Deep Immune Profiling of Multiple Myeloma at Diagnosis and under Lenalidomide Maintenance Therapy
Abstract
Simple Summary
Abstract
1. Introduction
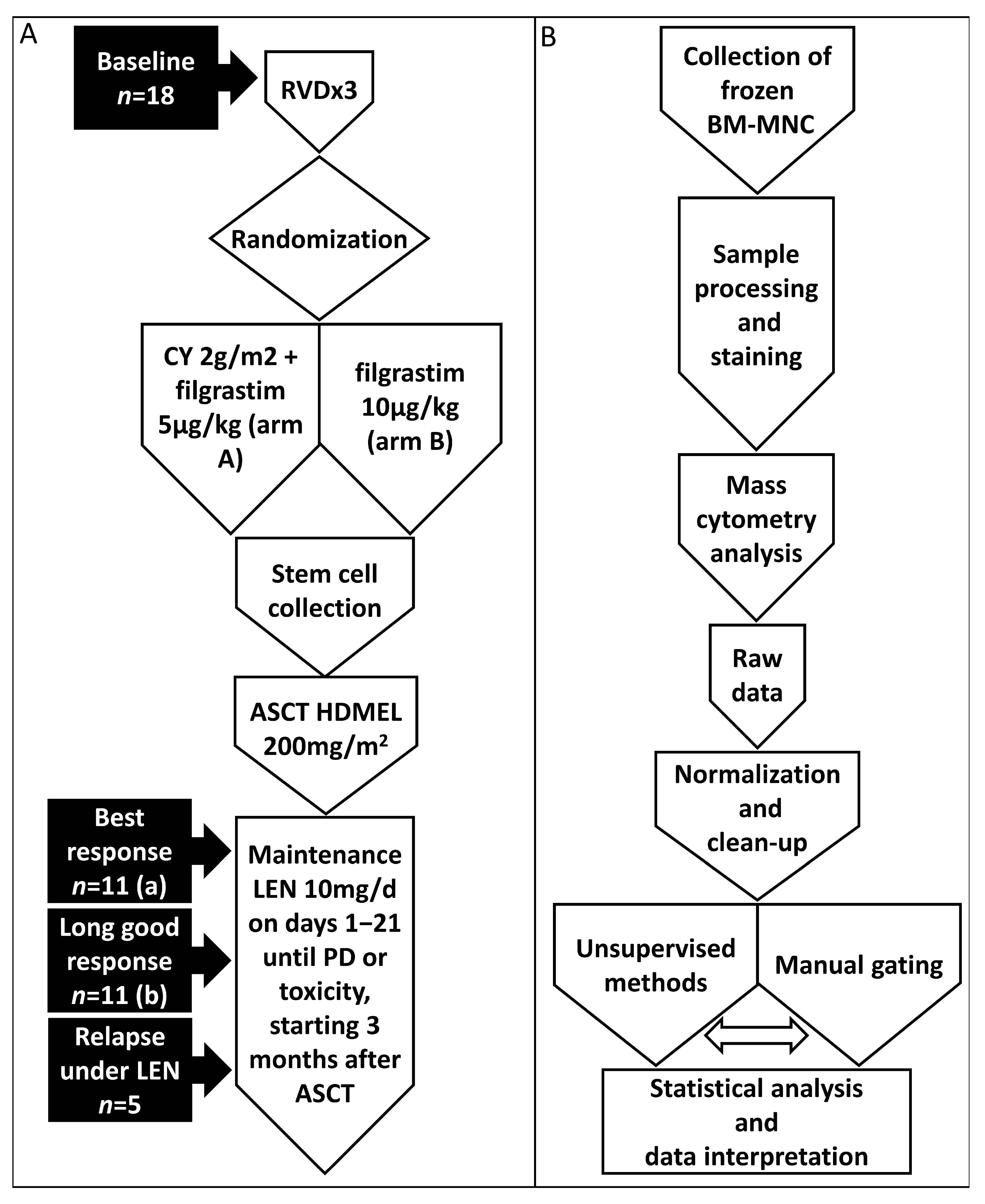
2. Materials and Methods
2.1. Patients and Samples
2.2. Cell Staining for CyTOF Analysis
2.3. Sample Acquisition and Data Normalization
2.4. Computational Analysis
2.5. Statistical Analysis
3. Results
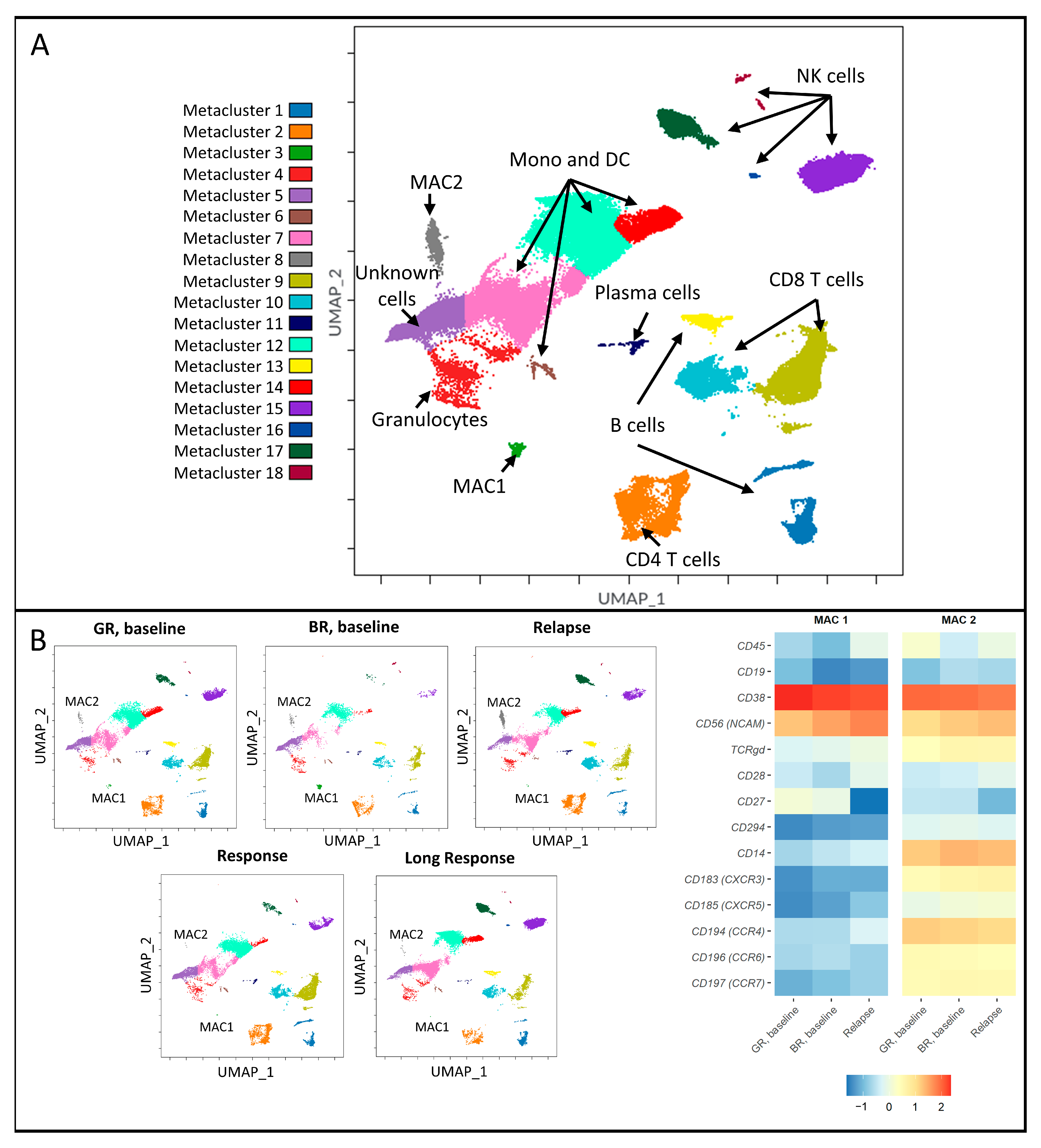
3.1. Distinct Immune Signatures Were Identified in Good and Bad Responders at Baseline
3.2. Malignancy-Associated Cell Populations in the Bad-Response Group
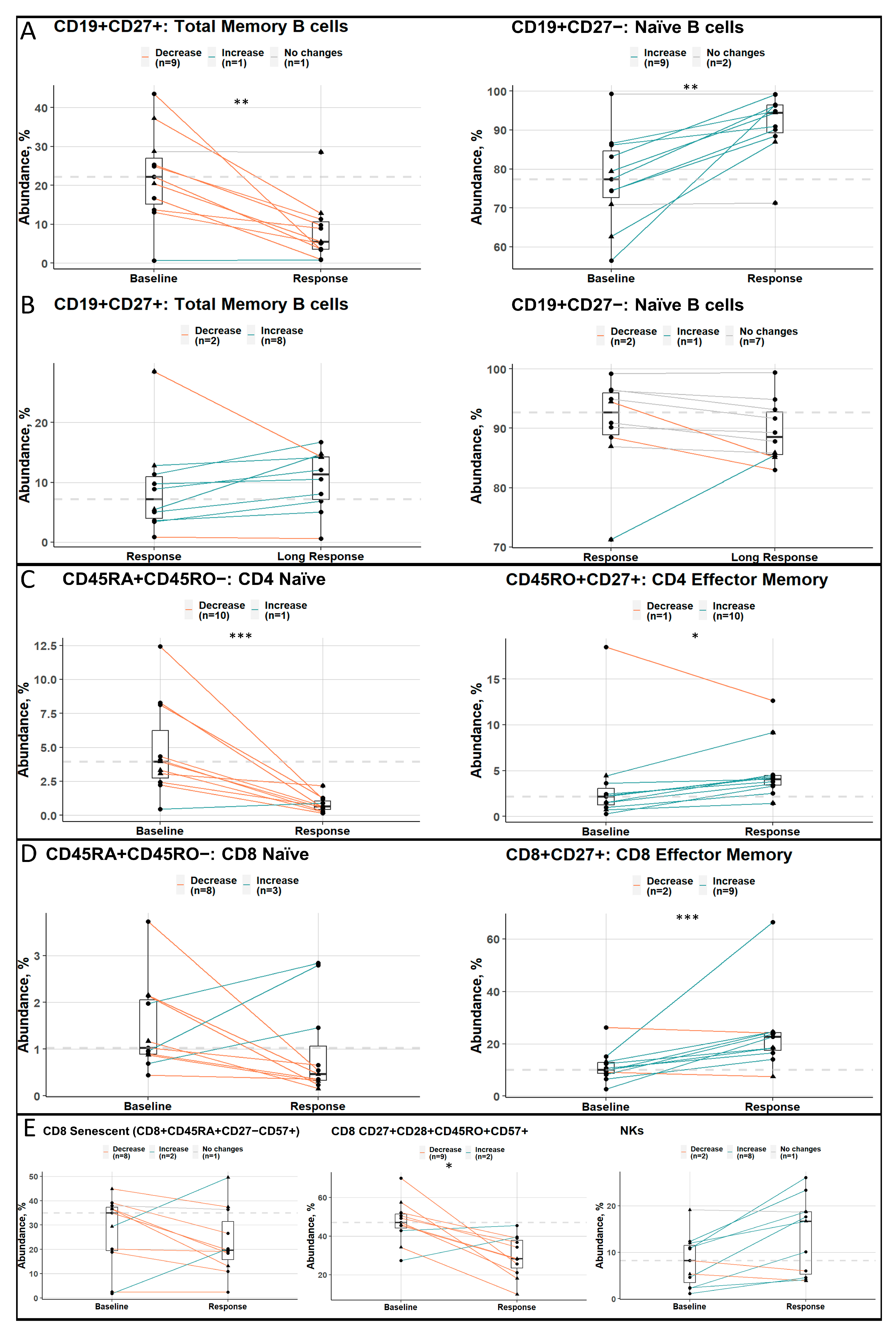
3.3. Good Responders Have More Cells with an Immunoreactive Phenotype
3.4. Longitudinal Analysis of Good Responders at Different Stages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lomas, O.C.; Tahri, S.; Ghobrial, I.M. The microenvironment in myeloma. Curr. Opin. Oncol. 2020, 32, 170–175. Available online: https://journals.lww.com/10.1097/CCO.0000000000000615 (accessed on 3 March 2020). [CrossRef]
- Ghobrial, I.M.; Detappe, A.; Anderson, K.C.; Steensma, D.P. The bone-marrow niche in MDS and MGUS: Implications for AML and MM. Nat. Rev. Clin. Oncol. 2018, 15, 219–233. Available online: http://www.nature.com/articles/nrclinonc.2017.197 (accessed on 9 April 2018). [CrossRef] [PubMed]
- Zavidij, O.; Haradhvala, N.J.; Mouhieddine, T.H.; Sklavenitis-Pistofidis, R.; Cai, S.; Reidy, M.; Rahmat, M.; Flaifel, A.; Ferland, B.; Su, N.K.; et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nat. Cancer 2020, 1, 493–506. Available online: http://www.nature.com/articles/s43018-020-0053-3 (accessed on 27 May 2020). [CrossRef] [PubMed]
- De Jong, M.M.E.; Kellermayer, Z.; Papazian, N.; Tahri, S.; Hofste op Bruinink, D.; Hoogenboezem, R.; Sanders, M.A.; van de Woestijne, P.C.; Bos, P.K.; Khandanpour, C.; et al. The multiple myeloma microenvironment is defined by an inflammatory stromal cell landscape. Nat. Immunol. 2021, 22, 769–780. Available online: http://www.nature.com/articles/s41590-021-00931-3 (accessed on 20 June 2021). [CrossRef]
- Coulson-Thomas, V.J.; Coulson-Thomas, Y.M.; Gesteira, T.F.; Kao, W.W.-Y. Extrinsic and Intrinsic Mechanisms by Which Mesenchymal Stem Cells Suppress the Immune System. Ocul. Surf. 2016, 14, 121–134. Available online: https://linkinghub.elsevier.com/retrieve/pii/S1542012415002074 (accessed on 11 April 2016). [CrossRef] [PubMed]
- Görgün, G.T.; Whitehill, G.; Anderson, J.L.; Hideshima, T.; Maguire, C.; Laubach, J.; Raje, N.; Munshi, N.C.; Richardson, P.G.; Anderson, K.C. Tumor-promoting immune-suppressive myeloid-derived suppressor cells in the multiple myeloma microenvironment in humans. Blood 2013, 121, 2975–2987. Available online: https://ashpublications.org/blood/article/121/15/2975/31384/Tumorpromoting-immunesuppressive-myeloidderived (accessed on 11 April 2013). [CrossRef] [PubMed]
- Prabhala, R.H.; Neri, P.; Bae, J.E.; Tassone, P.; Shammas, M.A.; Allam, C.K.; Daley, J.F.; Chauhan, D.; Blanchard, E.; Thatte, H.S.; et al. Dysfunctional T regulatory cells in multiple myeloma. Blood 2006, 107, 301–304. Available online: https://ashpublications.org/blood/article/107/1/301/21738/Dysfunctional-T-regulatory-cells-in-multiple (accessed on 1 January 2006). [CrossRef] [PubMed]
- Chauhan, D.; Singh, A.V.; Brahmandam, M.; Carrasco, R.; Bandi, M.; Hideshima, T.; Bianchi, G.; Podar, K.; Tai, Y.-T.; Mitsiades, C.; et al. Functional Interaction of Plasmacytoid Dendritic Cells with Multiple Myeloma Cells: A Therapeutic Target. Cancer Cell 2009, 16, 309–323. Available online: https://linkinghub.elsevier.com/retrieve/pii/S153561080900292X (accessed on 6 October 2009). [CrossRef] [PubMed]
- Zelle-Rieser, C.; Thangavadivel, S.; Biedermann, R.; Brunner, A.; Stoitzner, P.; Willenbacher, E.; Greil, R.; Jöhrer, K. T cells in multiple myeloma display features of exhaustion and senescence at the tumor site. J. Hematol. Oncol. 2016, 9, 116. Available online: http://jhoonline.biomedcentral.com/articles/10.1186/s13045-016-0345-3 (accessed on 3 December 2016). [CrossRef] [PubMed]
- Paiva, B.; Cedena, M.-T.; Puig, N.; Arana, P.; Vidriales, M.-B.; Cordon, L.; Flores-Montero, J.; Gutierrez, N.C.; Martín-Ramos, M.-L.; Martinez-Lopez, J.; et al. Minimal residual disease monitoring and immune profiling in multiple myeloma in elderly patients. Blood 2016, 127, 3165–3174. Available online: https://ashpublications.org/blood/article/127/25/3165/35183/Minimal-residual-disease-monitoring-and-immune (accessed on 23 June 2016). [CrossRef]
- Paiva, B.; Vídriales, M.-B.; Rosiñol, L.; Martínez-López, J.; Mateos, M.-V.; Ocio, E.M.; Montalbán, M.-Á.; Cordón, L.; Gutiérrez, N.C.; Corchete, L.; et al. A multiparameter flow cytometry immunophenotypic algorithm for the identification of newly diagnosed symptomatic myeloma with an MGUS-like signature and long-term disease control. Leukemia 2013, 27, 2056–2061. Available online: http://www.nature.com/articles/leu2013166 (accessed on 7 October 2013). [CrossRef] [PubMed]
- De Magalhães, R.J.P.; Vidriales, M.-B.; Paiva, B.; Fernandez-Gimenez, C.; García-Sanz, R.; Mateos, M.-V.; Gutierrez, N.C.; Lecrevisse, Q.; Blanco, J.F.; Hernández, J.; et al. Analysis of the immune system of multiple myeloma patients achieving long-term disease control by multidimensional flow cytometry. Haematologica 2013, 98, 79–86. Available online: http://www.haematologica.org/cgi/doi/10.3324/haematol.2012.067272 (accessed on 1 January 2013). [CrossRef] [PubMed]
- Österborg, A.; Nilsson, B.; Björkholm, M.; Holm, G.; Mellstedt, H. Natural killer cell activity in monoclonal gammopathies: Relation to disease activity. Eur. J. Haematol. 2009, 45, 153–157. Available online: https://onlinelibrary.wiley.com/doi/10.1111/j.1600-0609.1990.tb00443.x (accessed on 24 April 2009). [CrossRef] [PubMed]
- Yang, C.; Siebert, J.R.; Burns, R.; Gerbec, Z.J.; Bonacci, B.; Rymaszewski, A.; Rau, M.; Riese, M.J.; Rao, S.; Carlson, K.-S.; et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat. Commun. 2019, 10, 3931. Available online: http://www.nature.com/articles/s41467-019-11947-7 (accessed on 2 December 2019). [CrossRef]
- Jurisic, V.; Srdic, T.; Konjevic, G.; Markovic, O.; Colovic, M. Clinical stage-depending decrease of NK cell activity in multiple myeloma patients. Med. Oncol. 2007, 24, 312–317. Available online: http://link.springer.com/10.1007/s12032-007-0007-y (accessed on 22 August 2007). [CrossRef]
- Bryant, C.; Suen, H.; Brown, R.; Yang, S.; Favaloro, J.; Aklilu, E.; Gibson, J.; Ho, P.J.; Iland, H.; Fromm, P.; et al. Long-term survival in multiple myeloma is associated with a distinct immunological profile, which includes proliferative cytotoxic T-cell clones and a favourable Treg/Th17 balance. Blood Cancer J. 2013, 3, e148. Available online: http://www.nature.com/articles/bcj201334 (accessed on 13 September 2013). [CrossRef]
- Brown, R.D.; Spencer, A.; Ho, P.J.; Kennedy, N.; Kabani, K.; Yang, S.; Sze, D.M.; Aklilu, E.; Gibson, J.; Joshua, D.E. Prognostically significant cytotoxic T cell clones are stimulated after thalidomide therapy in patients with multiple myeloma. Leuk. Lymphoma 2009, 50, 1860–1864. Available online: http://www.tandfonline.com/doi/full/10.3109/10428190903216804 (accessed on 3 November 2009). [CrossRef]
- Pierceall, W.E.; Amatangelo, M.D.; Bahlis, N.J.; Siegel, D.S.; Rahman, A.; Van Oekelen, O.; Neri, P.; Young, M.; Chung, W.; Serbina, N.; et al. Immunomodulation in Pomalidomide, Dexamethasone, and Daratumumab-Treated Patients with Relapsed/Refractory Multiple Myeloma. Clin. Cancer Res. 2020, 26, 5895–5902. Available online: http://clincancerres.aacrjournals.org/lookup/doi/10.1158/1078-0432.CCR-20-1781 (accessed on 15 November 2020). [CrossRef]
- Kalff, A.; Khong, T.; Ramachandran, M.; Walker, P.; Schwarer, A.; Roberts, A.W.; Campbell, P.; Filshie, R.; Norton, S.; Reynolds, J.; et al. Cereblon pathway biomarkers and immune profiles in patients with myeloma receiving post-ASCT lenalidomide maintenance (LEOPARD). Leuk. Lymphoma 2021, 62, 2981–2991. Available online: https://www.tandfonline.com/doi/full/10.1080/10428194.2021.1948030 (accessed on 15 October 2021). [CrossRef]
- Parmar, H.; Gertz, M.; Anderson, E.I.; Kumar, S.; Kourelis, T.V. Microenvironment immune reconstitution patterns correlate with outcomes after autologous transplant in multiple myeloma. Blood Adv. 2021, 5, 1797–1804. Available online: https://ashpublications.org/bloodadvances/article/5/7/1797/475594/Microenvironment-immune-reconstitution-patterns (accessed on 13 April 2021). [CrossRef]
- Krönke, J.; Udeshi, N.D.; Narla, A.; Grauman, P.; Hurst, S.N.; McConkey, M.; Svinkina, T.; Heckl, D.; Comer, E.; Li, X.; et al. Lenalidomide Causes Selective Degradation of IKZF1 and IKZF3 in Multiple Myeloma Cells. Science 2014, 343, 301–305. Available online: https://www.science.org/doi/10.1126/science.1244851 (accessed on 17 January 2014). [CrossRef] [PubMed]
- Wu, L.; Parton, A.; Lu, L.; Adams, M.; Schafer, P.; Bartlett, J.B. Lenalidomide enhances antibody-dependent cellular cytotoxicity of solid tumor cells in vitro: Influence of host immune and tumor markers. Cancer Immunol. Immunother. 2011, 60, 61–73. Available online: http://link.springer.com/10.1007/s00262-010-0919-9 (accessed on 17 January 2011). [CrossRef] [PubMed]
- Davies, F.; Raje, N.; Hideshima, T.; Lentzsch, S.; Young, G.; Tai, Y.-T.; Lin, B.; Podar, K.; Gupta, D.; Chauhan, D.; et al. Thalidomide and immunomodulatory derivatives augment natural killer cell cytotoxicity in multiple myeloma. Blood 2001, 98, 210–216. Available online: https://ashpublications.org/blood/article/98/1/210/88908/Thalidomide-and-immunomodulatory-derivatives (accessed on 1 July 2001). [CrossRef] [PubMed]
- Silvennoinen, R.; Anttila, P.; Säily, M.; Lundan, T.; Heiskanen, J.; Siitonen, T.M.; Kakko, S.; Putkonen, M.; Ollikainen, H.; Terävä, V.; et al. A randomized phase II study of stem cell mobilization with cyclophosphamide + G-CSF or G-CSF alone after lenalidomide-based induction in multiple myeloma. Bone Marrow Transplant. 2016, 51, 372–376. Available online: http://www.nature.com/articles/bmt2015236 (accessed on 5 March 2016). [CrossRef]
- Luoma, S.; Anttila, P.; Säily, M.; Lundan, T.; Heiskanen, J.; Siitonen, T.; Kakko, S.; Putkonen, M.; Ollikainen, H.; Terävä, V.; et al. RVD induction and autologous stem cell transplantation followed by lenalidomide maintenance in newly diagnosed multiple myeloma: A phase 2 study of the Finnish Myeloma Group. Ann. Hematol. 2019, 98, 2781–2792. Available online: http://link.springer.com/10.1007/s00277-019-03815-7 (accessed on 31 December 2019). [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. Available online: http://arxiv.org/abs/1802.03426 (accessed on 9 February 2018).
- Singh, A.K.; Tripathi, P.; Cardell, S.L. Type II NKT Cells: An Elusive Population with Immunoregulatory Properties. Front. Immunol. 2018, 9, 1969. Available online: http://www.ncbi.nlm.nih.gov/pubmed/30210505 (accessed on 28 August 2018). [CrossRef]
- Takata, H.; Takiguchi, M. Three memory subsets of human CD8+ T cells differently expressing three cytolytic effector molecules. J. Immunol. 2006, 177, 4330–4340. Available online: http://www.ncbi.nlm.nih.gov/pubmed/16982867 (accessed on 1 October 2006). [CrossRef]
- Lowther, D.E.; Chong, D.L.; Ascough, S.; Ettorre, A.; Ingram, R.J.; Boyton, R.J.; Altmann, D.M. Th1 not Th17 cells drive spontaneous MS-like disease despite a functional regulatory T cell response. Acta Neuropathol. 2013, 126, 501–515. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23934116 (accessed on 15 October 2013). [CrossRef]
- Barski, A.; Cuddapah, S.; Kartashov, A.V.; Liu, C.; Imamichi, H.; Yang, W.; Peng, W.; Lane, H.C.; Zhao, K. Rapid Recall Ability of Memory T Cells Is Encoded in Their Epigenome. Sci. Rep. 2017, 7, 39785. Available online: http://www.ncbi.nlm.nih.gov/pubmed/28054639 (accessed on 5 January 2017). [CrossRef]
- Pellom, S.T.; Dudimah, D.F.; Thounaojam, M.C.; Sayers, T.J.; Shanker, A. Modulatory effects of bortezomib on host immune cell functions. Immunotherapy 2015, 7, 1011–1022. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26325610 (accessed on 1 September 2015). [CrossRef] [PubMed]
- Olsen, L.R.; Leipold, M.D.; Pedersen, C.B.; Maecker, H.T. The anatomy of single cell mass cytometry data. Cytom. Part A 2019, 95, 156–172. Available online: https://onlinelibrary.wiley.com/doi/10.1002/cyto.a.23621 (accessed on 2 February 2019). [CrossRef] [PubMed]
- Bagwell, C.B.; Hunsberger, B.; Hill, B.; Herbert, D.; Bray, C.; Selvanantham, T.; Li, S.; Villasboas, J.C.; Pavelko, K.; Strausbauch, M.; et al. Multi-site reproducibility of a human immunophenotyping assay in whole blood and peripheral blood mononuclear cells preparations using CyTOF technology coupled with Maxpar Pathsetter, an automated data analysis system. Cytom. Part B Clin. Cytom. 2020, 98, 146–160. Available online: https://onlinelibrary.wiley.com/doi/10.1002/cyto.b.21858 (accessed on 20 March 2020). [CrossRef] [PubMed]

| Characteristics | Patients with Good Response | Patients with Bad Response | p |
|---|---|---|---|
| N (%) | 11 (61) | 7 (39) | |
| Age at treatment start, years, median (range) | 61.5 (52.9–67.2) | 61.0 (56.0–70.0) | 0.536 |
| Gender male/female, n (%) | 10/1 (91/9) | 5/2 (71/29) | 0.528 |
| Myeloma subtype, n (%) | 0.050 | ||
| IgG kappa | 6 (55) | 2 (29) | |
| IgG lambda | 4 (36) | 0 (0) | |
| IgA kappa | 1 (9) | 2 (29) | |
| IgA lambda | 0 (0) | 1 (14) | |
| Kappa light chain | 0 (0) | 1 (14) | |
| Lambda light chain | 0 (0) | 1 (14) | |
| R-ISS stage, n (%) | 0.067 | ||
| I | 5 (45) | 0 (0) | |
| II | 6 (55) | 6 (86) | |
| III | 0 (0) | 1 (14) | |
| IMWG risk, n (%) | 0.748 | ||
| Low | 2 (18) | 0 (0) | |
| Standard | 8 (73) | 6 (86) | |
| High | 1 (9) | 1 (14) | |
| Cytogenetic risk, n (%) | 0.145 | ||
| Standard | 8 (73) | 2 (29) | |
| High(a) | 3 (27) | 5 (71) | |
| Median duration of lenalidomide maintenance, months, (range) | 72.9 (43.7–90.2) | 9.9 (1.5–33.9) | <0.001 |
| PFS, median, months, (range) | NR (74.2–96.8) | 17.7 (8.7–57.5) | <0.001 |
| OS, median, months, (range) | NR (76.1–96.8) | 34.5 (12.5–86.9) | <0.001 |
| MRD-negative at response, n (%) | 1.000 | ||
| Yes | 5 (50) | 1 (100) | |
| No | 1 (10) | 0 (0) | |
| Unknown | 4 (40) | 0 (0) | |
| MRD-negative at long response(b), n (%) | |||
| Yes | 6 (55) | ||
| No | 4 (36) | ||
| Unknown | 1 (9) | ||
| Sustained MRD-negative (c), n (%) | <0.001 | ||
| Yes | 7 (64) | 0 (0) | |
| No | 0 | 6 (86) | |
| Unknown | 4 (36) | 1 (14) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luoma, S.; Sergeev, P.; Javarappa, K.K.; Öhman, T.J.; Varjosalo, M.; Säily, M.; Anttila, P.; Sankelo, M.; Partanen, A.; Nihtinen, A.; et al. Deep Immune Profiling of Multiple Myeloma at Diagnosis and under Lenalidomide Maintenance Therapy. Cancers 2023, 15, 2604. https://doi.org/10.3390/cancers15092604
Luoma S, Sergeev P, Javarappa KK, Öhman TJ, Varjosalo M, Säily M, Anttila P, Sankelo M, Partanen A, Nihtinen A, et al. Deep Immune Profiling of Multiple Myeloma at Diagnosis and under Lenalidomide Maintenance Therapy. Cancers. 2023; 15(9):2604. https://doi.org/10.3390/cancers15092604
Chicago/Turabian StyleLuoma, Sini, Philipp Sergeev, Komal Kumar Javarappa, Tiina J. Öhman, Markku Varjosalo, Marjaana Säily, Pekka Anttila, Marja Sankelo, Anu Partanen, Anne Nihtinen, and et al. 2023. "Deep Immune Profiling of Multiple Myeloma at Diagnosis and under Lenalidomide Maintenance Therapy" Cancers 15, no. 9: 2604. https://doi.org/10.3390/cancers15092604
APA StyleLuoma, S., Sergeev, P., Javarappa, K. K., Öhman, T. J., Varjosalo, M., Säily, M., Anttila, P., Sankelo, M., Partanen, A., Nihtinen, A., Heckman, C. A., & Silvennoinen, R. (2023). Deep Immune Profiling of Multiple Myeloma at Diagnosis and under Lenalidomide Maintenance Therapy. Cancers, 15(9), 2604. https://doi.org/10.3390/cancers15092604







