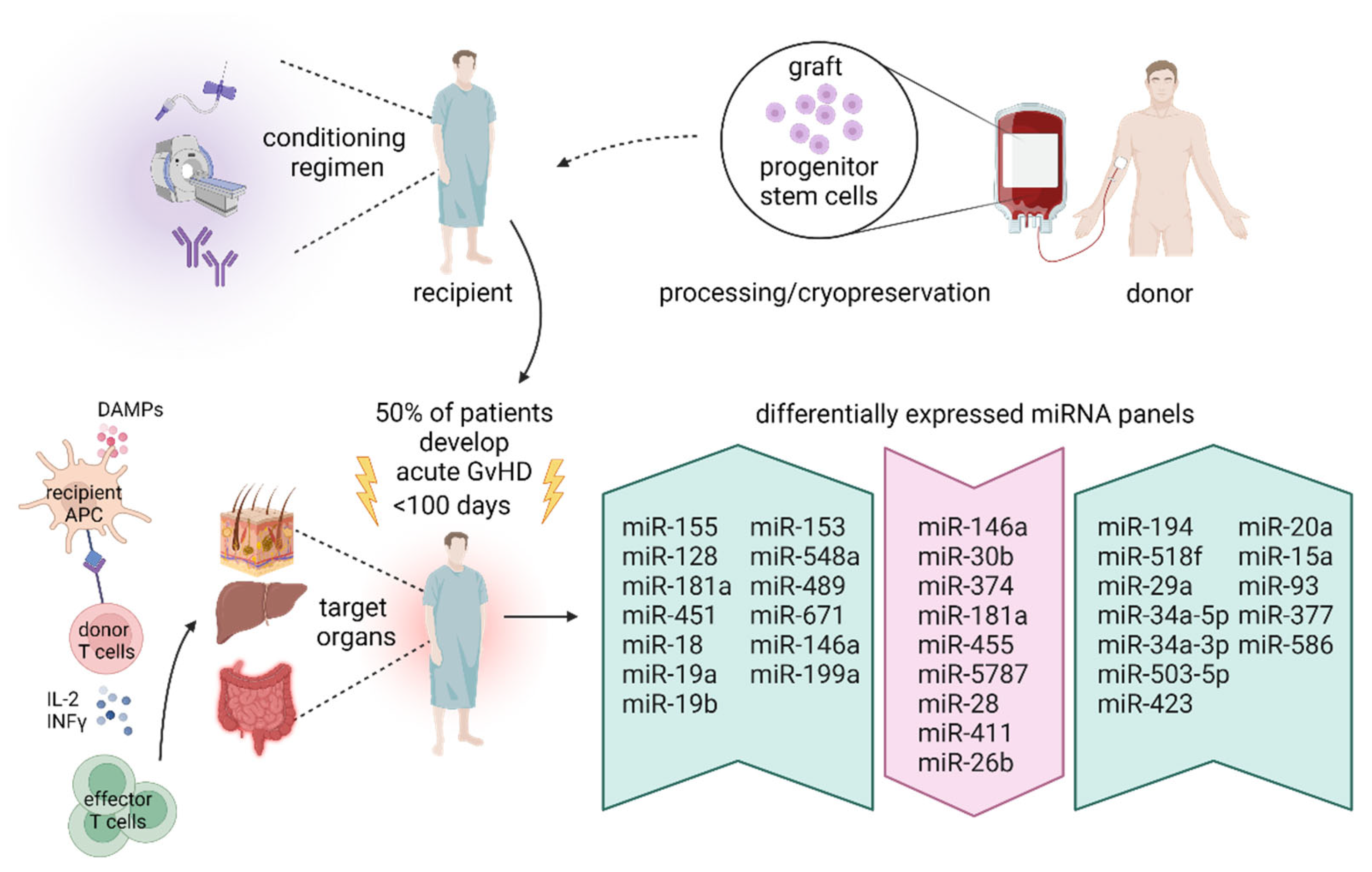
3. MicroRNA in Hematopoietic Stem Cell Transplantation
3.1. Hematopoietic Stem Cell Transplantation
3.2. MicroRNA and Graft-Versus-Host Disease
3.3. MicroRNA and Post-HSCT Patient Outcomes
4. Investigating the Therapeutical Potential of the miRNA-Based Approach in Hemato-Oncology
5. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.G.; Urazova, L.N.; Stegny, V.N. MicroRNAs and human cancer. Exp. Oncol. 2012, 34, 2–8. [Google Scholar] [PubMed]
- Han, T.S.; Hur, K.; Cho, H.S.; Ban, H.S. Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma. Cancers 2020, 12, 2622. [Google Scholar] [CrossRef] [PubMed]
- Anelli, L.; Zagaria, A.; Specchia, G.; Musto, P.; Albano, F. Dysregulation of miRNA in Leukemia: Exploiting miRNA Expression Profiles as Biomarkers. Int. J. Mol. Sci. 2021, 22, 7156. [Google Scholar] [CrossRef] [PubMed]
- Gounaris-Shannon, S.; Chevassut, T. The Role of miRNA in Haematological Malignancy. Bone Marrow Res. 2013, 2013, 269107. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Baltimore, D. MicroRNAs and hematopoietic cell development. Curr. Top. Dev. Biol. 2012, 99, 145–174. [Google Scholar] [PubMed]
- Jagannathan-Bogdan, M.; Zon, L.I. Hematopoiesis. Development 2013, 140, 2463–2467. [Google Scholar] [CrossRef] [PubMed]
- Kotaki, R.; Koyama-Nasu, R.; Yamakawa, N.; Kotani, A. miRNAs in Normal and Malignant Hematopoiesis. Int. J. Mol. Sci. 2017, 18, 1495. [Google Scholar] [CrossRef] [PubMed]
- Lazare, S.S.; Wojtowicz, E.E.; Bystrykh, L.V.; de Haan, G. microRNAs in hematopoiesis. Exp. Cell Res. 2014, 329, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Grinenko, T.; Eugster, A.; Thielecke, L.; Ramasz, B.; Kruger, A.; Dietz, S.; Glauche, I.; Gerbaulet, A.; von Bonin, M.; Basak, O.; et al. Hematopoietic stem cells can differentiate into restricted myeloid progenitors before cell division in mice. Nat. Commun. 2018, 9, 1898. [Google Scholar] [CrossRef] [PubMed]
- Akashi, K.; Traver, D.; Miyamoto, T.; Weissman, I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000, 404, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tanaka-Yano, M.; Meader, E.; Kinney, M.A.; Morris, V.; Lummertz da Rocha, E.; Liu, N.; Liu, T.; Zhu, Q.; Orkin, S.H.; et al. Developmental maturation of the hematopoietic system controlled by a Lin28b-let-7-Cbx2 axis. Cell Rep. 2022, 39, 110587. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, J.; Chen, H.; Fei, X.; Tang, Y.; Yan, Y.; Zhang, H.; Zhang, J. MiR-128-2 inhibits common lymphoid progenitors from developing into progenitor B cells. Oncotarget 2016, 7, 17520–17531. [Google Scholar] [CrossRef]
- Roberts, L.B.; Jowett, G.M.; Read, E.; Zabinski, T.; Berkachy, R.; Selkirk, M.E.; Jackson, I.; Niazi, U.; Anandagoda, N.; Araki, M.; et al. MicroRNA-142 Critically Regulates Group 2 Innate Lymphoid Cell Homeostasis and Function. J. Immunol. 2021, 206, 2725–2739. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Salim, U.; Kumar, A.; Kulshreshtha, R.; Vivekanandan, P. Biogenesis, characterization, and functions of mirtrons. Wiley Interdiscip. Rev. RNA 2022, 13, e1680. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Das, S. Profiling cell-free and circulating miRNA: A clinical diagnostic tool for different cancers. Tumor Biol. 2016, 37, 5705–5714. [Google Scholar] [CrossRef]
- Iftikhar, H.; Carney, G.E. Evidence and potential in vivo functions for biofluid miRNAs: From expression profiling to functional testing: Potential roles of extracellular miRNAs as indicators of physiological change and as agents of intercellular information exchange. Bioessays 2016, 38, 367–378. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Shi, K.; Yang, S.; Liu, J.; Zhou, Q.; Wang, G.; Song, J.; Li, Z.; Zhang, Z.; Yuan, W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer 2018, 17, 147. [Google Scholar] [CrossRef]
- Lawrie, C.H. MicroRNAs in hematological malignancies. Blood Rev. 2013, 27, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Rajasekhar, M.; Schmitz, U.; Flamant, S.; Wong, J.J.; Bailey, C.G.; Ritchie, W.; Holst, J.; Rasko, J.E.J. Identifying microRNA determinants of human myelopoiesis. Sci. Rep. 2018, 8, 7264. [Google Scholar] [CrossRef] [PubMed]
- Lechman, E.R.; Gentner, B.; Ng, S.W.; Schoof, E.M.; van Galen, P.; Kennedy, J.A.; Nucera, S.; Ciceri, F.; Kaufmann, K.B.; Takayama, N.; et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell 2016, 29, 214–228. [Google Scholar] [CrossRef]
- Herrera-Merchan, A.; Cerrato, C.; Luengo, G.; Dominguez, O.; Piris, M.A.; Serrano, M.; Gonzalez, S. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle 2010, 9, 3277–3285. [Google Scholar] [CrossRef]
- Ooi, A.G.; Sahoo, D.; Adorno, M.; Wang, Y.; Weissman, I.L.; Park, C.Y. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. USA 2010, 107, 21505–21510. [Google Scholar] [CrossRef] [PubMed]
- Ranganath, P. MicroRNA-155 and Its Role in Malignant Hematopoiesis. Biomark. Insights 2015, 10, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Bissels, U.; Bosio, A.; Wagner, W. MicroRNAs are shaping the hematopoietic landscape. Haematologica 2012, 97, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Pelosi, E.; Greco, P.; Racanicchi, S.; Testa, U.; Liuzzi, F.; Croce, C.M.; Brunetti, E.; Grignani, F.; Peschle, C. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 2007, 9, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Kim, K.S.; Oh, I.H. Concise review: Exploring miRNAs—Toward a better understanding of hematopoiesis. Stem Cells 2015, 33, 1–7. [Google Scholar] [CrossRef]
- El Gazzar, M. microRNAs as potential regulators of myeloid-derived suppressor cell expansion. Innate Immun. 2014, 20, 227–238. [Google Scholar] [CrossRef]
- Qin, Y.; Wu, L.; Ouyang, Y.; Zhou, P.; Zhou, H.; Wang, Y.; Ma, J.; Zhang, J.; Chen, Y.; Qian, J.; et al. MiR-125a Is a critical modulator for neutrophil development. PLoS Genet. 2017, 13, e1007027. [Google Scholar] [CrossRef]
- Undi, R.B.; Kandi, R.; Gutti, R.K. MicroRNAs as Haematopoiesis Regulators. Adv. Hematol. 2013, 2013, 695754. [Google Scholar] [CrossRef]
- Opalinska, J.B.; Bersenev, A.; Zhang, Z.; Schmaier, A.A.; Choi, J.; Yao, Y.; D’Souza, J.; Tong, W.; Weiss, M.J. MicroRNA expression in maturing murine megakaryocytes. Blood 2010, 116, e128–e138. [Google Scholar] [CrossRef]
- Weiss, C.N.; Ito, K. microRNA-22 promotes megakaryocyte differentiation through repression of its target, GFI1. Blood Adv. 2019, 3, 33–46. [Google Scholar] [CrossRef]
- Zini, R.; Rossi, C.; Norfo, R.; Pennucci, V.; Barbieri, G.; Ruberti, S.; Rontauroli, S.; Salati, S.; Bianchi, E.; Manfredini, R. miR-382-5p Controls Hematopoietic Stem Cell Differentiation through the Downregulation of MXD1. Stem Cells Dev. 2016, 25, 1433–1443. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Chen, M.T.; Zhang, X.H.; Yin, X.L.; Ning, H.M.; Su, R.; Lin, H.S.; Song, L.; Wang, F.; Ma, Y.N.; et al. The PU.1-Modulated MicroRNA-22 Is a Regulator of Monocyte/Macrophage Differentiation and Acute Myeloid Leukemia. PLoS Genet. 2016, 12, e1006259. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.F.; Wang, H.; Kong, F.X.; Xu, Q.Q.; Xiao, F.J.; Yang, Y.F.; Ge, R.L.; Wang, L.S. Exosomal miR-486 regulates hypoxia-induced erythroid differentiation of erythroleukemia cells through targeting Sirt1. Exp. Cell Res. 2017, 351, 74–81. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, Y.; Han, X.; Zhao, X.; Peng, Y.; Li, Y.; Peng, M.; Song, J.; Wu, K.; Sun, S.; et al. miR-150 inhibits terminal erythroid proliferation and differentiation. Oncotarget 2015, 6, 43033–43047. [Google Scholar] [CrossRef]
- Kotaki, R.; Kawashima, M.; Yamaguchi, A.; Suzuki, N.; Koyama-Nasu, R.; Ogiya, D.; Okuyama, K.; Yamamoto, Y.; Takamatsu, M.; Kurosaki, N.; et al. Overexpression of miR-669m inhibits erythroblast differentiation. Sci. Rep. 2020, 10, 13554. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wu, F.; Yang, L.; Wang, F.; Zhang, T.; Deng, X.; Zhang, X.; Yuan, X.; Yan, Y.; Li, Y.; et al. miR-144/451 inhibits c-Myc to promote erythroid differentiation. FASEB J. 2020, 34, 13194–13210. [Google Scholar] [CrossRef] [PubMed]
- Kretov, D.A.; Walawalkar, I.A.; Mora-Martin, A.; Shafik, A.M.; Moxon, S.; Cifuentes, D. Ago2-Dependent Processing Allows miR-451 to Evade the Global MicroRNA Turnover Elicited during Erythropoiesis. Mol. Cell 2020, 78, 317–328.e6. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Arthanari, H.; Akabayov, B.; Song, H.; Papadopoulos, E.; Qi, H.H.; Jedrychowski, M.; Guttler, T.; Guo, C.; Luna, R.E.; et al. eIF1A augments Ago2-mediated Dicer-independent miRNA biogenesis and RNA interference. Nat. Commun. 2015, 6, 7194. [Google Scholar] [CrossRef]
- Nguyen, T.; Rich, A.; Dahl, R. MiR-24 promotes the survival of hematopoietic cells. PLoS ONE 2013, 8, e55406. [Google Scholar] [CrossRef]
- Kong, K.Y.; Owens, K.S.; Rogers, J.H.; Mullenix, J.; Velu, C.S.; Grimes, H.L.; Dahl, R. MIR-23A microRNA cluster inhibits B-cell development. Exp. Hematol. 2010, 38, 629–640.e1. [Google Scholar] [CrossRef]
- Vasilatou, D.; Papageorgiou, S.; Pappa, V.; Papageorgiou, E.; Dervenoulas, J. The role of microRNAs in normal and malignant hematopoiesis. Eur. J. Haematol. 2010, 84, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Rosen, S.T.; Querfeld, C. Targeting microRNA in hematologic malignancies. Curr. Opin. Oncol. 2020, 32, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.N.; Ito, K. A Macro View of MicroRNAs: The Discovery of MicroRNAs and Their Role in Hematopoiesis and Hematologic Disease. Int. Rev. Cell Mol. Biol. 2017, 334, 99–175. [Google Scholar] [PubMed]
- Raghuwanshi, S.; Dahariya, S.; Kandi, R.; Gutti, U.; Undi, R.B.; Sharma, D.S.; Sahu, I.; Kovuru, N.; Yarla, N.S.; Saladi, R.G.V.; et al. Epigenetic Mechanisms: Role in Hematopoietic Stem Cell Lineage Commitment and Differentiation. Curr. Drug Targets 2018, 19, 1683–1695. [Google Scholar] [CrossRef]
- Agirre, X.; Vilas-Zornoza, A.; Jimenez-Velasco, A.; Martin-Subero, J.I.; Cordeu, L.; Garate, L.; San Jose-Eneriz, E.; Abizanda, G.; Rodriguez-Otero, P.; Fortes, P.; et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009, 69, 4443–4453. [Google Scholar] [CrossRef]
- De Leeuw, D.C.; Denkers, F.; Olthof, M.C.; Rutten, A.P.; Pouwels, W.; Schuurhuis, G.J.; Ossenkoppele, G.J.; Smit, L. Attenuation of microRNA-126 expression that drives CD34+38- stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 2014, 74, 2094–2105. [Google Scholar] [CrossRef]
- Maharry, S.E.; Walker, C.J.; Liyanarachchi, S.; Mehta, S.; Patel, M.; Bainazar, M.A.; Huang, X.; Lankenau, M.A.; Hoag, K.W.; Ranganathan, P.; et al. Dissection of the Major Hematopoietic Quantitative Trait Locus in Chromosome 6q23.3 Identifies miR-3662 as a Player in Hematopoiesis and Acute Myeloid Leukemia. Cancer Discov. 2016, 6, 1036–1051. [Google Scholar] [CrossRef]
- Lu, Y.; Zhong, L.; Luo, X.; Liu, C.; Dan, W.; Chu, X.; Wan, P.; Zhang, Z.; Wang, X.; Liu, Z.; et al. MiRNA-301b-3p induces proliferation and inhibits apoptosis in AML cells by targeting FOXF2 and regulating Wnt/beta-catenin axis. Mol. Cell Probes 2022, 63, 101805. [Google Scholar] [CrossRef]
- Jia, Q.; Sun, H.; Xiao, F.; Sai, Y.; Li, Q.; Zhang, X.; Yang, S.; Wang, H.; Wang, H.; Yang, Y.; et al. miR-17-92 promotes leukemogenesis in chronic myeloid leukemia via targeting A20 and activation of NF-kappaB signaling. Biochem. Biophys. Res. Commun. 2017, 487, 868–874. [Google Scholar] [CrossRef]
- Wen, F.; Cao, Y.X.; Luo, Z.Y.; Liao, P.; Lu, Z.W. LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem. Biophys. Res. Commun. 2018, 507, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.L.; Wang, X.; Mann, M.; Adamus, T.P.; Wang, D.; Moreira, D.F.; Zhang, Z.; Ouyang, C.; He, X.; Zhang, B.; et al. Myeloid cell-targeted miR-146a mimic inhibits NF-kappaB-driven inflammation and leukemia progression in vivo. Blood 2020, 135, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, Y.; Kitaura, J.; Hatakeyama, K.; Watanuki, J.; Akasaka, T.; Kato, N.; Shimanuki, M.; Nishimura, K.; Takahashi, M.; Taniwaki, M.; et al. Emu/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia 2011, 25, 1849–1856. [Google Scholar] [CrossRef]
- Ferdowsi, S.; Atarodi, K.; Amirizadeh, N.; Toogeh, G.; Azarkeivan, A.; Shirkoohi, R.; Faranoush, M.; Vaezi, M.; Alimoghaddam, K.; Ghavamzadeh, A.; et al. Expression analysis of microRNA-125 in patients with polycythemia vera and essential thrombocythemia and correlation with JAK2 allele burden and laboratory findings. Int. J. Lab. Hematol. 2015, 37, 661–667. [Google Scholar] [CrossRef]
- Bruchova, H.; Merkerova, M.; Prchal, J.T. Aberrant expression of microRNA in polycythemia vera. Haematologica 2008, 93, 1009–1016. [Google Scholar] [CrossRef]
- Gebauer, N.; Bernard, V.; Gebauer, W.; Feller, A.C.; Merz, H. MicroRNA expression and JAK2 allele burden in bone marrow trephine biopsies of polycythemia vera, essential thrombocythemia and early primary myelofibrosis. Acta Haematol. 2013, 129, 251–256. [Google Scholar] [CrossRef]
- Kuang, X.; Wei, C.; Zhang, T.; Yang, Z.; Chi, J.; Wang, L. miR-378 inhibits cell growth and enhances apoptosis in human myelodysplastic syndromes. Int. J. Oncol. 2016, 49, 1921–1930. [Google Scholar] [CrossRef]
- Leone, E.; Morelli, E.; Di Martino, M.T.; Amodio, N.; Foresta, U.; Gulla, A.; Rossi, M.; Neri, A.; Giordano, A.; Munshi, N.C.; et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin. Cancer Res. 2013, 19, 2096–2106. [Google Scholar] [CrossRef]
- Di Martino, M.T.; Gulla, A.; Cantafio, M.E.; Lionetti, M.; Leone, E.; Amodio, N.; Guzzi, P.H.; Foresta, U.; Conforti, F.; Cannataro, M.; et al. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget 2013, 4, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wong, K.Y.; Chan, G.C.; Chng, W.J.; Chim, C.S. Epigenetic silencing of EVL/miR-342 in multiple myeloma. Transl. Res. 2018, 192, 46–53. [Google Scholar] [CrossRef]
- Long, X.; Li, J.; Wen, F.; Cao, Y.; Luo, Z.; Luo, C. miR-140-3p attenuated the tumorigenesis of multiple myeloma via attenuating BZW2. Hematology 2022, 27, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Chen, D.; Xiao, T.; Lin, D.; Lin, D.; Lin, L.; Zhu, H.; Xu, J.; Huang, W.; Yang, T. DNA methylation-mediated silencing of microRNA-204 enhances T cell acute lymphoblastic leukemia by up-regulating MMP-2 and MMP-9 via NF-kappaB. J. Cell. Mol. Med. 2021, 25, 2365–2376. [Google Scholar] [CrossRef] [PubMed]
- Kasar, S.; Underbayev, C.; Yuan, Y.; Hanlon, M.; Aly, S.; Khan, H.; Chang, V.; Batish, M.; Gavrilova, T.; Badiane, F.; et al. Therapeutic implications of activation of the host gene (Dleu2) promoter for miR-15a/16-1 in chronic lymphocytic leukemia. Oncogene 2014, 33, 3307–3315. [Google Scholar] [CrossRef] [PubMed]
- Davari, N.; Ahmadpour, F.; Kiani, A.A.; Azadpour, M.; Asadi, Z.T. Evaluation of microRNA-223 and microRNA-125a expression association with STAT3 and Bcl2 genes in blood leukocytes of CLL patients: A case-control study. BMC Res. Notes 2021, 14, 21. [Google Scholar] [CrossRef]
- Wong, K.Y.; Yu, L.; Chim, C.S. DNA methylation of tumor suppressor miRNA genes: A lesson from the miR-34 family. Epigenomics 2011, 3, 83–92. [Google Scholar] [CrossRef]
- Sandhu, S.K.; Volinia, S.; Costinean, S.; Galasso, M.; Neinast, R.; Santhanam, R.; Parthun, M.R.; Perrotti, D.; Marcucci, G.; Garzon, R.; et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Emu-miR-155 transgenic mouse model. Proc. Natl. Acad. Sci. USA 2012, 109, 20047–20052. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef]
- Meyer, A.; Herkt, S.; Kunze-Schumacher, H.; Kohrs, N.; Ringleb, J.; Schneider, L.; Kuvardina, O.N.; Oellerich, T.; Haupl, B.; Krueger, A.; et al. The transcription factor TAL1 and miR-17-92 create a regulatory loop in hematopoiesis. Sci. Rep. 2020, 10, 21438. [Google Scholar] [CrossRef]
- Zhou, K.; Feng, X.; Wang, Y.; Liu, Y.; Tian, L.; Zuo, W.; Yi, S.; Wei, X.; Song, Y.; Qiu, L. miR-223 is repressed and correlates with inferior clinical features in mantle cell lymphoma through targeting SOX11. Exp. Hematol. 2018, 58, 27–34.e21. [Google Scholar] [CrossRef]
- Chen, P.; Price, C.; Li, Z.; Li, Y.; Cao, D.; Wiley, A.; He, C.; Gurbuxani, S.; Kunjamma, R.B.; Huang, H.; et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc. Natl. Acad. Sci. USA 2013, 110, 11511–11516. [Google Scholar] [CrossRef]
- Garzon, R.; Heaphy, C.E.; Havelange, V.; Fabbri, M.; Volinia, S.; Tsao, T.; Zanesi, N.; Kornblau, S.M.; Marcucci, G.; Calin, G.A.; et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 2009, 114, 5331–5341. [Google Scholar] [CrossRef]
- Leotta, M.; Biamonte, L.; Raimondi, L.; Ronchetti, D.; Di Martino, M.T.; Botta, C.; Leone, E.; Pitari, M.R.; Neri, A.; Giordano, A.; et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J. Cell. Physiol. 2014, 229, 2106–2116. [Google Scholar] [CrossRef]
- Rodriguez-Otero, P.; Roman-Gomez, J.; Vilas-Zornoza, A.; Jose-Eneriz, E.S.; Martin-Palanco, V.; Rifon, J.; Torres, A.; Calasanz, M.J.; Agirre, X.; Prosper, F. Deregulation of FGFR1 and CDK6 oncogenic pathways in acute lymphoblastic leukaemia harbouring epigenetic modifications of the MIR9 family. Br. J. Haematol. 2011, 155, 73–83. [Google Scholar] [CrossRef]
- Palacios, F.; Abreu, C.; Prieto, D.; Morande, P.; Ruiz, S.; Fernandez-Calero, T.; Naya, H.; Libisch, G.; Robello, C.; Landoni, A.I.; et al. Activation of the PI3K/AKT pathway by microRNA-22 results in CLL B-cell proliferation. Leukemia 2015, 29, 115–125. [Google Scholar] [CrossRef]
- Chim, C.S.; Wong, K.Y.; Qi, Y.; Loong, F.; Lam, W.L.; Wong, L.G.; Jin, D.Y.; Costello, J.F.; Liang, R. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis 2010, 31, 745–750. [Google Scholar] [CrossRef]
- Buhagiar, A.; Borg, J.; Ayers, D. Overview of current microRNA biomarker signatures as potential diagnostic tools for leukaemic conditions. Non-Coding RNA Res. 2020, 5, 22–26. [Google Scholar] [CrossRef]
- Moloudizargari, M.; Hekmatirad, S.; Mofarahe, Z.S.; Asghari, M.H. Exosomal microRNA panels as biomarkers for hematological malignancies. Curr. Probl. Cancer 2021, 45, 100726. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal miRNAs as Biomarkers for Prostate Cancer. Front. Genet. 2013, 4, 36. [Google Scholar] [CrossRef]
- Wang, H.; You, Y.; Zhu, X. The Role of Exosomes in the Progression and Therapeutic Resistance of Hematological Malignancies. Front. Oncol. 2022, 12, 887518. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, L.; Xiang, B.; Zhu, H.; Wu, Y.; Chen, M.; Guan, P.; Zou, X.; Valencia, C.A.; Dong, B.; et al. Potential role of exosome-associated microRNA panels and in vivo environment to predict drug resistance for patients with multiple myeloma. Oncotarget 2016, 7, 30876–30891. [Google Scholar] [CrossRef]
- Manier, S.; Liu, C.J.; Avet-Loiseau, H.; Park, J.; Shi, J.; Campigotto, F.; Salem, K.Z.; Huynh, D.; Glavey, S.V.; Rivotto, B.; et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017, 129, 2429–2436. [Google Scholar] [CrossRef]
- Bouyssou, J.M.; Liu, C.J.; Bustoros, M.; Sklavenitis-Pistofidis, R.; Aljawai, Y.; Manier, S.; Yosef, A.; Sacco, A.; Kokubun, K.; Tsukamoto, S.; et al. Profiling of circulating exosomal miRNAs in patients with Waldenstrom Macroglobulinemia. PLoS ONE 2018, 13, e0204589. [Google Scholar] [CrossRef]
- Jiang, L.; Deng, T.; Wang, D.; Xiao, Y. Elevated Serum Exosomal miR-125b Level as a Potential Marker for Poor Prognosis in Intermediate-Risk Acute Myeloid Leukemia. Acta Haematol. 2018, 140, 183–192. [Google Scholar] [CrossRef]
- Feng, Y.; Zhong, M.; Zeng, S.; Wang, L.; Liu, P.; Xiao, X.; Liu, Y. Exosome-derived miRNAs as predictive biomarkers for diffuse large B-cell lymphoma chemotherapy resistance. Epigenomics 2019, 11, 35–51. [Google Scholar] [CrossRef]
- Shahid, S.; Shahid, W.; Shaheen, J.; Akhtar, M.W.; Sadaf, S. Circulating miR-146a expression as a non-invasive predictive biomarker for acute lymphoblastic leukemia. Sci. Rep. 2021, 11, 22783. [Google Scholar] [CrossRef]
- El-Khazragy, N.; Noshi, M.A.; Abdel-Malak, C.; Zahran, R.F.; Swellam, M. miRNA-155 and miRNA-181a as prognostic biomarkers for pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2019, 120, 6315–6321. [Google Scholar] [CrossRef]
- Liang, C.; Li, Y.; Wang, L.N.; Zhang, X.L.; Luo, J.S.; Peng, C.J.; Tang, W.Y.; Huang, L.B.; Tang, Y.L.; Luo, X.Q. Up-regulated miR-155 is associated with poor prognosis in childhood acute lymphoblastic leukemia and promotes cell proliferation targeting ZNF238. Hematology 2021, 26, 16–25. [Google Scholar] [CrossRef]
- Ghodousi, E.S.; Rahgozar, S. MicroRNA-326 and microRNA-200c: Two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2018, 119, 6024–6032. [Google Scholar] [CrossRef]
- Zamani, A.; Fattahi Dolatabadi, N.; Houshmand, M.; Nabavizadeh, N. miR-324-3p and miR-508-5p expression levels could serve as potential diagnostic and multidrug-resistant biomarkers in childhood acute lymphoblastic leukemia. Leuk. Res. 2021, 109, 106643. [Google Scholar] [CrossRef]
- Schotte, D.; De Menezes, R.X.; Akbari Moqadam, F.; Khankahdani, L.M.; Lange-Turenhout, E.; Chen, C.; Pieters, R.; Den Boer, M.L. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica 2011, 96, 703–711. [Google Scholar] [CrossRef]
- Almeida, R.S.; Costa, E.S.M.; Coutinho, L.L.; Garcia Gomes, R.; Pedrosa, F.; Massaro, J.D.; Donadi, E.A.; Lucena-Silva, N. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol. Oncol. 2019, 37, 103–112. [Google Scholar] [CrossRef]
- Labib, H.A.; Elantouny, N.G.; Ibrahim, N.F.; Alnagar, A.A. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology 2017, 22, 392–397. [Google Scholar] [CrossRef]
- Amankwah, E.K.; Devidas, M.; Teachey, D.T.; Rabin, K.R.; Brown, P.A. Six Candidate miRNAs Associated with Early Relapse in Pediatric B-Cell Acute Lymphoblastic Leukemia. Anticancer Res. 2020, 40, 3147–3153. [Google Scholar] [CrossRef]
- Avigad, S.; Verly, I.R.; Lebel, A.; Kordi, O.; Shichrur, K.; Ohali, A.; Hameiri-Grossman, M.; Kaspers, G.J.; Cloos, J.; Fronkova, E.; et al. miR expression profiling at diagnosis predicts relapse in pediatric precursor B-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer 2016, 55, 328–339. [Google Scholar] [CrossRef]
- Mi, S.; Lu, J.; Sun, M.; Li, Z.; Zhang, H.; Neilly, M.B.; Wang, Y.; Qian, Z.; Jin, J.; Zhang, Y.; et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 19971–19976. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, N.; Wang, R.; Shao, T.; Feng, Y.; Yao, Y.; Wu, Q.; Zhu, S.; Cao, J.; Zhang, H.; et al. High expression of miR-363 predicts poor prognosis and guides treatment selection in acute myeloid leukemia. J. Transl. Med. 2019, 17, 106. [Google Scholar] [CrossRef]
- Li, S.M.; Zhao, Y.Q.; Hao, Y.L.; Liang, Y.Y. Upregulation of miR-504-3p is associated with favorable prognosis of acute myeloid leukemia and may serve as a tumor suppressor by targeting MTHFD2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1203–1213. [Google Scholar]
- Koutova, L.; Sterbova, M.; Pazourkova, E.; Pospisilova, S.; Svobodova, I.; Horinek, A.; Lysak, D.; Korabecna, M. The impact of standard chemotherapy on miRNA signature in plasma in AML patients. Leuk. Res. 2015, 39, 1389–1395. [Google Scholar] [CrossRef]
- Wang, X.; Zuo, D.; Yuan, Y.; Yang, X.; Hong, Z.; Zhang, R. MicroRNA-183 promotes cell proliferation via regulating programmed cell death 6 in pediatric acute myeloid leukemia. J. Cancer Res. Clin. Oncol. 2017, 143, 169–180. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, Y. MicroRNA-199a deficiency relates to higher bone marrow blasts, poor risk stratification and worse prognostication in pediatric acute myeloid leukemia patients. Pediatr. Hematol. Oncol. 2022, 39, 500–507. [Google Scholar] [CrossRef]
- Lin, X.; Wang, Z.; Wang, Y.; Feng, W. Serum MicroRNA-370 as a potential diagnostic and prognostic biomarker for pediatric acute myeloid leukemia. Int. J. Clin. Exp. Pathol. 2015, 8, 14658–14666. [Google Scholar]
- Zhu, R.; Lin, W.; Zhao, W.; Fan, F.; Tang, L.; Hu, Y. A 4-microRNA signature for survival prognosis in pediatric and adolescent acute myeloid leukemia. J. Cell. Biochem. 2019, 120, 3958–3968. [Google Scholar] [CrossRef]
- Rahimi, Z.; Ghorbani, Z.; Motamed, H.; Jalilian, N. Aberrant expression profile of miR-32, miR-98 and miR-374 in chronic lymphocytic leukemia. Leuk. Res. 2021, 111, 106691. [Google Scholar] [CrossRef]
- Bagheri, M.; Khansarinejad, B.; Mosayebi, G.; Moradabadi, A.; Mondanizadeh, M. Diagnostic Value of Plasma miR-145 and miR-185 as Targeting of the APRIL Oncogene in the B-cell Chronic Lymphocytic Leukemia. Asian Pac. J. Cancer Prev. 2021, 22, 111–117. [Google Scholar] [CrossRef]
- Klumper, T.; Bruckmueller, H.; Diewock, T.; Kaehler, M.; Haenisch, S.; Pott, C.; Bruhn, O.; Cascorbi, I. Expression differences of miR-142-5p between treatment-naive chronic myeloid leukemia patients responding and non-responding to imatinib therapy suggest a link to oncogenic ABL2, SRI, cKIT and MCL1 signaling pathways critical for development of therapy resistance. Exp. Hematol. Oncol. 2020, 9, 26. [Google Scholar]
- Habib, E.M.; Nosiar, N.A.; Eid, M.A.; Taha, A.M.; Sherief, D.E.; Hassan, A.E.; Abdel Ghafar, M.T. Circulating miR-146a expression predicts early treatment response to imatinib in adult chronic myeloid leukemia. J. Investig. Med. 2021, 69, 333–337. [Google Scholar] [CrossRef]
- Habib, E.M.; Nosiar, N.A.; Eid, M.A.; Taha, A.M.; Sherief, D.E.; Hassan, A.E.; Abdel Ghafar, M.T. MiR-150 Expression in Chronic Myeloid Leukemia: Relation to Imatinib Response. Lab. Med. 2022, 53, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Machova Polakova, K.; Lopotova, T.; Klamova, H.; Burda, P.; Trneny, M.; Stopka, T.; Moravcova, J. Expression patterns of microRNAs associated with CML phases and their disease related targets. Mol. Cancer 2011, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Ninawe, A.; Guru, S.A.; Yadav, P.; Masroor, M.; Samadhiya, A.; Bhutani, N.; Gupta, N.; Gupta, R.; Saxena, A. miR-486-5p: A Prognostic Biomarker for Chronic Myeloid Leukemia. ACS Omega 2021, 6, 7711–7718. [Google Scholar] [PubMed]
- Keramati, F.; Jafarian, A.; Soltani, A.; Javandoost, E.; Mollaei, M.; Fallah, P. Circulating miRNAs can serve as potential diagnostic biomarkers in chronic myelogenous leukemia patients. Leuk. Res. Rep. 2021, 16, 100257. [Google Scholar] [CrossRef] [PubMed]
- Ali Beg, M.M.; Guru, S.A.; Abdullah, S.M.; Ahmad, I.; Rizvi, A.; Akhter, J.; Goyal, Y.; Verma, A.K. Regulation of miR-126 and miR-122 Expression and Response of Imatinib Treatment on Its Expression in Chronic Myeloid Leukemia Patients. Oncol. Res. Treat. 2021, 44, 530–537. [Google Scholar] [CrossRef]
- Troppan, K.; Wenzl, K.; Pichler, M.; Pursche, B.; Schwarzenbacher, D.; Feichtinger, J.; Thallinger, G.G.; Beham-Schmid, C.; Neumeister, P.; Deutsch, A. miR-199a and miR-497 Are Associated with Better Overall Survival due to Increased Chemosensitivity in Diffuse Large B-Cell Lymphoma Patients. Int. J. Mol. Sci. 2015, 16, 18077–18095. [Google Scholar] [CrossRef]
- Hao, M.; Zang, M.; Wendlandt, E.; Xu, Y.; An, G.; Gong, D.; Li, F.; Qi, F.; Zhang, Y.; Yang, Y.; et al. Low serum miR-19a expression as a novel poor prognostic indicator in multiple myeloma. Int. J. Cancer 2015, 136, 1835–1844. [Google Scholar] [CrossRef]
- Corthals, S.L.; Sun, S.M.; Kuiper, R.; de Knegt, Y.; Broyl, A.; van der Holt, B.; Beverloo, H.B.; Peeters, J.K.; el Jarari, L.; Lokhorst, H.M.; et al. MicroRNA signatures characterize multiple myeloma patients. Leukemia 2011, 25, 1784–1789. [Google Scholar] [CrossRef]
- Papanota, A.M.; Karousi, P.; Kontos, C.K.; Artemaki, P.I.; Liacos, C.I.; Papadimitriou, M.A.; Bagratuni, T.; Eleutherakis-Papaiakovou, E.; Malandrakis, P.; Ntanasis-Stathopoulos, I.; et al. A Cancer-Related microRNA Signature Shows Biomarker Utility in Multiple Myeloma. Int. J. Mol. Sci. 2021, 22, 13144. [Google Scholar] [CrossRef]
- Yang, N.; Chen, J.; Zhang, H.; Wang, X.; Yao, H.; Peng, Y.; Zhang, W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017, 8, e2975. [Google Scholar] [CrossRef]
- Li, F.; Xu, Y.; Deng, S.; Li, Z.; Zou, D.; Yi, S.; Sui, W.; Hao, M.; Qiu, L. MicroRNA-15a/16-1 cluster located at chromosome 13q14 is down-regulated but displays different expression pattern and prognostic significance in multiple myeloma. Oncotarget 2015, 6, 38270–38282. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Agnelli, L.; Walker, B.A.; Todoerti, K.; Lionetti, M.; Johnson, D.C.; Kaiser, M.; Mirabella, F.; Wardell, C.; Gregory, W.M.; et al. Improved risk stratification in myeloma using a microRNA-based classifier. Br. J. Haematol. 2013, 162, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, X.; Wang, W.; He, W.; Wang, J.; Wang, Y. Expression of Peripheral Blood miRNA-720 and miRNA-1246 Can Be Used as a Predictor for Outcome in Multiple Myeloma Patients. Clin. Lymphoma Myeloma Leuk. 2017, 17, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, R.; Qu, X.; Zhao, M.; Zhang, S.; Wu, H.; Jianyong, L.; Chen, L. MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leuk. Res. 2012, 36, 1505–1509. [Google Scholar] [CrossRef]
- Chi, J.; Ballabio, E.; Chen, X.H.; Kusec, R.; Taylor, S.; Hay, D.; Tramonti, D.; Saunders, N.J.; Littlewood, T.; Pezzella, F.; et al. MicroRNA expression in multiple myeloma is associated with genetic subtype, isotype and survival. Biol. Direct 2011, 6, 23. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, L.; Xiang, P.; Zhang, J. Circulating miRNAs as Auxiliary Diagnostic Biomarkers for Multiple Myeloma: A Systematic Review, Meta-Analysis, and Recommendations. Front. Oncol. 2021, 11, 698197. [Google Scholar] [CrossRef]
- Xu, P.; Xia, T.; Ling, Y.; Chen, B. MiRNAs with prognostic significance in multiple myeloma: A systemic review and meta-analysis. Medicine 2019, 98, e16711. [Google Scholar] [CrossRef]
- Balassa, K.; Danby, R.; Rocha, V. Haematopoietic stem cell transplants: Principles and indications. Br. J. Hosp. Med. 2019, 80, 33–39. [Google Scholar] [CrossRef]
- Bazinet, A.; Popradi, G. A general practitioner’s guide to hematopoietic stem-cell transplantation. Curr. Oncol. 2019, 26, 187–191. [Google Scholar] [CrossRef]
- Khaddour, K.; Hana, C.K.; Mewawalla, P. Hematopoietic Stem Cell Transplantation. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Wingard, J.R.; Hsu, J.; Hiemenz, J.W. Hematopoietic stem cell transplantation: An overview of infection risks and epidemiology. Infect. Dis. Clin. N. Am. 2010, 24, 257–272. [Google Scholar] [CrossRef]
- Hosseini, E.; Schwarer, A.P.; Ghasemzadeh, M. Do human leukocyte antigen E polymorphisms influence graft-versus-leukemia after allogeneic hematopoietic stem cell transplantation? Exp. Hematol. 2015, 43, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Passweg, J.R.; Baldomero, H.; Bader, P.; Bonini, C.; Cesaro, S.; Dreger, P.; Duarte, R.F.; Dufour, C.; Kuball, J.; Farge-Bancel, D.; et al. Hematopoietic stem cell transplantation in Europe 2014: More than 40,000 transplants annually. Bone Marrow Transplant. 2016, 51, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.E.; Barker, J.N.; DeFor, T.E.; Baker, K.S.; Blazar, B.R.; Eide, C.; Goldman, A.; Kersey, J.; Krivit, W.; MacMillan, M.L.; et al. Transplantation of unrelated donor umbilical cord blood in 102 patients with malignant and nonmalignant diseases: Influence of CD34 cell dose and HLA disparity on treatment-related mortality and survival. Blood 2002, 100, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Copelan, E.A. Hematopoietic stem-cell transplantation. N. Engl. J. Med. 2006, 354, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, J.L.; Levine, J.E.; Reddy, P.; Holler, E. Graft-versus-host disease. Lancet 2009, 373, 1550–1561. [Google Scholar] [CrossRef]
- Toubai, T.; Sun, Y.; Reddy, P. GVHD pathophysiology: Is acute different from chronic? Best Pract. Res. Clin. Haematol. 2008, 21, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Ball, L.M.; Egeler, R.M.; EBMT Paediatric Working Party. Acute GvHD: Pathogenesis and classification. Bone Marrow Transplant. 2008, 41 (Suppl. S2), S58–S64. [Google Scholar] [CrossRef]
- Jagasia, M.H.; Greinix, H.T.; Arora, M.; Williams, K.M.; Wolff, D.; Cowen, E.W.; Palmer, J.; Weisdorf, D.; Treister, N.S.; Cheng, G.S.; et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group report. Biol. Blood Marrow Transplant. 2015, 21, 389–401.e1. [Google Scholar] [CrossRef]
- Zecca, M.; Prete, A.; Rondelli, R.; Lanino, E.; Balduzzi, A.; Messina, C.; Fagioli, F.; Porta, F.; Favre, C.; Pession, A.; et al. Chronic graft-versus-host disease in children: Incidence, risk factors, and impact on outcome. Blood 2002, 100, 1192–1200. [Google Scholar] [CrossRef]
- Lee, S.J.; Vogelsang, G.; Flowers, M.E. Chronic graft-versus-host disease. Biol. Blood Marrow Transplant. 2003, 9, 215–233. [Google Scholar] [CrossRef]
- Fall-Dickson, J.M.; Pavletic, S.Z.; Mays, J.W.; Schubert, M.M. Oral Complications of Chronic Graft-Versus-Host Disease. J. Natl. Cancer Inst. Monogr. 2019, 2019, lgz007. [Google Scholar] [CrossRef] [PubMed]
- Holler, E. Risk assessment in haematopoietic stem cell transplantation: GvHD prevention and treatment. Best Pract. Res. Clin. Haematol. 2007, 20, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Wolff, D.; Radojcic, V.; Lafyatis, R.; Cinar, R.; Rosenstein, R.K.; Cowen, E.W.; Cheng, G.S.; Sheshadri, A.; Bergeron, A.; Williams, K.M.; et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: IV. The 2020 Highly morbid forms report. Transplant. Cell. Ther. 2021, 27, 817–835. [Google Scholar] [CrossRef] [PubMed]
- Aladag, E.; Kelkitli, E.; Goker, H. Acute Graft-Versus-Host Disease: A Brief Review. Turk. J. Haematol. 2020, 37, 1–4. [Google Scholar] [CrossRef]
- Ali, A.M.; DiPersio, J.F.; Schroeder, M.A. The Role of Biomarkers in the Diagnosis and Risk Stratification of Acute Graft-versus-Host Disease: A Systematic Review. Biol. Blood Marrow Transplant. 2016, 22, 1552–1564. [Google Scholar] [CrossRef]
- Newmarch, M.; Kostantin, E.; Tsongalis, G.J.; Guimond, M.; Roy, J.; De Guire, V.; Ahmad, I. MicroRNAs in graft-versus-host disease: A review of the latest data. Bone Marrow Transplant. 2020, 55, 1014–1020. [Google Scholar] [CrossRef]
- Vajari, M.K.; Moradinasab, S.; Yousefi, A.M.; Bashash, D. Noncoding RNAs in diagnosis and prognosis of graft-versus-host disease (GVHD). J. Cell. Physiol. 2022, 237, 3480–3495. [Google Scholar] [CrossRef]
- Xie, L.N.; Zhou, F.; Liu, X.M.; Fang, Y.; Yu, Z.; Song, N.X.; Kong, F.S. Serum microRNA155 is increased in patients with acute graft-versus-host disease. Clin. Transplant. 2014, 28, 314–323. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, X.; Hong, M.; Luo, T.; Zhao, M.; Shen, H.; Fang, J.; Li, X.; Zang, S.; Chen, P.; et al. Endothelial microparticles delivering microRNA-155 into T lymphocytes are involved in the initiation of acute graft-versus-host disease following allogeneic hematopoietic stem cell transplantation. Oncotarget 2017, 8, 23360–23375. [Google Scholar] [CrossRef]
- Stickel, N.; Prinz, G.; Pfeifer, D.; Hasselblatt, P.; Schmitt-Graeff, A.; Follo, M.; Thimme, R.; Finke, J.; Duyster, J.; Salzer, U.; et al. MiR-146a regulates the TRAF6/TNF-axis in donor T cells during GVHD. Blood 2014, 124, 2586–2595. [Google Scholar] [CrossRef]
- Stickel, N.; Hanke, K.; Marschner, D.; Prinz, G.; Kohler, M.; Melchinger, W.; Pfeifer, D.; Schmitt-Graeff, A.; Brummer, T.; Heine, A.; et al. MicroRNA-146a reduces MHC-II expression via targeting JAK/STAT signaling in dendritic cells after stem cell transplantation. Leukemia 2017, 31, 2732–2741. [Google Scholar] [CrossRef] [PubMed]
- Atarod, S.; Ahmed, M.M.; Lendrem, C.; Pearce, K.F.; Cope, W.; Norden, J.; Wang, X.N.; Collin, M.; Dickinson, A.M. miR-146a and miR-155 Expression Levels in Acute Graft-Versus-Host Disease Incidence. Front. Immunol. 2016, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Crossland, R.E.; Norden, J.; Ghimire, S.; Juric, M.K.; Pearce, K.F.; Lendrem, C.; Collin, M.; Mischak-Weissinger, E.; Holler, E.; Greinix, H.T.; et al. Profiling Tissue and Biofluid miR-155-5p, miR-155*, and miR-146a-5p Expression in Graft vs. Host Disease. Front. Immunol. 2021, 12, 639171. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Wang, Y.; Li, W.; Baker, M.; Guo, J.; Corbet, K.; Tsalik, E.L.; Li, Q.J.; Palmer, S.M.; Woods, C.W.; et al. Plasma microRNA signature as a noninvasive biomarker for acute graft-versus-host disease. Blood 2013, 122, 3365–3375. [Google Scholar] [CrossRef] [PubMed]
- Crossland, R.E.; Norden, J.; Kralj Juric, M.; Pearce, K.F.; Lendrem, C.; Bibby, L.A.; Collin, M.; Greinix, H.T.; Dickinson, A.M. Serum and Extracellular Vesicle MicroRNAs miR-423, miR-199, and miR-93* As Biomarkers for Acute Graft-versus-Host Disease. Front. Immunol. 2017, 8, 1446. [Google Scholar] [CrossRef]
- Crossland, R.E.; Norden, J.; Juric, M.K.; Green, K.; Pearce, K.F.; Lendrem, C.; Greinix, H.T.; Dickinson, A.M. Expression of Serum microRNAs is Altered During Acute Graft-versus-Host Disease. Front. Immunol. 2017, 8, 308. [Google Scholar] [CrossRef]
- Xie, L.; Zhou, F.; Liu, X.; Fang, Y.; Yu, Z.; Song, N.; Kong, F. Serum microRNA181a: Correlates with the intracellular cytokine levels and a potential biomarker for acute graft-versus-host disease. Cytokine 2016, 85, 37–44. [Google Scholar] [CrossRef]
- Gimondi, S.; Dugo, M.; Vendramin, A.; Bermema, A.; Biancon, G.; Cavane, A.; Corradini, P.; Carniti, C. Circulating miRNA panel for prediction of acute graft-versus-host disease in lymphoma patients undergoing matched unrelated hematopoietic stem cell transplantation. Exp. Hematol. 2016, 44, 624–634.e1. [Google Scholar] [CrossRef]
- Ranganathan, P.; Ngankeu, A.; Zitzer, N.C.; Leoncini, P.; Yu, X.; Casadei, L.; Challagundla, K.; Reichenbach, D.K.; Garman, S.; Ruppert, A.S.; et al. Serum miR-29a Is Upregulated in Acute Graft-versus-Host Disease and Activates Dendritic Cells through TLR Binding. J. Immunol. 2017, 198, 2500–2512. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, X.; Ye, X.; Luo, H.; Zhao, T.; Diao, Y.; Zhang, H.; Lv, M.; Zhang, W.; Huang, X.; et al. Plasma microRNA-586 is a new biomarker for acute graft-versus-host disease. Ann. Hematol. 2015, 94, 1505–1514. [Google Scholar] [CrossRef]
- Zhao, X.S.; Wang, Y.N.; Lv, M.; Kong, Y.; Luo, H.X.; Ye, X.Y.; Wu, Q.; Zhao, T.F.; Hu, Y.H.; Zhang, H.Y.; et al. miR-153-3p, a new bio-target, is involved in the pathogenesis of acute graft-versus-host disease via inhibition of indoleamine- 2,3-dioxygenase. Oncotarget 2016, 7, 48321–48334. [Google Scholar] [CrossRef] [PubMed]
- Motaei, J.; Kerachian, M.A.; Mousavi, S.A.; Alimoghadam, K.; Ghavamzadeh, A.; Manoochehrabadi, S.; Ahmadvand, M.; Yaghmaie, M. Circulating miR-455-3p, miR-5787, and miR-548a-3p as potential noninvasive biomarkers in the diagnosis of acute graft-versus-host disease: A validation study. Ann. Hematol. 2021, 100, 2621–2631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Bai, N.; Huang, W.; Zhang, P.; Luo, Y.; Men, S.; Wen, T.; Tong, H.; Wang, S.; Tian, Y.P. The predictive value of selected serum microRNAs for acute GVHD by TaqMan MicroRNA arrays. Ann. Hematol. 2016, 95, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, S.; Umezu, T.; Saitoh, Y.; Gotoh, M.; Akahane, D.; Kobayashi, C.; Ohyashiki, J.H.; Ohyashiki, K. Exosomal miRNA Signatures for Late-Onset Acute Graft-Versus-Host Disease in Allogenic Hematopoietic Stem Cell Transplantation. Int. J. Mol. Sci. 2018, 19, 2493. [Google Scholar] [CrossRef] [PubMed]
- Atarod, S.; Norden, J.; Bibby, L.A.; Janin, A.; Ratajczak, P.; Lendrem, C.; Pearce, K.F.; Wang, X.N.; O’Reilly, S.; Van Laar, J.M.; et al. Differential MicroRNA Expression Levels in Cutaneous Acute Graft-Versus-Host Disease. Front. Immunol. 2018, 9, 1485. [Google Scholar] [CrossRef]
- Juric, M.K.; Shevtsov, M.; Mozes, P.; Ogonek, J.; Crossland, R.E.; Dickinson, A.M.; Greinix, H.T.; Holler, E.; Weissinger, E.M.; Multhoff, G. B-Cell-Based and Soluble Biomarkers in Body Liquids for Predicting Acute/Chronic Graft-versus-Host Disease after Allogeneic Hematopoietic Stem Cell Transplantation. Front. Immunol. 2016, 7, 660. [Google Scholar] [CrossRef]
- Reikvam, H.; Vo, A.K.; Johansen, S.; Hemsing, A.L.; Solheim, M.H.; Mosevoll, K.A.; Tvedt, T.H.A.; Hatfield, K.J. MicroRNA serum profiles and chronic graft-versus-host disease. Blood Adv. 2022, 6, 5295–5306. [Google Scholar] [CrossRef]
- Lacina, P.; Crossland, R.E.; Wielinska, J.; Czyz, A.; Szeremet, A.; Ussowicz, M.; Wrobel, T.; Dickinson, A.M.; Bogunia-Kubik, K. Differential expression of miRNAs from extracellular vesicles in chronic graft-versus-host disease: A preliminary study. Adv. Clin. Exp. Med. 2022. [Google Scholar] [CrossRef]
- Sorror, M.L.; Gooley, T.A.; Maclean, K.H.; Hubbard, J.; Marcondes, M.A.; Torok-Storb, B.J.; Tewari, M. Pre-transplant expressions of microRNAs, comorbidities, and post-transplant mortality. Bone Marrow Transplant. 2019, 54, 973–979. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhou, L.; Hu, K.; Dai, Y.; Pang, Y.; Zhao, H.; Wu, S.; Qin, T.; Han, Y.; Hu, N.; et al. Prognostic significance of microRNA-99a in acute myeloid leukemia patients undergoing allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2018, 53, 1089–1095. [Google Scholar] [CrossRef]
- Yang, C.; Shao, T.; Zhang, H.; Zhang, N.; Shi, X.; Liu, X.; Yao, Y.; Xu, L.; Zhu, S.; Cao, J.; et al. MiR-425 expression profiling in acute myeloid leukemia might guide the treatment choice between allogeneic transplantation and chemotherapy. J. Transl. Med. 2018, 16, 267. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.C.; Soneji, S.; Palmason, R.; Lenhoff, S.; Laurell, T.; Scheding, S. Development of acoustically isolated extracellular plasma vesicles for biomarker discovery in allogeneic hematopoietic stem cell transplantation. Biomark. Res. 2021, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Pontoppidan, P.L.; Jordan, K.; Carlsen, A.L.; Uhlving, H.H.; Kielsen, K.; Christensen, M.; Ifversen, M.; Nielsen, C.H.; Sangild, P.; Heegaard, N.H.; et al. Associations between gastrointestinal toxicity, micro RNA and cytokine production in patients undergoing myeloablative allogeneic stem cell transplantation. Int. Immunopharmacol. 2015, 25, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, M.; Szemraj, J.; Wierzbowska, A.; Misiewicz, M.; Malachowski, R.; Pluta, A.; Grzybowska-Izydorczyk, O.; Robak, T.; Szmigielska-Kaplon, A. miRNA-15a, miRNA-16, miRNA-126, miRNA-146a, and miRNA-223 expressions in autologous hematopoietic stem cell transplantation and their impact on engraftment. Eur. J. Haematol. 2018, 100, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Rafiee, M.; Farsani, M.A.; Amiri, V.; Hajifathali, A.; Gharehbaghian, A.; Mohammadi, M.H. Circulatory miR-155 correlation with platelet and neutrophil recovery after autologous hematopoietic stem cell transplantation, a multivariate analysis. Int. J. Hematol. 2021, 114, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Park, S.S.; Lim, J.Y.; Kim, T.W.; Ko, Y.H.; Jeon, W.J.; Lee, S.Y.; Lee, J.H.; Min, C.K. Predictive impact of circulating microRNA-193a-5p on early relapse after autologous stem cell transplantation in patients with multiple myeloma. Br. J. Haematol. 2020, 189, 518–523. [Google Scholar] [CrossRef]
- Innao, V.; Allegra, A.; Pulvirenti, N.; Allegra, A.G.; Musolino, C. Therapeutic potential of antagomiRs in haematological and oncological neoplasms. Eur. J. Cancer Care 2020, 29, e13208. [Google Scholar] [CrossRef]
- Dorrance, A.M.; Neviani, P.; Ferenchak, G.J.; Huang, X.; Nicolet, D.; Maharry, K.S.; Ozer, H.G.; Hoellarbauer, P.; Khalife, J.; Hill, E.B.; et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia 2015, 29, 2143–2153. [Google Scholar] [CrossRef]
- Wang, W.Z.; Lin, X.H.; Pu, Q.H.; Liu, M.Y.; Li, L.; Wu, L.R.; Wu, Q.Q.; Mao, J.W.; Zhu, J.Y.; Jin, X.B. Targeting miR-21 sensitizes Ph+ ALL Sup-b15 cells to imatinib-induced apoptosis through upregulation of PTEN. Biochem. Biophys. Res. Commun. 2014, 454, 423–428. [Google Scholar] [CrossRef]
- Zhang, L.; Nguyen, L.X.T.; Chen, Y.C.; Wu, D.; Cook, G.J.; Hoang, D.H.; Brewer, C.J.; He, X.; Dong, H.; Li, S.; et al. Targeting miR-126 in inv(16) acute myeloid leukemia inhibits leukemia development and leukemia stem cell maintenance. Nat. Commun. 2021, 12, 6154. [Google Scholar] [CrossRef]
- Velu, C.S.; Chaubey, A.; Phelan, J.D.; Horman, S.R.; Wunderlich, M.; Guzman, M.L.; Jegga, A.G.; Zeleznik-Le, N.J.; Chen, J.; Mulloy, J.C.; et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J. Clin. Investig. 2014, 124, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin. Cancer Res. 2021, 27, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Zhang, C.; Lu, T.; Liao, E.J.; Huang, H.; Wei, S. Roles of circRNAs in hematological malignancies. Biomark. Res. 2022, 10, 50. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Barta, T.; Peskova, L.; Hampl, A. miRNAsong: A web-based tool for generation and testing of miRNA sponge constructs in silico. Sci. Rep. 2016, 6, 36625. [Google Scholar] [CrossRef]
- Han, F.; Zhong, C.; Li, W.; Wang, R.; Zhang, C.; Yang, X.; Ji, C.; Ma, D. hsa_circ_0001947 suppresses acute myeloid leukemia progression via targeting hsa-miR-329-5p/CREBRF axis. Epigenomics 2020, 12, 935–953. [Google Scholar] [CrossRef]
- Wu, Z.J.; Sun, Q.; Gu, D.L.; Wang, L.Q.; Li, J.Y.; Jin, H. [Expression of circ-KEL in acute myeloid leukemia and its regulatory mechanisms in leukemic cells]. Zhonghua Xue Ye Xue Za Zhi 2021, 42, 230–237. [Google Scholar]
- Cao, H.X.; Miao, C.F.; Sang, L.N.; Huang, Y.M.; Zhang, R.; Sun, L.; Jiang, Z.X. Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 2020, 243, 117255. [Google Scholar] [CrossRef]
- Wang, D.; Ming, X.; Xu, J.; Xiao, Y. Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR-5195-3p/GRB10 axis. Hematol. Oncol. 2021, 39, 390–400. [Google Scholar] [CrossRef]
- Chen, F.; Wang, X.; Fu, S.; Wang, S.; Fu, Y.; Liu, Z.; Zhang, J. Effect of the Up-Regulation of Circular RNA Hsa_circ_0069767 Derived from C-KIT on the Biological Behavior of Multiple Myeloma Cells. Cancer Manag. Res. 2020, 12, 11321–11331. [Google Scholar] [CrossRef]
- Xu, L.; Xu, Y.; Jing, Z.; Wang, X.; Zha, X.; Zeng, C.; Chen, S.; Yang, L.; Luo, G.; Li, B.; et al. Altered expression pattern of miR-29a, miR-29b and the target genes in myeloid leukemia. Exp. Hematol. Oncol. 2014, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mao, M.; Liu, H.; Wang, X.; Kou, Z.; Nie, Y.; Wang, Y.; Wang, Z.; Huang, Q.; Lang, T.; et al. miR-34a and miR-29b as indicators for prognosis of treatment-free survival of chronic lymphocytic leukemia patients in Chinese Uygur and Han populations. Mol. Cell. Probes 2019, 47, 101436. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef] [PubMed]
- Blum, W.; Klisovic, R.B.; Hackanson, B.; Liu, Z.; Liu, S.; Devine, H.; Vukosavljevic, T.; Huynh, L.; Lozanski, G.; Kefauver, C.; et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J. Clin. Oncol. 2007, 25, 3884–3891. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, Y.; Song, X.; Zhou, H.; Li, N.; Miao, Y.; Jia, L. Upregulation of miR-181c inhibits chemoresistance by targeting ST8SIA4 in chronic myelocytic leukemia. Oncotarget 2016, 7, 60074–60086. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Schwind, S.; Yu, B.; Santhanam, R.; Wang, H.; Hoellerbauer, P.; Mims, A.; Klisovic, R.; Walker, A.R.; Chan, K.K.; et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: A novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. 2013, 19, 2355–2367. [Google Scholar] [CrossRef]
- Huang, X.; Schwind, S.; Santhanam, R.; Eisfeld, A.K.; Chiang, C.L.; Lankenau, M.; Yu, B.; Hoellerbauer, P.; Jin, Y.; Tarighat, S.S.; et al. Targeting the RAS/MAPK pathway with miR-181a in acute myeloid leukemia. Oncotarget 2016, 7, 59273–59286. [Google Scholar] [CrossRef]
- Marcucci, G.; Radmacher, M.D.; Maharry, K.; Mrozek, K.; Ruppert, A.S.; Paschka, P.; Vukosavljevic, T.; Whitman, S.P.; Baldus, C.D.; Langer, C.; et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008, 358, 1919–1928. [Google Scholar] [CrossRef]
| Malignancy | miRNA | miRNA Expression Level | Target Genes | Study Type | miRNA Function/Clinical Consequence | Reference |
|---|---|---|---|---|---|---|
| AML | miR-3662 | DOWN | IKBKB | AM, PS (N = 20), TC | Acceleration of the growth and colony formation of HP cells, survival of leukemic cells | [58] |
| miR-9 | UP | RHOH RYBP + 15 other potential targets | AM CL PS (N = 85) | Increasing cell survival and decreasing apoptosis | [82] | |
| miR-29b | DOWN | MCL-1, CXXC6, CDK6 | AM, CL, PS (N = 100) | Cell growth and anti-apoptotic activity | [83] | |
| miR-126 | UP | SLC9A7, ABCG1, MEF2C, RBMPS, LYZ, CSTA, HAL | AM, CL, PS (N = 6) | Cell growth and anti-apoptotic activity | [57] | |
| miR-301b | UP | FOXF2 | CL | Cell proliferation and decreasing apoptosis | [59] | |
| CML | miR-19b (miR-17-92 cluster) | UP | A20 | AM, BM, CL | Cell proliferation, cell cycle, and decreasing apoptosis | [60] |
| miR-328 | DOWN | PIM-1, TCF-4 | CL | Cell proliferation, survival | [61] | |
| MDS | miR-378 | DOWN | CDC40 | CL, PS (BM) (N = 20) | Inducing the apoptosis and blocking the cell cycle of MDS cells | [69] |
| MM | miR-21 | UP | PTEN, Rho-B, BTG2 | AM, CL | Growth and anti-apoptotic activity | [70] |
| miR-221/222 | UP | p27Kip1, PUMA, PTEN, p57Kip2 | AM, CL | Proliferation and cell survival | [71] | |
| miR-342-3p | DOWN | FOXQ1 RAP2B CDC42 | CL, PS diagnostic and relapsed MM (N = 93) | Methylation-derived silencing of miR-342-3p might be an early event in MM pathogenesis | [72] | |
| miR-140-3p | DOWN | BZW2 | AM, CL | Cell proliferation, decreasing the apoptosis in MM cells | [73] | |
| miR-125a-5p | UP | TP53 | CL | Cell proliferation, cell growth decreasing apoptosis of cancer cells | [84] | |
| miR-9 | DOWN | FGFR1 CDK6 | CL PS (N = 200) | MiR-9 hypermethylation lead to the activation of oncogenic pathways and represents a prognostic factor for survival | [85] | |
| ALL | miR-124a | DOWN | CDK6 | AM, CL, PS (N = 353) | Cell proliferation and growth of ALL cells | [56] |
| T-ALL | miR-204 | DOWN | IRAK1, NF-kB | AM, CL, PS (N = 32) | T-ALL growth and metastasis by increased IRAK1 and activation of NF-kB signaling pathway and targets | [74] |
| CLL | miR-125a, miR-223 | DOWN | BCL2, STAT3 | PS (N = 30) | Control of white blood cell production | [76] |
| miR-22 | UP | PTEN | PS (N = 22) | B-CLL cell proliferation | [86] | |
| miR-15a, miR-16-1 | DOWN | DLEU2 | AM, PBMC | Decreasing apoptosis and cell cycle of malignant B-cells | [75] | |
| NHL | miR-34a | DOWN | TP53, CDK6 | CL, PS (N = 32) | MiR-34a is preferentially hypermethylated in NHL, the role of miR-34a in lymphomagenesis | [87] |
| miR-29 | DOWN | IGF-1R, CDK6 | CL | Cell survival and growth regulation in MCLs | [79] | |
| DLBCL | miR-155 | UP | HDAC4 | AM, CL | Block the development of B cells at the immature stage and induce cell proliferation | [78] |
| Malignancy | MiRNA | Sample Type | Expression Pattern | No. of Patients | Diagnosis | Prognosis | Therapy Response | Ref. |
|---|---|---|---|---|---|---|---|---|
| ALL | miR-146a | Plasma | UP | N = 66 | Diagnostic marker pediatric and adult ALL | DOWN after CHT | [99] | |
| ALL pediatric | miR-155a | BM | UP | N = 45 | MRD, poor prognosis | DOWN after CHT | [100] | |
| miR-155 | BM | UP | N = 42 | Poor outcome | [101] | |||
| miR-200c and miR-326 | BM | DOWN | N = 46 | Drug resistance | [102] | |||
| miR-324-3p and miR-508-5p | BM | DOWN | N = 50 | Drug resistance | [103] | |||
| let-7b, miR-511, and miR-708 miR-196a, miR-383, and miR-542-5p | BM, PB | DOWN UP | N = 81 | Genetic subtype discrimination | [104] | |||
| miR-99a, miR-100, and miR-125b | UP | Drug resistance | ||||||
| miR-10a, miR-33, miR-134, miR -214, miR -215, miR-369-5p, miR -484, miR-496, miR-518d, miR-572, miR-580, miR-599, miR-624, and miR-627 | UP or DOWN | Clinical outcome | ||||||
| miR-151a-5p, miR-151b, miR-195-5p, miR-371b-5p, miR-425-5p, miR-455-5p, miR-497-5p, miR-574-5p, miR-708-5p, and miR-1266-5p | BM, PB | DOWN | N = 16 | T-ALL and B-ALL discrimination | [105] | |||
| miR-29c-5p, miR-424-5p, miR-450a-5p miR-450b-5p, miR-542-5p, and miR-629-5p | UP | |||||||
| B-ALL pediatric | miR-21 | BM, PB | UP | N = 75 | Shorter DFS and OS | [106] | ||
| miR-101-3p, miR-631, miR-922, miR-1324, miR-4699-5p, and miR-4774-5p | BM | UP | N = 40 | Prediction of early relapse | [107] | |||
| Precursor B-ALL pediatric | miR-151-5p and mR-451 | BM | DOWN | N = 189 | Shorter RFS | [108] | ||
| miR-1290 | UP | |||||||
| ALL and AML | miR-128a and miR-128b | BM | UP in ALL | N = 136 | ALL and AML discrimination | [109] | ||
| let-7b and miR-223 | DOWN in ALL | |||||||
| AML | miR-125b | Exosomes | UP | N = 154 | Higher risk of relapse and overall death | [97] | ||
| miR-363 | PB | UP | N = 162 | Shorter EFS and OS | Preference allo-HCST to CHT | [110] | ||
| miR-504-3p | Serum | DOWN | N = 134 | Shorter OS | [111] | |||
| miR-199b-5p, miR-301b, miR-326, miR-361-5p, miR-625, and miR-655 | Plasma | UP | N = 8 | DOWN after CHT | [112] | |||
| AML pediatric | miR-183 | BM, serum | UP | N = 106 | Shorter PFS and OS | [113] | ||
| miR-199a | BM | DOWN | N = 71 | Higher BM blasts | Shorter EFS | Lower therapy response | [114] | |
| miR-370 | BM, serum | DOWN | N = 106 | Shorter RFS and OS | [115] | |||
| miR-146a, miR-509, miR-542, and miR-3667 | unknown | UP | N = 229 * | Shorter OS | [116] | |||
| CLL | miR-32-5p, miR-98-5p, and miR-374b-5p | PB | DOWN | N = 32 | Early diagnosis | [117] | ||
| B-CLL | miR-145-5p and miR-185-5p | Plasma | DOWN | N = 40 | B-CLL detection | [118] | ||
| CML | miR-142-5p | BM, PB | DOWN | N = 45 | Drug resistance | [119] | ||
| miR-146a | Plasma | DOWN | N = 60 | Prediction of imatinib response | [120] | |||
| miR-150 | Plasma | DOWN | N = 60 | Prediction of imatinib response, UP after CHT | [121] | |||
| miR-150 | PB | DOWN | N = 24 | Potential marker for blast crisis and hematologic relapses | [122] | |||
| miR-486-5p | PB | DOWN | N = 36 | Early diagnosis | Prediction of imatinib response, UP after CHT | [123] | ||
| miR-20, miR-106, and miR-222, | Plasma | UP | N = 50 | Potential markers for therapy response | [124] | |||
| miR-122 and miR-126 | PB | DOWN | N = 100 | Prediction of imatinib response, UP after CHT | [125] | |||
| DLBCL | miR-99a-5p and miR-125b-5p | Exosomes | UP | Shorter PFS | CHT resistance | [98] | ||
| miR-199a and miR-497 | LN biopsies | UP | N = 63 | Longer OS | Drug sensitivity DOWN after CHT | [126] | ||
| MM | miR-19a | Serum | DOWN | N = 108 | Shorter PFS and OS | Bortezomib sensitivity | [127] | |
| miR-194 | BM | UP | N = 44 | Longer OS | [128] | |||
| miR-223-3p | BM | DOWN | N = 94 | Shorter OS | [129] | |||
| miR-410 | BM | UP | N = 97 | Shorter PFS and OS | [130] | |||
| miR-15a | BM | DOWN | N = 117 | Shorter PFS and OS | [131] | |||
| miR-16-1 | MM detection | |||||||
| let-7b and miR-18a | Exosomes | DOWN | N = 156 | Shorter PFS and OS | [95] | |||
| miR-17 and miR-885-5p | BM | UP | N = 163 | Risk stratification | [132] | |||
| miR-720 and miR-1246 | PB | UP | N = 60 | Shorter PFS | [133] | |||
| miR-15a-5p, miR-16-5p, miR-17-5p, and miR-20a-5p | Exosomes | DOWN | N = 330 | Bortezomib resistance | [94] | |||
| miR-15a, miR-16-1, miR-17, miR-20a, and miR-92-1 | Plasma | UP | N = 85 | Shorter PFS | [134] | |||
| miR-153, miR-296, miR-490, miR-455, miR-500, and miR-642 | BM | DOWN | N = 33 | Shorter EFS | [135] | |||
| miR-373, miR-548d, miR-554, and miR-888 | UP | |||||||
| miR-4254 | Serum, plasma ** | UP | N = 627 ‡ | Potential marker for MM | [136] | |||
| miR-92a | Mainly serum ** | UP | N = 1214 ‡‡ | Shorter PFS and OS | [137] | |||
| let-7e, miR-15a, miR-16, miR-25, and miR-744 | DOWN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sevcikova, A.; Fridrichova, I.; Nikolaieva, N.; Kalinkova, L.; Omelka, R.; Martiniakova, M.; Ciernikova, S. Clinical Significance of microRNAs in Hematologic Malignancies and Hematopoietic Stem Cell Transplantation. Cancers 2023, 15, 2658. https://doi.org/10.3390/cancers15092658
Sevcikova A, Fridrichova I, Nikolaieva N, Kalinkova L, Omelka R, Martiniakova M, Ciernikova S. Clinical Significance of microRNAs in Hematologic Malignancies and Hematopoietic Stem Cell Transplantation. Cancers. 2023; 15(9):2658. https://doi.org/10.3390/cancers15092658
Chicago/Turabian StyleSevcikova, Aneta, Ivana Fridrichova, Nataliia Nikolaieva, Lenka Kalinkova, Radoslav Omelka, Monika Martiniakova, and Sona Ciernikova. 2023. "Clinical Significance of microRNAs in Hematologic Malignancies and Hematopoietic Stem Cell Transplantation" Cancers 15, no. 9: 2658. https://doi.org/10.3390/cancers15092658
APA StyleSevcikova, A., Fridrichova, I., Nikolaieva, N., Kalinkova, L., Omelka, R., Martiniakova, M., & Ciernikova, S. (2023). Clinical Significance of microRNAs in Hematologic Malignancies and Hematopoietic Stem Cell Transplantation. Cancers, 15(9), 2658. https://doi.org/10.3390/cancers15092658





