A Network of MicroRNAs and mRNAs Involved in Melanosome Maturation and Trafficking Defines the Lower Response of Pigmentable Melanoma Cells to Targeted Therapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Drugs
2.3. Oligos
2.4. Generation of 501Mel Cells That Stably Express TYR-mCherry Fluorescent Protein
2.5. Transfection of miRNA Mimics and LNAs
2.6. Melanin Content Evaluation
2.7. Transmission Electron Microscope Analysis
2.8. Quantification of Released Melanosomes
2.9. Clonogenicity Assay
2.10. Co-Colture Assay
2.11. Xenograft in Zebrafish Embryos
2.12. RNA Extraction and Quantification
2.13. DNAse Treatment and Retrotranscription
2.14. Real-Time PCR
2.15. Protein Extraction and Western Blot Analysis
- -
- anti-DCT (#sc-74439, Santa Cruz Biotechnology, Dallas, TX, USA; mouse monoclonal antibody, dilution 1:1000 in 3% milk in TBST);
- -
- anti-GAPDH (#2118, Cell Signaling, Danvers, MA, USA; rabbit polyclonal antibody, dilution 1:3000 in 3% milk in TBST);
- -
- anti-PMEL17 (#sc-377325, Santa Cruz Biotechnology, Dallas, TX, USA; mouse monoclonal antibody, dilution 1:1000 in 3% milk in TBST);
- -
- anti-TYR (#sc-20035, Santa Cruz Biotechnology, Dallas, TX, USA; mouse monoclonal antibody, dilution 1:1000 in 3% milk in TBST);
- -
- anti-TYRP1 (#sc-166857, Santa Cruz Biotechnology, Dallas, TX, USA; mouse monoclonal antibody, dilution 1:1000 in 3% milk in TBST).
2.16. Statistical Analysis
2.17. Deep Sequencing
2.17.1. Sample Preparation
2.17.2. Library Generation and Sequencing
2.17.3. Primary Analysis, Clustering, and Differential Expression Analysis
2.18. clusterProfiler Analysis
2.19. WGCNA Analysis
2.19.1. Construction of Weighted Gene Co-Expression Network and Identification of Hub Modules
2.19.2. clusterProfiler Analysis of the Genes Belonging to WGCNA Modules
2.20. SWIMmeR Analysis
2.20.1. miRNAs-Module Association
2.20.2. Functional Enrichment Analysis
2.21. Gene Correlations Analysis
3. Results and Discussion
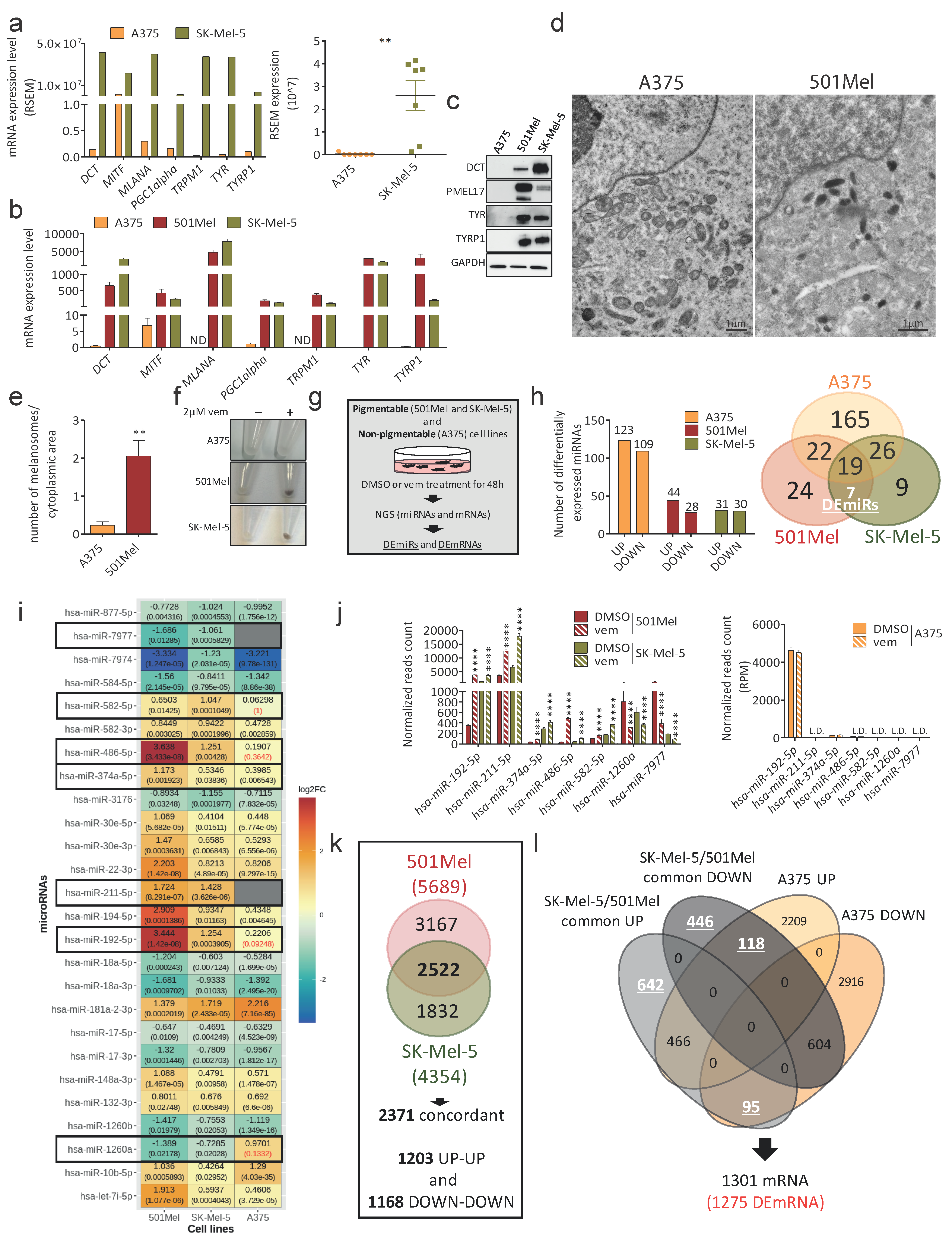
3.1. Identification of Differentially Expressed miRNAs and mRNAs in Pigmentable vs. Non-Pigmentable Cell Lines upon Vem Treatment
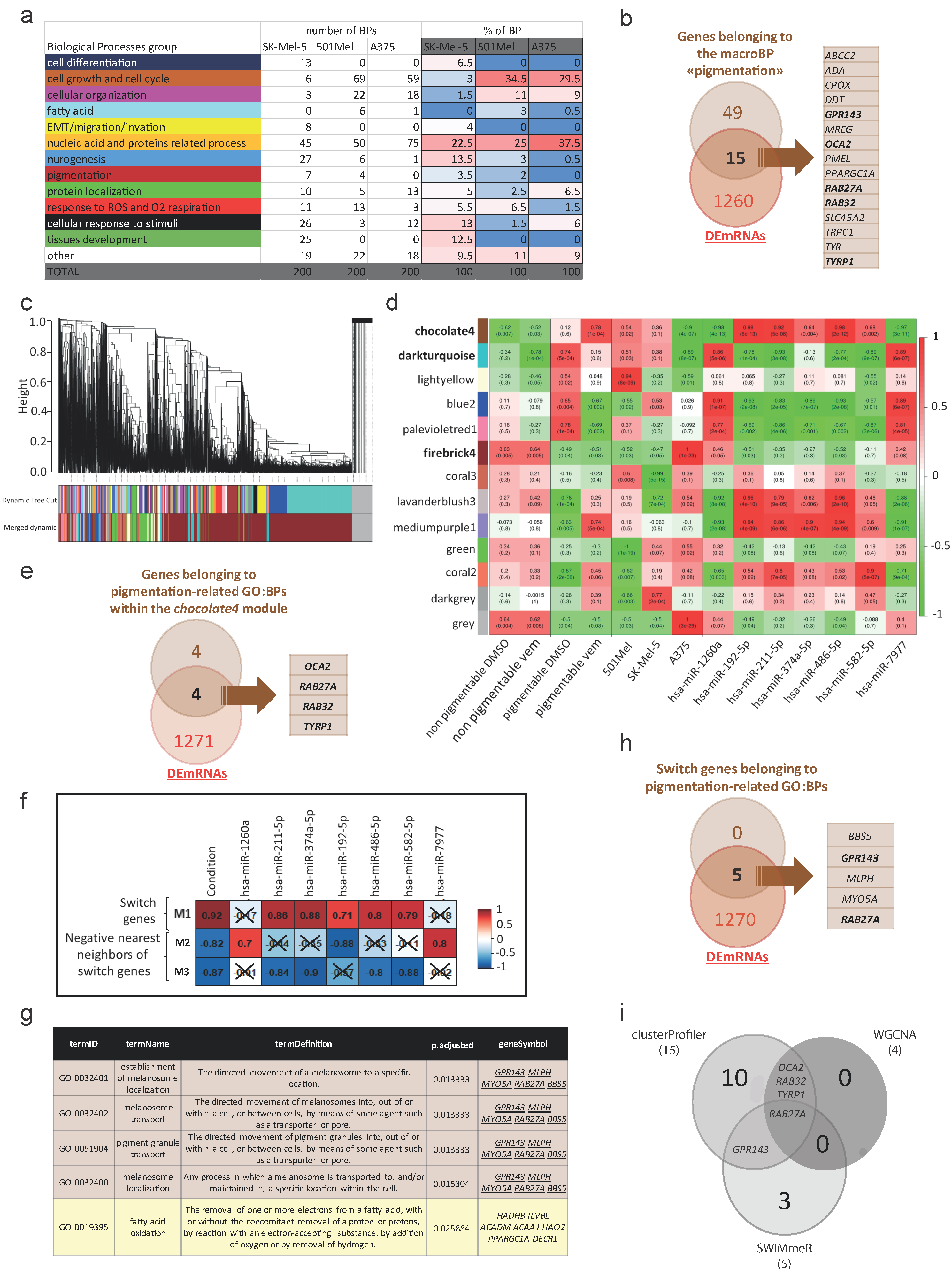
3.2. Identification of Biological Processes Selectively Enriched in Pigmentable vs. Non-Pigmentable Cell Lines upon Vem Treatment
3.2.1. Enriched Functional Profiles Analysis by clusterProfiler
3.2.2. Modules Analysis by WGCNA
3.2.3. Switch Genes Analysis by SWIMmeR
3.3. Validation of DEmiRs and DEmRNAs Identified by Bioinformatic Analyses
3.4. Validation of the Pigmentation-Related Biological Processes Identified by Bioinformatic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Subbiah, V.; Baik, C.; Kirkwood, J.M. Clinical Development of BRAF plus MEK Inhibitor Combinations. Trends Cancer 2020, 6, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Saei, A.; Eichhorn, P.J.A. Adaptive Responses as Mechanisms of Resistance to BRAF Inhibitors in Melanoma. Cancers 2019, 11, 1176. [Google Scholar] [CrossRef] [PubMed]
- Marine, J.-C.; Dawson, S.-J.; Dawson, M.A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 2020, 20, 743–756. [Google Scholar] [CrossRef]
- Roesch, A.; Vultur, A.; Bogeski, I.; Wang, H.; Zimmermann, K.M.; Speicher, D.; Körbel, C.; Laschke, M.W.; Gimotty, P.A.; Philipp, S.E.; et al. Overcoming Intrinsic Multidrug Resistance in Melanoma by Blocking the Mitochondrial Respiratory Chain of Slow-Cycling JARID1Bhigh Cells. Cancer Cell 2013, 23, 811–825. [Google Scholar] [CrossRef]
- Chartrain, M.; Riond, J.; Stennevin, A.; Vandenberghe, I.; Gomes, B.; Lamant, L.; Meyer, N.; Gairin, J.E.; Guilbaud, N.; Annereau, J.P. Melanoma Chemotherapy Leads to the Selection of ABCB5-Expressing Cells. PLoS ONE 2012, 7, e36762. [Google Scholar] [CrossRef]
- Rambow, F.; Rogiers, A.; Marin-Bejar, O.; Aibar, S.; Femel, J.; Dewaele, M.; Karras, P.; Brown, D.; Chang, Y.H.; Debiec-Rychter, M.; et al. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell 2018, 174, 843–855.e19. [Google Scholar] [CrossRef] [PubMed]
- Basile, K.J.; Abel, E.V.; Aplin, A.E. Adaptive upregulation of FOXD3 and resistance to PLX4032/4720-induced cell death in mutant B-RAF melanoma cells. Oncogene 2012, 31, 2471–2479. [Google Scholar] [CrossRef]
- Tsoi, J.; Robert, L.; Paraiso, K.; Galvan, C.; Sheu, K.M.; Lay, J.; Wong, D.J.; Atefi, M.; Shirazi, R.; Wang, X.; et al. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell 2018, 33, 890–904.e5. [Google Scholar] [CrossRef]
- Marin-Bejar, O.; Rogiers, A.; Dewaele, M.; Femel, J.; Karras, P.; Pozniak, J.; Bervoets, G.; Van Raemdonck, N.; Pedri, D.; Swings, T.; et al. Evolutionary predictability of genetic versus nongenetic resistance to anticancer drugs in melanoma. Cancer Cell 2021, 39, 1135–1149.e8. [Google Scholar] [CrossRef]
- Smith, M.P.; Brunton, H.; Rowling, E.J.; Ferguson, J.; Arozarena, I.; Miskolczi, Z.; Lee, J.L.; Girotti, M.R.; Marais, R.; Levesque, M.P.; et al. Inhibiting drivers of non-mutational drug tolerance is a salvage strategy for targeted melanoma therapy. Cancer Cell 2016, 29, 270–284. [Google Scholar] [CrossRef]
- Su, Y.; Ko, M.E.; Cheng, H.; Zhu, R.; Xue, M.; Wang, J.; Lee, J.W.; Frankiw, L.; Xu, A.; Wong, S.; et al. Multi-omic single-cell snapshots reveal multiple independent trajectories to drug tolerance in a melanoma cell line. Nat. Commun. 2020, 11, 2345. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Delgado-Goni, T.; Galobart, T.C.; Wantuch, S.; Jamin, Y.; Leach, M.O.; Robinson, S.P.; Bamber, J.; Beloueche-Babari, M. Detecting human melanoma cell re-differentiation following BRAF or heat shock protein 90 inhibition using photoacoustic and magnetic resonance imaging. Sci. Rep. 2017, 7, 8215. [Google Scholar] [CrossRef] [PubMed]
- Fufa, T.D.; Program, N.C.S.; Baxter, L.L.; Wedel, J.C.; Gildea, D.E.; Loftus, S.K.; Pavan, W.J. MEK inhibition remodels the active chromatin landscape and induces SOX10 genomic recruitment in BRAF(V600E) mutant melanoma cells. Epigenet. Chromatin 2019, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Haq, R.; Shoag, J.; Andreu-Perez, P.; Yokoyama, S.; Edelman, H.; Rowe, G.C.; Frederick, D.T.; Hurley, A.D.; Nellore, A.; Kung, A.L.; et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 2013, 23, 302–315. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23477830 (accessed on 31 August 2022). [CrossRef] [PubMed]
- Zhang, G.; Frederick, D.T.; Wu, L.; Wei, Z.; Krepler, C.; Srinivasan, S.; Chae, Y.C.; Xu, X.; Choi, H.; Dimwamwa, E.; et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Investig. 2016, 126, 1834–1856. [Google Scholar] [CrossRef]
- Kugel, C.H., 3rd; Hartsough, E.J.; Davies, M.A.; Setiady, Y.Y.; Aplin, A.E. Function-blocking ERBB3 antibody inhibits the adaptive response to RAF inhibitor. Cancer Res. 2014, 74, 4122–4132. [Google Scholar] [CrossRef]
- Du, J.; Miller, A.J.; Widlund, H.R.; Horstmann, M.A.; Ramaswamy, S.; Fisher, D.E. MLANA/MART1 and SILV/PMEL17/GP100 Are Transcriptionally Regulated by MITF in Melanocytes and Melanoma. Am. J. Pathol. 2003, 163, 333–343. [Google Scholar] [CrossRef]
- Vetrini, F.; Auricchio, A.; Du, J.; Angeletti, B.; Fisher, D.E.; Ballabio, A.; Marigo, V. The Microphthalmia Transcription Factor (Mitf) Controls Expression of the Ocular Albinism Type 1 Gene: Link between Melanin Synthesis and Melanosome Biogenesis. Mol. Cell. Biol. 2004, 24, 6550–6559. [Google Scholar] [CrossRef]
- Kawakami, A.; Fisher, D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 2017, 97, 649–656. [Google Scholar] [CrossRef]
- Zhiqi, S.; Soltani, M.H.; Bhat, K.M.R.; Sangha, N.; Fang, D.; Hunter, J.J.; Setaluri, V. Human melastatin 1 (TRPM1) is regulated by MITF and produces multiple polypeptide isoforms in melanocytes and melanoma. Melanoma Res. 2004, 14, 509–516. [Google Scholar] [CrossRef]
- Aida, S.; Sonobe, Y.; Tanimura, H.; Oikawa, N.; Yuhki, M.; Sakamoto, H.; Mizuno, T. MITF suppression improves the sensitivity of melanoma cells to a BRAF inhibitor. Cancer Lett. 2017, 409, 116–124. [Google Scholar] [CrossRef]
- Yang, Y.; Jang, G.-B.; Yang, X.; Wang, Q.; He, S.; Li, S.; Quach, C.; Zhao, S.; Li, F.; Yuan, Z.; et al. Central role of autophagic UVRAG in melanogenesis and the suntan response. Proc. Natl. Acad. Sci. USA 2018, 115, E7728–E7737. [Google Scholar] [CrossRef] [PubMed]
- Gaudel, C.; Soysouvanh, F.; Leclerc, J.; Bille, K.; Husser, C.; Montcriol, F.; Bertolotto, C.; Ballotti, R. Regulation of Melanogenesis by the Amino Acid Transporter SLC7A5. J. Investig. Dermatol. 2020, 140, 2253–2259.e4. [Google Scholar] [CrossRef] [PubMed]
- Chiaverini, C.; Beuret, L.; Flori, E.; Busca, R.; Abbe, P.; Bille, K.; Bahadoran, P.; Ortonne, J.-P.; Bertolotto, C.; Ballotti, R. Microphthalmia-associated Transcription Factor Regulates RAB27A Gene Expression and Controls Melanosome Transport. J. Biol. Chem. 2008, 283, 12635–12642. [Google Scholar] [CrossRef]
- Vitiello, M.; Tuccoli, A.; D’Aurizio, R.; Sarti, S.; Giannecchini, L.; Lubrano, S.; Marranci, A.; Evangelista, M.; Peppicelli, S.; Ippolito, C.; et al. Context-dependent miR-204 and miR-211 affect the biological properties of amelanotic and melanotic melanoma cells. Oncotarget 2017, 8, 25395–25417. [Google Scholar] [CrossRef]
- Vitiello, M.; D’Aurizio, R.; Poliseno, L. Biological role of miR-204 and miR-211 in melanoma. Oncoscience 2018, 5, 248–251. [Google Scholar] [CrossRef]
- Marranci, A.; Tuccoli, A.; Vitiello, M.; Mercoledi, E.; Sarti, S.; Lubrano, S.; Evangelista, M.; Fogli, A.; Valdes, C.; Russo, F.; et al. Identification of BRAF 3′UTR Isoforms in Melanoma. J. Investig. Dermatol. 2015, 135, 1694–1697. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wang, W.-C.; Lin, F.-M.; Chang, W.-C.; Lin, K.-Y.; Huang, H.-D.; Lin, N.-S. miRExpress: Analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinform. 2009, 10, 328. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Paci, P.; Colombo, T.; Fiscon, G.; Gurtner, A.; Pavesi, G.; Farina, L. SWIM: A computational tool to unveiling crucial nodes in complex biological networks. Sci. Rep. 2017, 7, 44797. [Google Scholar] [CrossRef] [PubMed]
- Paci, P.; Fiscon, G. SWIMmeR: An R-based software to unveiling crucial nodes in complex biological networks. Bioinformatics 2021, 38, 586–588. [Google Scholar] [CrossRef] [PubMed]
- Falcone, R.; Conte, F.; Fiscon, G.; Pecce, V.; Sponziello, M.; Durante, C.; Farina, L.; Filetti, S.; Paci, P.; Verrienti, A. BRAFV600E-mutant cancers display a variety of networks by SWIM analysis: Prediction of vemurafenib clinical response. Endocrine 2019, 64, 406–413. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Licursi, V.; Nasi, S.; Paci, P. Computational identification of specific genes for glioblastoma stem-like cells identity. Sci. Rep. 2018, 8, 7769. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Paci, P. SWIM tool application to expression data of glioblastoma stem-like cell lines, corresponding primary tumors and conventional glioma cell lines. BMC Bioinform. 2018, 19, 436. [Google Scholar] [CrossRef]
- Paci, P.; Fiscon, G.; Conte, F.; Licursi, V.; Morrow, J.; Hersh, C.; Cho, M.; Castaldi, P.; Glass, K.; Silverman, E.K.; et al. Integrated transcriptomic correlation network analysis identifies COPD molecular determinants. Sci. Rep. 2020, 10, 3361. [Google Scholar] [CrossRef]
- Paci, P.; Fiscon, G.; Conte, F.; Wang, R.-S.; Farina, L.; Loscalzo, J. Gene co-expression in the interactome: Moving from correlation toward causation via an integrated approach to disease module discovery. NPJ Syst. Biol. Appl. 2021, 7, 3. [Google Scholar] [CrossRef]
- Palumbo, M.C.; Zenoni, S.; Fasoli, M.; Massonnet, M.; Farina, L.; Castiglione, F.; Pezzotti, M.; Paci, P. Integrated Network Analysis Identifies Fight-Club Nodes as a Class of Hubs Encompassing Key Putative Switch Genes That Induce Major Transcriptome Repro-gramming during Grapevine Development. Plant Cell. 2015, 26, 4617–4635. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., III; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Rusan, M.; Li, K.; Li, Y.; Christensen, C.L.; Abraham, B.J.; Kwiatkowski, N.; Buczkowski, K.A.; Bockorny, B.; Chen, T.; Li, S.; et al. Suppression of Adaptive Responses to Targeted Cancer Therapy by Transcriptional Repression. Cancer Discov. 2018, 8, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Theodosakis, N.; Held, M.A.; Marzuka-Alcala, A.; Meeth, K.M.; Micevic, G.; Long, G.V.; Scolyer, R.A.; Stern, D.F.; Bosenberg, M.W. BRAF Inhibition Decreases Cellular Glucose Uptake in Melanoma in Association with Reduction in Cell Volume. Mol. Cancer Ther. 2015, 14, 1680–1692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, R.; Li, B.; Xin, Z.; Ling, Z.; Zhu, W.; Li, X.; Zhang, P.; Fu, Y.; Chen, J.; et al. LncRNA NEAT1 controls the lineage fates of BMSCs during skeletal aging by impairing mitochondrial function and pluripotency maintenance. Cell Death Differ. 2021, 29, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhang, D. Down-regulated expression of miR-582 predicts poor prognosis and facilitates melanoma progression by targeting FOXC1. Arch. Dermatol. Res. 2021, 314, 1–8. [Google Scholar] [CrossRef]
- Tseng, H.-W.; Li, S.-C.; Tsai, K.-W. Metformin Treatment Suppresses Melanoma Cell Growth and Motility through Modulation of microRNA Expression. Cancers 2019, 11, 209. [Google Scholar] [CrossRef] [PubMed]
- Tittarelli, A.; Navarrete, M.; Lizana, M.; Hofmann-Vega, F.; Salazar-Onfray, F. Hypoxic Melanoma Cells Deliver microRNAs to Dendritic Cells and Cytotoxic T Lymphocytes through Connexin-43 Channels. Int. J. Mol. Sci. 2020, 21, 7567. [Google Scholar] [CrossRef]
- Latchana, N.; DiVincenzo, M.J.; Regan, K.; Abrams, Z.; Zhang, X.; Jacob, N.K.; Gru, A.A.; Fadda, P.; Markowitz, J.; Howard, J.H.; et al. Alterations in patient plasma microRNA expression profiles following resection of metastatic melanoma. J. Surg. Oncol. 2018, 118, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Marranci, A.; D’Aurizio, R.; Vencken, S.; Mero, S.; Guzzolino, E.; Rizzo, M.; Pitto, L.; Pellegrini, M.; Chiorino, G.; Greene, C.M.; et al. Systematic evaluation of the microRNAome through miR-CATCHv2.0 identifies positive and negative regulators of BRAF-X1 mRNA. RNA Biol. 2019, 16, 865–878. [Google Scholar] [CrossRef]
- Lunavat, T.R.; Cheng, L.; Einarsdottir, B.O.; Bagge, R.O.; Muralidharan, S.V.; Sharples, R.A.; Lässer, C.; Gho, Y.S.; Hill, A.F.; Nilsson, J.A.; et al. BRAFV600 inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc. Natl. Acad. Sci. USA 2017, 114, E5930–E5939. [Google Scholar] [CrossRef] [PubMed]
- Dror, S.; Sander, L.; Schwartz, H.; Sheinboim, D.; Barzilai, A.; Dishon, Y.; Apcher, S.; Golan, T.; Greenberger, S.; Barshack, I.; et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nature 2016, 18, 1006–1017. [Google Scholar] [CrossRef]
- Sahoo, A.; Sahoo, S.K.; Joshi, P.; Lee, B.; Perera, R.J. MicroRNA-211 Loss Promotes Metabolic Vulnerability and BRAF Inhibitor Sensitivity in Melanoma. J. Investig. Dermatol. 2019, 139, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Sahoo, A.; Sawada, J.; Marchica, J.; Sahoo, S.; Layng, F.I.; Finlay, D.; Mazar, J.; Joshi, P.; Komatsu, M.; et al. MicroRNA-211 Modulates the DUSP6-ERK5 Signaling Axis to Promote BRAFV600E-Driven Melanoma Growth In Vivo and BRAF/MEK Inhibitor Resistance. J. Investig. Dermatol. 2020, 141, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Chen, S.; Qu, L.; Wang, Y.; Chen, H.; He, C. Identification of a five-miRNA signature predicting survival in cutaneous melanoma cancer patients. Peer J. 2019, 7, e7831. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Rao, C.; Li, H.; Chen, Y.; Fan, L.; Geng, H.; Li, S.; Qu, J.; Hou, L. Regulation of pigmentation by microRNAs: MITF-dependent microRNA-211 targets TGF- β receptor 2. Pigment Cell Melanoma Res. 2015, 28, 217–222. [Google Scholar] [CrossRef] [PubMed]
- el Hajj, P.; Journe, F.; Wiedig, M.; Laios, I.; Salès, F.; Galibert, M.-D.; Van Kempen, L.C.; Spatz, A.; Badran, B.; Larsimont, D.; et al. Tyrosinase-related protein 1 mRNA expression in lymph node metastases predicts overall survival in high-risk melanoma patients. Br. J. Cancer 2013, 108, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- el Hajj, P.; Gilot, D.; Migault, M.; Theunis, A.; van Kempen, L.C.; Salés, F.; Fayyad-Kazan, H.; Badran, B.; Larsimont, D.; Awada, A.; et al. SNPs at miR-155 binding sites of TYRP1 explain discrepancy between mRNA and protein and refine TYRP1 prognostic value in melanoma. Br. J. Cancer 2015, 113, 91–98. [Google Scholar] [CrossRef]
- Bueschbell, B.; Manga, P.; Schiedel, A.C. The Many Faces of G Protein-Coupled Receptor 143, an Atypical Intracellular Receptor. Front. Mol. Biosci. 2022, 9, 873777. [Google Scholar] [CrossRef]
- Falletta, P.; Bagnato, P.; Bono, M.; Monticone, M.; Schiaffino, M.V.; Bennett, D.C.; Goding, C.R.; Tacchetti, C.; Valetti, C. Melanosome-autonomous regulation of size and number: The OA1 receptor sustains PMEL expression. Pigment Cell Melanoma Res. 2014, 27, 565–579. [Google Scholar] [CrossRef]
- Bai, J.; Xie, X.; Lei, Y.; An, G.; He, L.; Lv, X. Ocular albinism type 1-induced melanoma cell migration is mediated through the RAS/RAF/MEK/ERK signaling pathway. Mol. Med. Rep. 2014, 10, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Ostojić, J.; Yoon, Y.-S.; Sonntag, T.; Nguyen, B.; Vaughan, J.M.; Shokhirev, M.; Montminy, M. Transcriptional co-activator regulates melanocyte differentiation and oncogenesis by integrating cAMP and MAPK/ERK pathways. Cell Rep. 2021, 35, 109136. [Google Scholar] [CrossRef] [PubMed]
- Gautron, A.; Migault, M.; Bachelot, L.; Corre, S.; Galibert, M.; Gilot, D. Human TYRP1: Two functions for a single gene. Pigment Cell Melanoma Res. 2021, 34, 836–852. [Google Scholar] [CrossRef]
- Gilot, D.; Migault, M.; Bachelot, L.; Journé, F.; Rogiers, A.; Donnou-Fournet, E.; Mogha, A.; Mouchet, N.; Pinel-Marie, M.-L.; Mari, B.; et al. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat. Cell Biol. 2017, 19, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Ohbayashi, N.; Fukuda, M.; Kanaho, Y. Rab32 subfamily small GTPases: Pleiotropic Rabs in endosomal trafficking. J. Biochem. 2017, 162, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Wasmeier, C.; Romao, M.; Plowright, L.; Bennett, D.; Raposo, G.; Seabra, M.C. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol. 2006, 175, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M. Rab GTPases: Key players in melanosome biogenesis, transport, and transfer. Pigment Cell Melanoma Res. 2020, 34, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Kuroda, T.S.; Mikoshiba, K. Slac2-a/Melanophilin, the Missing Link between Rab27 and Myosin Va. J. Biol. Chem. 2002, 277, 12432–12436. [Google Scholar] [CrossRef]
- Moreiras, H.; Seabra, M.; Barral, D. Melanin Transfer in the Epidermis: The Pursuit of Skin Pigmentation Control Mechanisms. Int. J. Mol. Sci. 2021, 22, 4466. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fang, R.; Fang, J.; He, S.; Liu, T. Functional implications of Rab27 GTPases in Cancer. Cell Commun. Signal. 2018, 16, 1–8. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.G.; Valencia, J.C.; Lai, B.; Zhang, G.; Paterson, J.K.; Rouzaud, F.; Berens, W.; Wincovitch, S.M.; Garfield, S.H.; Leapman, R.D.; et al. Melanosomal sequestration of cytotoxic drugs contributes to the intractability of malignant melanomas. Proc. Natl. Acad. Sci. USA 2006, 103, 9903–9907. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Del-Campo, L.; Montenegro, M.F.; Cabezas-Herrera, J.; Rodríguez-López, J.N. The critical role of alpha-folate receptor in the resistance of melanoma to methotrexate. Pigment Cell Melanoma Res. 2009, 22, 588–600. [Google Scholar] [CrossRef]
- Makino-Okamura, C.; Niki, Y.; Takeuchi, S.; Nishigori, C.; Declercq, L.; Yaroch, D.B.; Saito, N. Heparin inhibits melanosome uptake and inflammatory response coupled with phagocytosis through blocking PI3k/Akt and MEK/ERK signaling pathways in human epidermal keratinocytes. Pigment Cell Melanoma Res. 2014, 27, 1063–1074. [Google Scholar] [CrossRef]
- Kim, B.; Lee, J.-Y.; Lee, H.-Y.; Nam, K.-Y.; Park, J.; Lee, S.M.; Kim, J.E.; Lee, J.D.; Hwang, J.S. Hesperidin Suppresses Melanosome Transport by Blocking the In-teraction of Rab27A-Melanophilin. Biomol. Ther. 2013, 21, 343–348. [Google Scholar] [CrossRef]
- Qian, W.; Liu, W.; Zhu, D.; Cao, Y.; Tang, A.; Gong, G.; Su, H. Natural skin-whitening compounds for the treatment of melanogenesis (Review). Exp. Ther. Med. 2020, 20, 173–185. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, J.; Liu, J.; Zhang, G.; Lu, A. Advances in the discovery of exosome inhibitors in cancer. J. Enzym. Inhib. Med. Chem. 2020, 35, 1322–1330. [Google Scholar] [CrossRef]

| Type | Gene | Sense (5′-3′) | Antisense (5′-3′) | PMID |
|---|---|---|---|---|
| qRT-PCR primers | ATPA1 | CTCAGATGTGTCCAAGCAAG | GTCAGTGCCCAAGTCAATG | 28445987 |
| DCT | CCTTTCTTCCCTCCAGTGAC | AGCCAACAGCACAAAAAGAC | 28445987 | |
| GAPDH | CGCTCTCTGCTCCTCCTGTT | CCATGGTGTCTGAGCGATGT | 28445987 | |
| MITF | TGACCGCATTAAAGAACTAGG | GTGCTCCAGTTTCTTCTGTCG | 28445987 | |
| MLANA | CTCTTACACCACGGCTGAA | AGACTCCCAGGATCACT | 28445987 | |
| PBGD | TCCAAGCGGAGCCATGTCTG | AGAATCTTGTCCCCTGTGGTGGA | 28445987 | |
| PGC1alpha | GTCACCACCCAAATCCTTAT | CGGTGTCTGTAGTGGCTTGA | 28445987 | |
| SDHA | CCACTCGCTATTGCACACC | CACTCCCCATTCTCCATCA | 28445987 | |
| TRPM1 | TGCGAAGGCTGCTGGAAA | CAAGACGATGGACACCACGTTAGG | 28445987 | |
| TYR | GATGAGTACATGGGAGGTCAGC | GTACTCCTCCAATCGGCTACAG | 28445987 | |
| TYRP1 | GGACCAGCTTTTCTCACAT | GAATCAAAGTTGCTTCTGGA | 28445987 | |
| LNAs | LNA-CT | GTGTAACACGTCTATACGCCCA | 28445987 | |
| LNA-211 | AGGCGAAGGATGACAAAGGGAA | 28445987 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitiello, M.; Mercatanti, A.; Podda, M.S.; Baldanzi, C.; Prantera, A.; Sarti, S.; Rizzo, M.; Salvetti, A.; Conte, F.; Fiscon, G.; et al. A Network of MicroRNAs and mRNAs Involved in Melanosome Maturation and Trafficking Defines the Lower Response of Pigmentable Melanoma Cells to Targeted Therapy. Cancers 2023, 15, 894. https://doi.org/10.3390/cancers15030894
Vitiello M, Mercatanti A, Podda MS, Baldanzi C, Prantera A, Sarti S, Rizzo M, Salvetti A, Conte F, Fiscon G, et al. A Network of MicroRNAs and mRNAs Involved in Melanosome Maturation and Trafficking Defines the Lower Response of Pigmentable Melanoma Cells to Targeted Therapy. Cancers. 2023; 15(3):894. https://doi.org/10.3390/cancers15030894
Chicago/Turabian StyleVitiello, Marianna, Alberto Mercatanti, Maurizio Salvatore Podda, Caterina Baldanzi, Antonella Prantera, Samanta Sarti, Milena Rizzo, Alessandra Salvetti, Federica Conte, Giulia Fiscon, and et al. 2023. "A Network of MicroRNAs and mRNAs Involved in Melanosome Maturation and Trafficking Defines the Lower Response of Pigmentable Melanoma Cells to Targeted Therapy" Cancers 15, no. 3: 894. https://doi.org/10.3390/cancers15030894
APA StyleVitiello, M., Mercatanti, A., Podda, M. S., Baldanzi, C., Prantera, A., Sarti, S., Rizzo, M., Salvetti, A., Conte, F., Fiscon, G., Paci, P., & Poliseno, L. (2023). A Network of MicroRNAs and mRNAs Involved in Melanosome Maturation and Trafficking Defines the Lower Response of Pigmentable Melanoma Cells to Targeted Therapy. Cancers, 15(3), 894. https://doi.org/10.3390/cancers15030894








