Artificial Intelligence in CT and MR Imaging for Oncological Applications
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Highlights
- o
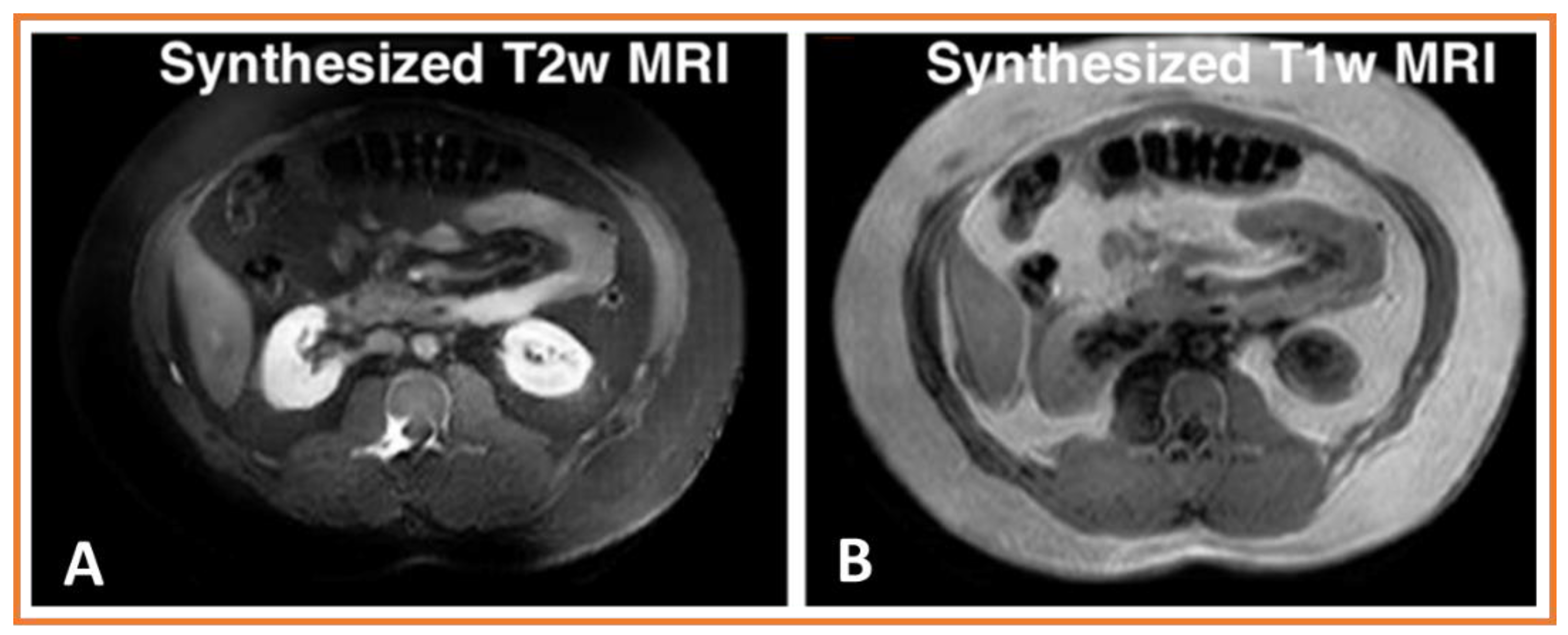
- Deep learning methods can be used to synthesize different contrast modality images for many purposes, including training networks for multi-modality segmentation, image harmonization, and missing modality synthesis.
- o
- AI-based auto-segmentation for discerning abdominal organs is presented here. Deep learning methods can leverage different modalities with more information (e.g., higher contrast from MRI or many experts segmented labeled datasets such as from CT) to improve tumor segmentation performance in a different modality without requiring paired image sets.
- o
- Deep learning reconstruction algorithms are illustrated with examples for both CT and MRI. Such approaches improve image quality, which aids in better tumor detection, segmentation, and monitoring of response.
- o
- It is emphasized that large quantities of data are requirements for AI development, and this has created opportunities for collaboration, open team science, and knowledge sharing.
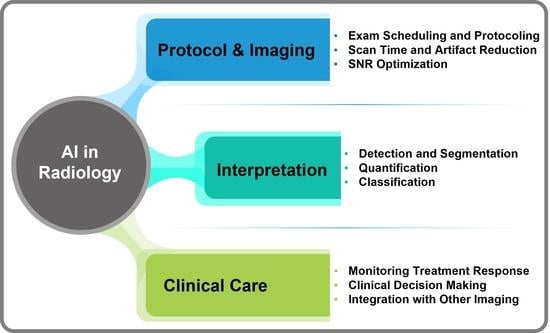
1.2. AI in CT and MRI for Oncological Imaging
2. Specific-Narrow Tasks Developed Using AI for Radiological Workflow
3. Major Challenges with Solutions for Radiological Image Analysis
3.1. Variability in Imaging Acquisition Pose Challenges for Large-Scale Radiomics Analysis Studies
3.2. Volumetric Segmentation of Tumor Volumes and Longitudinal Tracking of Tumor Volume Response
3.3. Optimization of Dose and Image Quality Improvement in CT Scans
3.4. Optimization of Image Quality in MRI Scans
3.5. Bias in AI Models
4. Discussion
5. Future Directions
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Histed, S.N.; Lindenberg, M.L.; Mena, E.; Turkbey, B.; Choyke, P.L.; Kurdziel, K.A. Review of functional/anatomical imaging in oncology. Nucl. Med. Commun. 2012, 33, 349–361. [Google Scholar] [CrossRef]
- Beaton, L.; Bandula, S.; Gaze, M.N.; Sharma, R.A. How rapid advances in imaging are defining the future of precision radiation oncology. Br. J. Cancer 2019, 120, 779–790. [Google Scholar] [CrossRef]
- Meyer, H.J.; Purz, S.; Sabri, O.; Surov, A. Relationships between histogram analysis of ADC values and complex 18F-FDG-PET parameters in head and neck squamous cell carcinoma. PLoS ONE 2018, 13, e0202897. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, K.S.; Ohno, Y.; Van Beek, E.J.; Biederer, J. PET/CT versus MRI for diagnosis, staging, and follow-up of lung cancer. J. Magn. Reason. Imaging 2015, 42, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.H.; Litière, S.; de Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1-Update and clarification: From the RECIST committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef]
- Tacher, V.; Lin, M.; Chao, M.; Gjesteby, L.; Bhagat, N.; Mahammedi, A.; Ardon, R.; Mory, B.; Geschwind, J.F. Semiautomatic volumetric tumor segmentation for hepatocellular carcinoma: Comparison between C-arm cone beam computed tomography and MRI. Acad. Radiol. 2013, 20, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Primakov, S.P.; Ibrahim, A.; van Timmeren, J.E.; Wu, G.; Keek, S.A.; Beuque, M.; Granzier, R.W.Y.; Lavrova, E.; Scrivener, M.; Sanduleanu, S.; et al. Automated detection and segmentation of non-small cell lung cancer computed tomography images. Nat. Commun. 2022, 13, 3423. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Thrall, J.H.; Li, X.; Li, Q.; Cruz, C.; Do, S.; Dreyer, K.; Brink, J. Artificial intelligence and machine learning in radiology: Opportunities, challenges, pitfalls, and criteria for success. J. Am. Coll. Radiol. 2018, 15, 504–508. [Google Scholar] [CrossRef]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef]
- Dercle, L.; McGale, J.; Sun, S.; Marabelle, A.; Yeh, R.; Deutsch, E.; Mokrane, F.Z.; Farwell, M.; Ammari, S.; Schoder, H.; et al. Artificial intelligence and radiomics: Fundamentals, applications, and challenges in immunotherapy. J. Immunother. Cancer 2022, 10, e005292. [Google Scholar] [CrossRef]
- Abdar, M.; Pourpanah, F.; Hussain, S.; Rezazadegan, D.; Liu, L.; Ghavamzadeh, M.; Fieguth, P.; Cao, X.; Khosravi, A.; Acharya, U.R. A review of uncertainty quantification in deep learning: Techniques, applications and challenges. Inf. Fusion 2021, 76, 243–297. [Google Scholar] [CrossRef]
- Diaz, O.; Kushibar, K.; Osuala, R.; Linardos, A.; Garrucho, L.; Igual, L.; Radeva, P.; Prior, F.; Gkontra, P.; Lekadir, K. Data preparation for artificial intelligence in medical imaging: A comprehensive guide to open-access platforms and tools. Phys. Med. 2021, 83, 25–37. [Google Scholar] [CrossRef]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef] [PubMed]
- Koh, D.-M.; Papanikolaou, N.; Bick, U.; Illing, R.; Kahn, C.E.; Kalpathi-Cramer, J.; Matos, C.; Martí-Bonmatí, L.; Miles, A.; Mun, S.K.; et al. Artificial intelligence and machine learning in cancer imaging. Commun. Med. 2022, 2, 133. [Google Scholar] [CrossRef]
- Abdel Razek, A.A.K.; Alksas, A.; Shehata, M.; AbdelKhalek, A.; Abdel Baky, K.; El-Baz, A.; Helmy, E. Clinical applications of artificial intelligence and radiomics in neuro-oncology imaging. Insights Imaging 2021, 12, 152. [Google Scholar] [CrossRef]
- Lin, M.; Wynne, J.F.; Zhou, B.; Wang, T.; Lei, Y.; Curran, W.J.; Liu, T.; Yang, X. Artificial intelligence in tumor subregion analysis based on medical imaging: A review. J. Appl. Clin. Med. Phys. 2021, 22, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Petry, M.; Lansky, C.; Chodakiewitz, Y.; Maya, M.; Pressman, B. Decreased Hospital Length of Stay for ICH and PE after Adoption of an Artificial Intelligence-Augmented Radiological Worklist Triage System. Radiol. Res. Pract. 2022, 2022, 2141839. [Google Scholar] [CrossRef] [PubMed]
- Recht, M.P.; Dewey, M.; Dreyer, K.; Langlotz, C.; Niessen, W.; Prainsack, B.; Smith, J.J. Integrating artificial intelligence into the clinical practice of radiology: Challenges and recommendations. Eur. Radiol. 2020, 30, 3576–3584. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef]
- Mahmood, U.; Shrestha, R.; Bates, D.D.B.; Mannelli, L.; Corrias, G.; Erdi, Y.E.; Kanan, C. Detecting Spurious Correlations with Sanity Tests for Artificial Intelligence Guided Radiology Systems. Front. Digit. Health 2021, 3, 671015. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Hu, Y.-C.; Tyagi, N.; Zhang, P.; Rimner, A.; Mageras, G.S.; Deasy, J.O.; Veeraraghavan, H. Tumor-Aware, Adversarial Domain Adaptation from CT to MRI for Lung Cancer Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2018, Cham, Switzerland, 26 September 2018; pp. 777–785. [Google Scholar]
- Wang, T.; Lei, Y.; Tian, Z.; Dong, X.; Liu, Y.; Jiang, X.; Curran, W.J.; Liu, T.; Shu, H.-K.; Yang, X. Deep learning-based image quality improvement for low-dose computed tomography simulation in radiation therapy. J. Med. Imaging 2019, 6, 043504. [Google Scholar] [CrossRef] [PubMed]
- Davatzikos, C.; Barnholtz-Sloan, J.S.; Bakas, S.; Colen, R.; Mahajan, A.; Quintero, C.B.; Capellades Font, J.; Puig, J.; Jain, R.; Sloan, A.E.; et al. AI-based prognostic imaging biomarkers for precision neuro-oncology: The ReSPOND consortium. Neuro. Oncol. 2020, 22, 886–888. [Google Scholar] [CrossRef] [PubMed]
- Lipkova, J.; Chen, R.J.; Chen, B.; Lu, M.Y.; Barbieri, M.; Shao, D.; Vaidya, A.J.; Chen, C.; Zhuang, L.; Williamson, D.F.K.; et al. Artificial intelligence for multimodal data integration in oncology. Cancer cell 2022, 40, 1095–1110. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.F.; Mardis, E.R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 2018, 15, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Dlamini, Z.; Francies, F.Z.; Hull, R.; Marima, R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput. Struct Biotechnol. J. 2020, 18, 2300–2311. [Google Scholar] [CrossRef]
- Jacobs, C.; Setio, A.A.A.; Scholten, E.T.; Gerke, P.K.; Bhattacharya, H.; FA, M.H.; Brink, M.; Ranschaert, E.; de Jong, P.A.; Silva, M.; et al. Deep Learning for Lung Cancer Detection on Screening CT Scans: Results of a Large-Scale Public Competition and an Observer Study with 11 Radiologists. Radiol. Artif. Intell. 2021, 3, e210027. [Google Scholar] [CrossRef]
- Turkbey, B.; Haider, M.A. Artificial Intelligence for Automated Cancer Detection on Prostate MRI: Opportunities and Ongoing Challenges, From the AJR Special Series on AI Applications. Am. J. Roentgenol. 2021, 219, 188–194. [Google Scholar] [CrossRef]
- Shin, I.; Kim, H.; Ahn, S.S.; Sohn, B.; Bae, S.; Park, J.E.; Kim, H.S.; Lee, S.-K. Development and Validation of a Deep Learning–Based Model to Distinguish Glioblastoma from Solitary Brain Metastasis Using Conventional MR Images. Am. J. NeuroRadiol. 2021, 42, 838–844. [Google Scholar] [CrossRef]
- Portnoi, T.; Yala, A.; Schuster, T.; Barzilay, R.; Dontchos, B.; Lamb, L.; Lehman, C. Deep Learning Model to Assess Cancer Risk on the Basis of a Breast MR Image Alone. AJR Am. J. Roentgenol. 2019, 213, 227–233. [Google Scholar] [CrossRef]
- Bahl, M. Harnessing the Power of Deep Learning to Assess Breast Cancer Risk. Radiology 2020, 294, 273–274. [Google Scholar] [CrossRef] [PubMed]
- Diamant, A.; Chatterjee, A.; Vallières, M.; Shenouda, G.; Seuntjens, J. Deep learning in head & neck cancer outcome prediction. Sci. Rep. 2019, 9, 2764. [Google Scholar] [PubMed]
- Nikolov, S.; Blackwell, S.; Zverovitch, A.; Mendes, R.; Livne, M.; De Fauw, J.; Patel, Y.; Meyer, C.; Askham, H.; Romera-Paredes, B. Clinically applicable segmentation of head and neck anatomy for radiotherapy: Deep learning algorithm development and validation study. J. Med. Internet Res. 2021, 23, e26151. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, D.; Tsuneda, M.; Ozawa, S.; Okamoto, H.; Nakamura, M.; Nishio, T.; Nagata, Y. Deep learning-based auto segmentation using generative adversarial network on magnetic resonance images obtained for head and neck cancer patients. J. Appl. Clin. Med. Phys. 2022, 23, e13579. [Google Scholar] [CrossRef]
- Silva, M.; Schaefer-Prokop, C.M.; Jacobs, C.; Capretti, G.; Ciompi, F.; van Ginneken, B.; Pastorino, U.; Sverzellati, N. Detection of Subsolid Nodules in Lung Cancer Screening: Complementary Sensitivity of Visual Reading and Computer-Aided Diagnosis. Investig. Radiol. 2018, 53, 441–449. [Google Scholar] [CrossRef]
- Matchett, K.B.; Lynam-Lennon, N.; Watson, R.W.; Brown, J.A.L. Advances in Precision Medicine: Tailoring Individualized Therapies. Cancers 2017, 9, 146. [Google Scholar] [CrossRef]
- Cheung, H.; Rubin, D. Challenges and opportunities for artificial intelligence in oncological imaging. Clin. Radiol. 2021, 76, 728–736. [Google Scholar] [CrossRef]
- Fitzmaurice, C.; Abate, D.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdel-Rahman, O.; Abdelalim, A.; Abdoli, A.; Abdollahpour, I.; Abdulle, A.S.M.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017 A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef]
- Chakrabarty, S.; Sotiras, A.; Milchenko, M.; LaMontagne, P.; Hileman, M.; Marcus, D. MRI-based Identification and Classification of Major Intracranial Tumor Types by Using a 3D Convolutional Neural Network: A Retrospective Multi-institutional Analysis. Radiol. Artif. Intell. 2021, 3, e200301. [Google Scholar] [CrossRef]
- Kawka, M.; Dawidziuk, A.; Jiao, L.R.; Gall, T.M.H. Artificial intelligence in the detection, characterisation and prediction of hepatocellular carcinoma: A narrative review. Transl. Gastroenterol. Hepatol. 2022, 7, 41. [Google Scholar] [CrossRef]
- Ramón, Y.C.S.; Sesé, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellví, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol Med. 2020, 98, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Tong, E.; McCullagh, K.L.; Iv, M. Advanced Imaging of Brain Metastases: From Augmenting Visualization and Improving Diagnosis to Evaluating Treatment. Adv. Neuroimaging Brain Metastases 2021, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Gang, Y.; Chen, X.; Li, H.; Wang, H.; Li, J.; Guo, Y.; Zeng, J.; Hu, Q.; Hu, J.; Xu, H. A comparison between manual and artificial intelligence-based automatic positioning in CT imaging for COVID-19 patients. Eur. Radiol. 2021, 31, 6049–6058. [Google Scholar] [CrossRef] [PubMed]
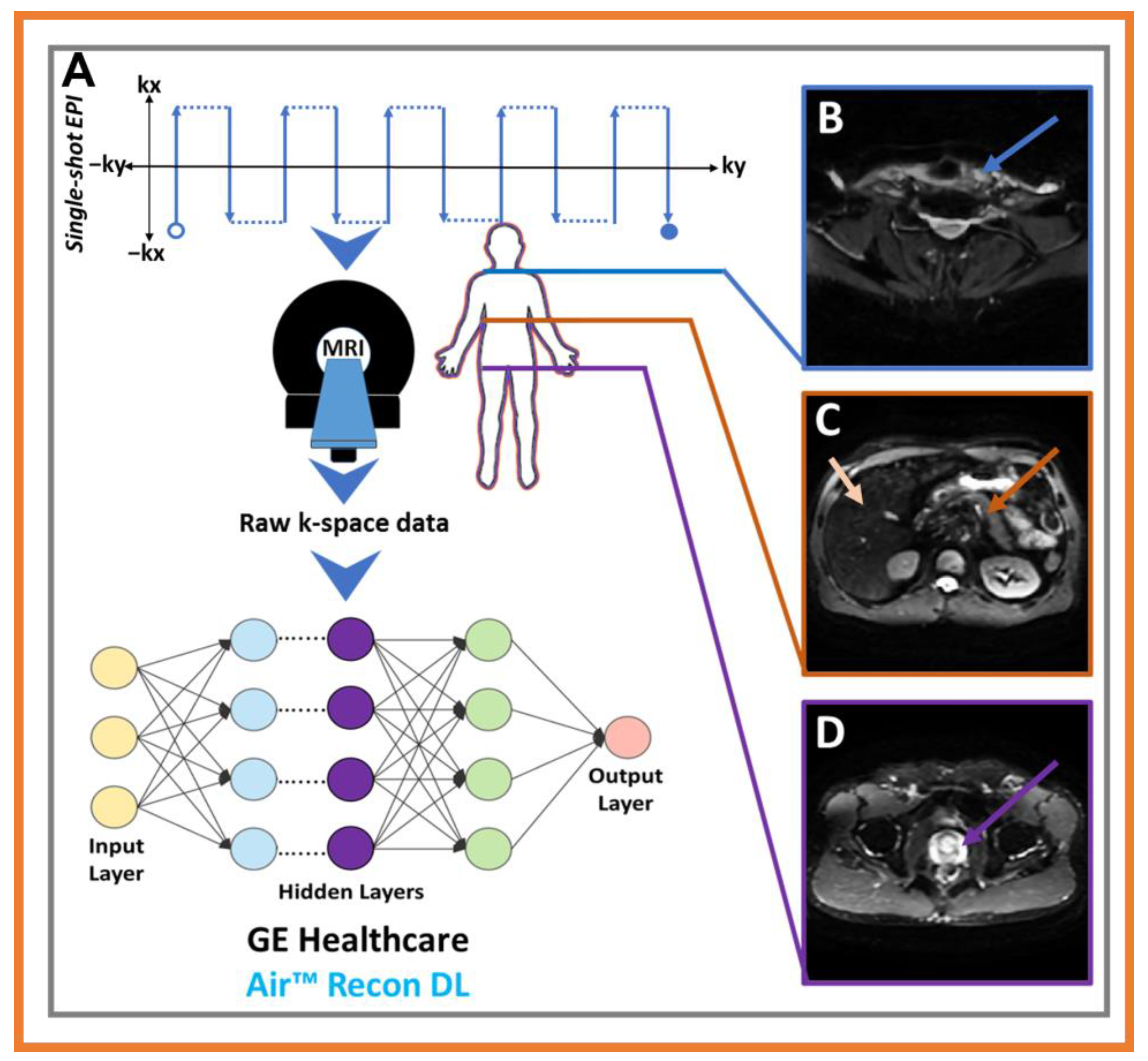
- Lin, D.J.; Johnson, P.M.; Knoll, F.; Lui, Y.W. Artificial Intelligence for MR Image Reconstruction: An Overview for Clinicians. J. Magn. Reson Imaging 2021, 53, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Yaqub, M.; Jinchao, F.; Arshid, K.; Ahmed, S.; Zhang, W.; Nawaz, M.Z.; Mahmood, T. Deep learning-based image reconstruction for different medical imaging modalities. Comput. Math. Methods Med. 2022, 2022, 8750648. [Google Scholar] [CrossRef] [PubMed]
- Akagi, M.; Nakamura, Y.; Higaki, T.; Narita, K.; Honda, Y.; Zhou, J.; Yu, Z.; Akino, N.; Awai, K. Deep learning reconstruction improves image quality of abdominal ultra-high-resolution CT. Eur. Radiol. 2019, 29, 6163–6171. [Google Scholar] [CrossRef]
- Wei, L.; El Naqa, I. Artificial Intelligence for Response Evaluation With PET/CT. Semin. Nucl. Med. 2021, 51, 157–169. [Google Scholar] [CrossRef]
- Lebel, R.M. Performance characterization of a novel deep learning-based MR image reconstruction pipeline. arXiv 2020, arXiv:200806559. [Google Scholar] [CrossRef]
- Gassenmaier, S.; Afat, S.; Nickel, D.; Mostapha, M.; Herrmann, J.; Othman, A.E. Deep learning–accelerated T2-weighted imaging of the prostate: Reduction of acquisition time and improvement of image quality. Eur. J. Radiol. 2021, 137, 109600. [Google Scholar] [CrossRef]
- Iseke, S.; Zeevi, T.; Kucukkaya, A.S.; Raju, R.; Gross, M.; Haider, S.P.; Petukhova-Greenstein, A.; Kuhn, T.N.; Lin, M.; Nowak, M.; et al. Machine Learning Models for Prediction of Posttreatment Recurrence in Early-Stage Hepatocellular Carcinoma Using Pretreatment Clinical and MRI Features: A Proof-of-Concept Study. AJR Am. J. Roentgenol. 2023, 220, 245–255. [Google Scholar] [CrossRef]
- Kaus, M.R.; Warfield, S.K.; Nabavi, A.; Black, P.M.; Jolesz, F.A.; Kikinis, R. Automated segmentation of MR images of brain tumors. Radiology 2001, 218, 586–591. [Google Scholar] [CrossRef]
- Arimura, H.; Soufi, M.; Kamezawa, H.; Ninomiya, K.; Yamada, M. Radiomics with artificial intelligence for precision medicine in radiation therapy. J. Radiat. Res. 2019, 60, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Rogers, W.; Thulasi Seetha, S.; Refaee, T.A.G.; Lieverse, R.I.Y.; Granzier, R.W.Y.; Ibrahim, A.; Keek, S.A.; Sanduleanu, S.; Primakov, S.P.; Beuque, M.P.L.; et al. Radiomics: From qualitative to quantitative imaging. Br. J. Radiol. 2020, 93, 20190948. [Google Scholar] [CrossRef]
- Torrente, M.; Sousa, P.A.; Hernández, R.; Blanco, M.; Calvo, V.; Collazo, A.; Guerreiro, G.R.; Núñez, B.; Pimentao, J.; Sánchez, J.C.; et al. An Artificial Intelligence-Based Tool for Data Analysis and Prognosis in Cancer Patients: Results from the Clarify Study. Cancers 2022, 14, 4041. [Google Scholar] [CrossRef] [PubMed]
- Razek, A.A.K.A.; Khaled, R.; Helmy, E.; Naglah, A.; AbdelKhalek, A.; El-Baz, A. Artificial intelligence and deep learning of head and neck cancer. Magn. Reson. Imaging Clin. 2022, 30, 81–94. [Google Scholar] [CrossRef] [PubMed]
- McCollough, C.; Leng, S. Use of artificial intelligence in computed tomography dose optimisation. Ann. ICRP 2020, 49, 113–125. [Google Scholar] [CrossRef] [PubMed]
- McLeavy, C.; Chunara, M.; Gravell, R.; Rauf, A.; Cushnie, A.; Talbot, C.S.; Hawkins, R. The future of CT: Deep learning reconstruction. Clin. Radiol. 2021, 76, 407–415. [Google Scholar] [CrossRef]
- Jiang, J.; Hu, Y.C.; Tyagi, N.; Zhang, P.; Rimner, A.; Deasy, J.O.; Veeraraghavan, H. Cross-modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small MR datasets. Med. Phys. 2019, 46, 4392–4404. [Google Scholar] [CrossRef]
- Venkadesh, K.V.; Setio, A.A.; Schreuder, A.; Scholten, E.T.; Chung, K.W.; Wille, M.M.; Saghir, Z.; van Ginneken, B.; Prokop, M.; Jacobs, C. Deep learning for malignancy risk estimation of pulmonary nodules detected at low-dose screening CT. Radiology 2021, 300, 438–447. [Google Scholar] [CrossRef]
- Liu, K.-L.; Wu, T.; Chen, P.-T.; Tsai, Y.M.; Roth, H.; Wu, M.-S.; Liao, W.-C.; Wang, W. Deep learning to distinguish pancreatic cancer tissue from non-cancerous pancreatic tissue: A retrospective study with cross-racial external validation. Lancet Digit. Health 2020, 2, e303–e313. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Hoebel, K.V.; Patel, J.B.; Beers, A.L.; Chang, K.; Singh, P.; Brown, J.M.; Pinho, M.C.; Batchelor, T.T.; Gerstner, E.R.; Rosen, B.R.; et al. Radiomics Repeatability Pitfalls in a Scan-Rescan MRI Study of Glioblastoma. Radiol. Artif. Intell. 2021, 3, e190199. [Google Scholar] [CrossRef] [PubMed]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef] [PubMed]
- Shaham, U.; Stanton, K.P.; Zhao, J.; Li, H.; Raddassi, K.; Montgomery, R.; Kluger, Y. Removal of batch effects using distribution-matching residual networks. Bioinformatics 2017, 33, 2539–2546. [Google Scholar] [CrossRef]
- Horng, H.; Singh, A.; Yousefi, B.; Cohen, E.A.; Haghighi, B.; Katz, S.; Noël, P.B.; Shinohara, R.T.; Kontos, D. Generalized ComBat harmonization methods for radiomic features with multi-modal distributions and multiple batch effects. Sci. Rep. 2022, 12, 4493. [Google Scholar] [CrossRef] [PubMed]
- Andrearczyk, V.; Depeursinge, A.; Müller, H. Neural network training for cross-protocol radiomic feature standardization in computed tomography. J. Med. Imaging 2019, 6, 024008. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Maiti, P.; Thomopoulos, S.; Zhu, A.; Chai, Y.; Kim, H.; Jahanshad, N. Style transfer using generative adversarial networks for multi-site mri harmonization. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 18–22 September 2021; pp. 313–322. [Google Scholar]
- Li, Y.; Han, G.; Wu, X.; Li, Z.H.; Zhao, K.; Zhang, Z.; Liu, Z.; Liang, C. Normalization of multicenter CT radiomics by a generative adversarial network method. Phys. Med. Biol. 2021, 66, 055030. [Google Scholar] [CrossRef] [PubMed]
- Bashyam, V.M.; Doshi, J.; Erus, G.; Srinivasan, D.; Abdulkadir, A.; Singh, A.; Habes, M.; Fan, Y.; Masters, C.L.; Maruff, P. Deep Generative Medical Image Harmonization for Improving Cross-Site Generalization in Deep Learning Predictors. J. Magn. Reson Imaging 2022, 55, 908–916. [Google Scholar] [CrossRef]
- Alexander, A.; Jiang, A.; Ferreira, C.; Zurkiya, D. An intelligent future for medical imaging: A market outlook on artificial intelligence for medical imaging. J. Am. Coll. Radiol. 2020, 17, 165–170. [Google Scholar] [CrossRef]
- Armanious, K.; Jiang, C.; Fischer, M.; Küstner, T.; Hepp, T.; Nikolaou, K.; Gatidis, S.; Yang, B. MedGAN: Medical image translation using GANs. Comput. Med. Imaging Graph. 2020, 79, 101684. [Google Scholar] [CrossRef]
- Chen, J.; Wei, J.; Li, R. TarGAN: Target-aware generative adversarial networks for multi-modality medical image translation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Part VI 24. pp. 24–33. [Google Scholar]
- Emami, H.; Dong, M.; Nejad-Davarani, S.P.; Glide-Hurst, C.K. SA-GAN: Structure-aware GAN for organ-preserving synthetic CT generation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Part VI 24. pp. 471–481. [Google Scholar]
- Jiang, J.; Veeraraghavan, H. Unified cross-modality feature disentangler for unsupervised multi-domain MRI abdomen organs segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 347–358. [Google Scholar]
- Zeng, P.; Zhou, L.; Zu, C.; Zeng, X.; Jiao, Z.; Wiu, X.; Zhou, J.; Shen, D.; Wang, Y. 3D Convolutional Vision Transformer-GAN for PET Reconstructio. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2022: 25th International Conference, Singapore, 17 September 2022; pp. 516–526. [Google Scholar]
- Zhao, J.; Li, D.; Kassam, Z.; Howey, J.; Chong, J.; Chen, B.; Li, S. Tripartite-GAN: Synthesizing liver contrast-enhanced MRI to improve tumor detection. Med. Image Anal. 2020, 63, 101667. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Hu, Y.-C.; Tyagi, N.; Rimner, A.; Lee, N.; Deasy, J.O.; Berry, S.; Veeraraghavan, H. PSIGAN: Joint probabilistic segmentation and image distribution matching for unpaired cross-modality adaptation-based MRI segmentation. IEEE T Med. Imaging 2020, 39, 4071–4084. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Rimner, A.; Deasy, J.O.; Veeraraghavan, H. Unpaired cross-modality educed distillation (CMEDL) for medical image segmentation. IEEE T Med. Imaging 2021, 41, 1057–1068. [Google Scholar] [CrossRef]
- Jiang, J.; Tyagi, N.; Tringale, K.; Crane, C.; Veeraraghavan, H. Self-supervised 3D anatomy segmentation using self-distilled masked image transformer (SMIT). arXiv 2022, arXiv:2205.10342. [Google Scholar]
- Fei, Y.; Zu, C.; Jiao, Z.; Wu, X.; Zhou, J.; Shen, D.; Wang, Y. Classification-Aided High-Quality PET Image Synthesis via Bidirectional Contrastive GAN with Shared Information Maximization. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2022: 25th International Conference, Singapore, 18–22 September 2022; Part VI. pp. 527–537. [Google Scholar]
- Wu, P.-W.; Lin, Y.-J.; Chang, C.-H.; Chang, E.Y.; Liao, S.-W. Relgan: Multi-domain image-to-image translation via relative attributes. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27–28 October 2019; pp. 5914–5922. [Google Scholar]
- Fave, X.; Zhang, L.; Yang, J.; Mackin, D.; Balter, P.; Gomez, D.; Followill, D.; Jones, A.K.; Stingo, F.; Liao, Z. Delta-radiomics features for the prediction of patient outcomes in non–small cell lung cancer. Sci. Rep. 2017, 7, 588. [Google Scholar] [CrossRef]
- Shi, L.; Rong, Y.; Daly, M.; Dyer, B.; Benedict, S.; Qiu, J.; Yamamoto, T. Cone-beam computed tomography-based delta-radiomics for early response assessment in radiotherapy for locally advanced lung cancer. Phys. Med. Biol. 2020, 65, 015009. [Google Scholar] [CrossRef] [PubMed]
- Sutton, E.J.; Onishi, N.; Fehr, D.A.; Dashevsky, B.Z.; Sadinski, M.; Pinker, K.; Martinez, D.F.; Brogi, E.; Braunstein, L.; Razavi, P. A machine learning model that classifies breast cancer pathologic complete response on MRI post-neoadjuvant chemotherapy. Breast Cancer Res. 2020, 22, 57. [Google Scholar] [CrossRef]
- Nardone, V.; Reginelli, A.; Grassi, R.; Boldrini, L.; Vacca, G.; D’Ippolito, E.; Annunziata, S.; Farchione, A.; Belfiore, M.P.; Desideri, I. Delta radiomics: A systematic review. La Radiol. Med. 2021, 126, 1571–1583. [Google Scholar] [CrossRef]
- Jin, C.; Yu, H.; Ke, J.; Ding, P.; Yi, Y.; Jiang, X.; Duan, X.; Tang, J.; Chang, D.T.; Wu, X. Predicting treatment response from longitudinal images using multi-task deep learning. Nat. Commun. 2021, 12, 1851. [Google Scholar] [CrossRef]
- Beers, A.; Brown, J.; Chang, K.; Hoebel, K.; Patel, J.; Ly, K.I.; Tolaney, S.M.; Brastianos, P.; Rosen, B.; Gerstner, E.R. DeepNeuro: An open-source deep learning toolbox for neuroimaging. Neuroinformatics 2021, 19, 127–140. [Google Scholar] [CrossRef]
- Mehrtash, A.; Pesteie, M.; Hetherington, J.; Behringer, P.A.; Kapur, T.; Wells, W.M., III; Rohling, R.; Fedorov, A.; Abolmaesumi, P. DeepInfer: Open-source deep learning deployment toolkit for image-guided therapy. In Proceedings of the Medical Imaging 2017: Image-Guided Procedures, Robotic Interventions, and Modeling, Orlando, FL, USA, 3 March 2017; pp. 410–416. [Google Scholar]
- Ma, N.; Li, W.; Brown, R.; Wang, Y.; Gorman, B.; Behrooz; Johnson, H.; Yang, I.; Kerfoot, E.; Li, Y.; et al. Project-MONAI/MONAI: 0.5.0. 2021. Available online: https://zenodo.org/record/4679866#.ZD9KOXZByUk (accessed on 30 January 2023).
- Haibe-Kains, B.; Adam, G.A.; Hosny, A.; Khodakarami, F.; Shraddha, T.; Kusko, R.; Sansone, S.-A.; Tong, W.; Wolfinger, R.D.; Mason, C.E.; et al. Transparency and reproducibility in artificial intelligence. Nature 2020, 586, E14–E16. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Wu, M.; Chen, L.; Bai, W.; Yang, M.; Lyu, S.; Zhao, Q. Using Guided Self-Attention with Local Information for Polyp Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; pp. 629–638. [Google Scholar]
- Zheng, Y. Cross-modality medical image detection and segmentation by transfer learning of shapel priors. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 424–427. [Google Scholar]
- Li, K.; Yu, L.; Wang, S.; Heng, P.-A. Towards cross-modality medical image segmentation with online mutual knowledge distillation. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; pp. 775–783. [Google Scholar]
- Li, K.; Wang, S.; Yu, L.; Heng, P.-A. Dual-teacher: Integrating intra-domain and inter-domain teachers for annotation-efficient cardiac segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 418–427. [Google Scholar]
- Gan, W.; Wang, H.; Gu, H.; Duan, Y.; Shao, Y.; Chen, H.; Feng, A.; Huang, Y.; Fu, X.; Ying, Y. Automatic segmentation of lung tumors on CT images based on a 2D & 3D hybrid convolutional neural network. Br. J. Radiol. 2021, 94, 20210038. [Google Scholar] [CrossRef]
- Jiang, J.; Hu, Y.C.; Liu, C.J.; Halpenny, D.; Hellmann, M.D.; Deasy, J.O.; Mageras, G.; Veeraraghavan, H. Multiple Resolution Residually Connected Feature Streams for Automatic Lung Tumor Segmentation from CT Images. IEEE Trans. Med. Imaging 2019, 38, 134–144. [Google Scholar] [CrossRef]
- Just, N. Improving tumour heterogeneity MRI assessment with histograms. Br. J. Cancer 2014, 111, 2205–2213. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Singh, T.; Soni, S. Pre-operative Assessment of Ablation Margins for Variable Blood Perfusion Metrics in a Magnetic Resonance Imaging Based Complex Breast Tumour Anatomy: Simulation Paradigms in Thermal Therapies. Comput. Methods Programs Biomed. 2021, 198, 105781. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, C.E.; Beadle, B.M.; Garden, A.S.; Skinner, H.D.; Yang, J.; Rhee, D.J.; McCarroll, R.E.; Netherton, T.J.; Gay, S.S.; Zhang, L. Generating high-quality lymph node clinical target volumes for head and neck cancer radiation therapy using a fully automated deep learning-based approach. Int. J. Radiat. Oncol. Biol. Phys. 2021, 109, 801–812. [Google Scholar] [CrossRef]
- Cardenas, C.E.; McCarroll, R.E.; Court, L.E.; Elgohari, B.A.; Elhalawani, H.; Fuller, C.D.; Kamal, M.J.; Meheissen, M.A.; Mohamed, A.S.; Rao, A. Deep learning algorithm for auto-delineation of high-risk oropharyngeal clinical target volumes with built-in dice similarity coefficient parameter optimization function. Int. J. Radiat. Oncol. Biol. Phys. 2018, 101, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Kang, K.; Wang, Y.; Khandekar, M.J.; Willers, H.; Keane, F.K.; Bortfeld, T.R. Automated clinical target volume delineation using deep 3D neural networks in radiation therapy of Non-small Cell Lung Cancer. Phys. Imaging Radiat. Oncol. 2021, 19, 131–137. [Google Scholar] [CrossRef]
- McCollough, C.H. Computed tomography technology—And dose—In the 21st century. Health Phys. 2019, 116, 157–162. [Google Scholar] [CrossRef]
- Arndt, C.; Güttler, F.; Heinrich, A.; Bürckenmeyer, F.; Diamantis, I.; Teichgräber, U. Deep Learning CT Image Reconstruction in Clinical Practice. RoFo Fortschr. Auf Dem Geb. Der Rontgenstrahlen Und Der Nukl. 2021, 193, 252–261. [Google Scholar] [CrossRef]
- Willemink, M.J.; Noël, P.B. The evolution of image reconstruction for CT—From filtered back projection to artificial intelligence. Eur. Radiol. 2019, 29, 2185–2195. [Google Scholar] [CrossRef]
- Hsieh, S.S.; Leng, S.; Rajendran, K.; Tao, S.; McCollough, C.H.; Sciences, P.M. Photon counting CT: Clinical applications and future developments. IEEE Trans. Radiat. Plasma Med. Sci. 2020, 5, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Kwan, A.C.; Pourmorteza, A.; Stutman, D.; Bluemke, D.A.; Lima, J.A. Next-generation hardware advances in CT: Cardiac applications. Radiology 2021, 298, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Mileto, A.; Guimaraes, L.S.; McCollough, C.H.; Fletcher, J.G.; Yu, L. State of the art in abdominal CT: The limits of iterative reconstruction algorithms. Radiology 2019, 293, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Liu, E.; Nett, B.; Tang, J.; Thibault, J.-B.; Sahney, S. A New Era of Image Reconstruction: TrueFidelity™; White Paper; GE Healthcare: Chicago, IL, USA, 2019. [Google Scholar]
- Laurent, G.; Villani, N.; Hossu, G.; Rauch, A.; Noël, A.; Blum, A.; Gondim Teixeira, P.A. Full model-based iterative reconstruction (MBIR) in abdominal CT increases objective image quality, but decreases subjective acceptance. Eur. Radiol. 2019, 29, 4016–4025. [Google Scholar] [CrossRef]
- Chen, M.M.; Terzic, A.; Becker, A.S.; Johnson, J.M.; Wu, C.C.; Wintermark, M.; Wald, C.; Wu, J. Artificial intelligence in oncologic imaging. Eur. J. Radiol. Open 2022, 9, 100441. [Google Scholar] [CrossRef]
- Boedeker, K. AiCE Deep Learning Reconstruction: Bringing the Power of Ultra-High Resolution CT to Routine Imaging; Canon Medical Systems Corporation: Ohtawara, Japan, 2019. [Google Scholar]
- Jensen, C.T.; Liu, X.; Tamm, E.P.; Chandler, A.G.; Sun, J.; Morani, A.C.; Javadi, S.; Wagner-Bartak, N.A. Image quality assessment of abdominal CT by use of new deep learning image reconstruction: Initial experience. Am. J. Roentgenol. 2020, 215, 50–57. [Google Scholar] [CrossRef]
- Ricci Lara, M.A.; Echeveste, R.; Ferrante, E. Addressing fairness in artificial intelligence for medical imaging. Nat. Commun. 2022, 13, 4581. [Google Scholar] [CrossRef]
- Szczykutowicz, T.P.; Toia, G.V.; Dhanantwari, A.; Nett, B. A Review of Deep Learning CT Reconstruction: Concepts, Limitations, and Promise in Clinical Practice. Curr. Radiol. Rep. 2022, 10, 101–115. [Google Scholar] [CrossRef]
- Noda, Y.; Kawai, N.; Nagata, S.; Nakamura, F.; Mori, T.; Miyoshi, T.; Suzuki, R.; Kitahara, F.; Kato, H.; Hyodo, F. Deep learning image reconstruction algorithm for pancreatic protocol dual-energy computed tomography: Image quality and quantification of iodine concentration. Eur. Radiol. 2022, 32, 384–394. [Google Scholar] [CrossRef]
- Jensen, C.T.; Gupta, S.; Saleh, M.M.; Liu, X.; Wong, V.K.; Salem, U.; Qiao, W.; Samei, E.; Wagner-Bartak, N.A. Reduced-dose deep learning reconstruction for abdominal CT of liver metastases. Radiology 2022, 303, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. Radiographics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [PubMed]
- Argentieri, E.; Zochowski, K.; Potter, H.; Shin, J.; Lebel, R.; Sneag, D. Performance of a Deep Learning-Based MR Reconstruction Algorithm for the Evaluation of Peripheral Nerves. In Proceedings of the RSNA, Chicago, IL, USA, 1–6 December 2019. [Google Scholar]
- Le Bihan, D. Molecular diffusion, tissue microdynamics and microstructure. NMR Biomed. 1995, 8, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, C.; Chen, W.; Wang, F.; Yang, Z.; Xu, S.; Wang, H. Deep learning based multiplexed sensitivity-encoding (DL-MUSE) for high-resolution multi-shot DWI. NeuroImage 2021, 244, 118632. [Google Scholar] [CrossRef]
- Gadjimuradov, F.; Benkert, T.; Nickel, M.D.; Führes, T.; Saake, M.; Maier, A. Deep learning–guided weighted averaging for signal dropout compensation in DWI of the liver. Magn. Reson. Med. 2022, 88, 2679–2693. [Google Scholar] [CrossRef]
- Ueda, T.; Ohno, Y.; Yamamoto, K.; Murayama, K.; Ikedo, M.; Yui, M.; Hanamatsu, S.; Tanaka, Y.; Obama, Y.; Ikeda, H. Deep Learning Reconstruction of Diffusion-weighted MRI Improves Image Quality for Prostatic Imaging. Radiology 2022, 303, 373–381. [Google Scholar] [CrossRef]
- Langlotz, C.P.; Allen, B.; Erickson, B.J.; Kalpathy-Cramer, J.; Bigelow, K.; Cook, T.S.; Flanders, A.E.; Lungren, M.P.; Mendelson, D.S.; Rudie, J.D. A roadmap for foundational research on artificial intelligence in medical imaging: From the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology 2019, 291, 781. [Google Scholar] [CrossRef]
- Schork, N.J. Artificial intelligence and personalized medicine. In Precision Medicine in Cancer Therapy; Springer: Berlin/Heidelberg, Germany, 2019; pp. 265–283. [Google Scholar]
- Chung, C.; Kalpathy-Cramer, J.; Knopp, M.V.; Jaffray, D.A. In the era of deep learning, why reconstruct an image at all? J. Am. Coll. Radiol. 2021, 18, 170–173. [Google Scholar] [CrossRef]
- Li, G.; Zhang, X.; Song, X.; Duan, L.; Wang, G.; Xiao, Q.; Li, J.; Liang, L.; Bai, L.; Bai, S. Machine learning for predicting accuracy of lung and liver tumor motion tracking using radiomic features. Quant. Imaging Med. Surg. 2023, 13, 1605–1618. [Google Scholar] [CrossRef]


| Study | Narrow-Specific Tasks | Design: Title | Objective | Advantages/Recommendations | Limitations |
|---|---|---|---|---|---|
| Hosny, A. et al. [8] | Medical Imaging (MI) | Review: Artificial Intelligence (AI) in radiology | To establish a general understanding of AI methods, particularly those pertaining to image-based tasks. The AI methods could impact multiple facets of radiology, with a general focus on applications in oncology, and demonstrate how these methods are advancing the field. | There is a need to understand that AI is unlike human intelligence in many ways. Excelling in one task does not necessarily imply excellence in others. The roles of radiologists will expand as they have access to better tools. The data to train AI on a massive scale will enable a robust AI that is generalizable across different patient demographics, geographic regions, diseases, and standards of care. | Not Applicable (NA) |
| Koh, D.M. et al. [15] | MI | Review: Artificial Intelligence and machine learning in cancer imaging | To foster interdisciplinary communication because many technological solutions are being developed in isolation and may struggle to achieve routine clinical use. Hence, it is important to work together, including with commercial partners (as appropriate) to drive innovations and developments. | There is a need for systematic evaluation of new software, which often undergoes only limited testing prior to release. | NA |
| Razek, A.A.K.A. et al. [56] | MI | Review: Artificial Intelligence and deep learning of head and neck cancer | To summarize the clinical applications of AI in head and neck cancer, including differentiation, grading, staging, prognosis, genetic profile, and monitoring after treatment. | AI studies are required to establish a powerful methodology and coupling of genetic and radiologic profiles to be validated in clinical use. | NA |
| McCollough, C.H. et al. [57] | MI | Review: Use of Artificial Intelligence in computed tomography dose optimization | To illustrate the promise of AI in the processes involved in a CT examination, from setting up the patient on the scanner table to the reconstruction of final images. | AI could be a part of CT imaging in the future, and both manufacturers and users must proceed cautiously because it is not yet clear how these AI algorithms can be evaluated in the clinical setting. | NA |
| Lin, D.J. et al. [45] | Image reconstruction and registration (IRR) | Review: Artificial Intelligence for MR Image Reconstruction: An Overview for Clinicians | To cover how deep learning algorithms transform raw k-space data into image data and examine accelerated imaging and artifact suppression. | Future research needs continued sharing of image and raw k space datasets to expand access and allow for model comparisons, defining the best clinically relevant loss functions and/or quality metrics by which to judge a model’s performance, examining perturbations in model performance relating to acquisition parameters, and validating high-performing models in new scenarios to determine generalizability. | NA |
| McLeavy, C.M. et al. [58] | IRR | Review: The future of CT: deep learning reconstruction | To emphasize the advantages of deep learning reconstruction (DLR) over other reconstruction methods regarding dose reduction, image quality, and tailoring protocols to specific clinical situations. | DLR is the future of CT technology and should be considered when procuring new CT scanners. | NA |
| Jiang J. et al. [59] | Lesion segmentation, detection, and characterization (LSDC) | Original Research: Cross-modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small MR datasets | To develop a cross-modality (MR-CT) deep learning segmentation approach that augments training data using pseudo-MR images produced by transforming expert-segmented CT images. | The advantage of this model is that it is learned as a deep generative adversarial network and transforms expert segmented CT into pseudo-MR images with expert segmentations. | A minor limitation is the number of test datasets, particularly for longitudinal analysis, due to the lack of additional recruitment of patients. |
| Venkadesh, K.V. et al. [60] | LSDC | Original Research: Deep Learning for Malignancy Risk Estimation of Pulmonary Nodules Detected at Low-Dose Screening CT | To develop and validate a deep learning (DL) algorithm for malignancy risk estimation of pulmonary nodules detected at screening CT. | The DL algorithm has the potential to provide reliable and reproducible malignancy risk scores for clinicians from low-dose screening CT, leading to better management in lung cancer. | A minor limitation, the group did not assess how the algorithm would affect the radiologists’ assessment. |
| Bi, W.L. et al. [10] | Clinical Applications in Oncology (CAO) | Review: Artificial Intelligence in cancer imaging: Clinical challenges and applications | Highlights AI applied to medical imaging of lung, brain, breast, and prostate cancer and illustrates how clinical problems are being addressed using imaging/radiomic feature types. | AI applications in oncological imaging need to be vigorously validated for reproducibility and generalizability. | NA |
| Huang, S. et al. [20] | CAO | Review: Artificial Intelligence in cancer diagnosis and prognosis: Opportunities and challenges | Highlights how AI assists in cancer diagnosis and prognosis, specifically about its unprecedented accuracy, which is even higher than that of general statistical applications in oncology. | The use of AI-based applications in clinical cancer research represents a paradigm shift in cancer treatment, leading to a dramatic improvement in patient survival due to enhanced prediction rates. | NA |
| Diamant, A. et al. [33] | CAO | Original research: Deep learning in head & neck cancer outcome prediction | To apply convolutional neural network (CNN) to predict treatment outcomes of patients with head & neck cancer using pretreatment CT images. | The work identifies traditional radiomic features derived from CT images that can be visualized and used to perform accurate outcome prediction in head & neck cancers. However, future work could be done to further investigate the difference between the two representations. | There is no major limitation mentioned by the authors. However, they do mention that the framework used here considers the central slice, and the results could have been further improved by incorporating the entire tumor. |
| Liu, K.L. et al. [61] | CAO | Original research: Deep learning to distinguish pancreatic cancer tissue from noncancerous pancreatic tissue: a retrospective study with cross-racial external validation | To investigate whether CNNs can distinguish individuals with and without pancreatic cancer on CT, compared with radiologist interpretation. | CNNs can accurately distinguish pancreatic cancer on CT, with acceptable generalizability to images of patients from various races and ethnicities. Additionally, CNNs can supplement radiologist interpretation. | A minor limitation is the modest sample size. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paudyal, R.; Shah, A.D.; Akin, O.; Do, R.K.G.; Konar, A.S.; Hatzoglou, V.; Mahmood, U.; Lee, N.; Wong, R.J.; Banerjee, S.; et al. Artificial Intelligence in CT and MR Imaging for Oncological Applications. Cancers 2023, 15, 2573. https://doi.org/10.3390/cancers15092573
Paudyal R, Shah AD, Akin O, Do RKG, Konar AS, Hatzoglou V, Mahmood U, Lee N, Wong RJ, Banerjee S, et al. Artificial Intelligence in CT and MR Imaging for Oncological Applications. Cancers. 2023; 15(9):2573. https://doi.org/10.3390/cancers15092573
Chicago/Turabian StylePaudyal, Ramesh, Akash D. Shah, Oguz Akin, Richard K. G. Do, Amaresha Shridhar Konar, Vaios Hatzoglou, Usman Mahmood, Nancy Lee, Richard J. Wong, Suchandrima Banerjee, and et al. 2023. "Artificial Intelligence in CT and MR Imaging for Oncological Applications" Cancers 15, no. 9: 2573. https://doi.org/10.3390/cancers15092573
APA StylePaudyal, R., Shah, A. D., Akin, O., Do, R. K. G., Konar, A. S., Hatzoglou, V., Mahmood, U., Lee, N., Wong, R. J., Banerjee, S., Shin, J., Veeraraghavan, H., & Shukla-Dave, A. (2023). Artificial Intelligence in CT and MR Imaging for Oncological Applications. Cancers, 15(9), 2573. https://doi.org/10.3390/cancers15092573