Role of Secreted Frizzled-Related Protein 1 in Early Breast Carcinogenesis and Breast Cancer Aggressiveness
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Analyses of Public Datasets
2.2. Mice Organoid Culture and Data Collection
2.3. Cell Lines
2.4. Lentiviral Production and Cell Infection
2.5. 2D Proliferation Assays
2.6. Spheroids Assays
2.7. Migration Assays
2.8. RNA Extraction, Purification, and Reverse Transcription
2.9. Quantitative Real Time Polymerase Chain Reaction (qPCR)
2.10. Statistical Analyses
3. Results
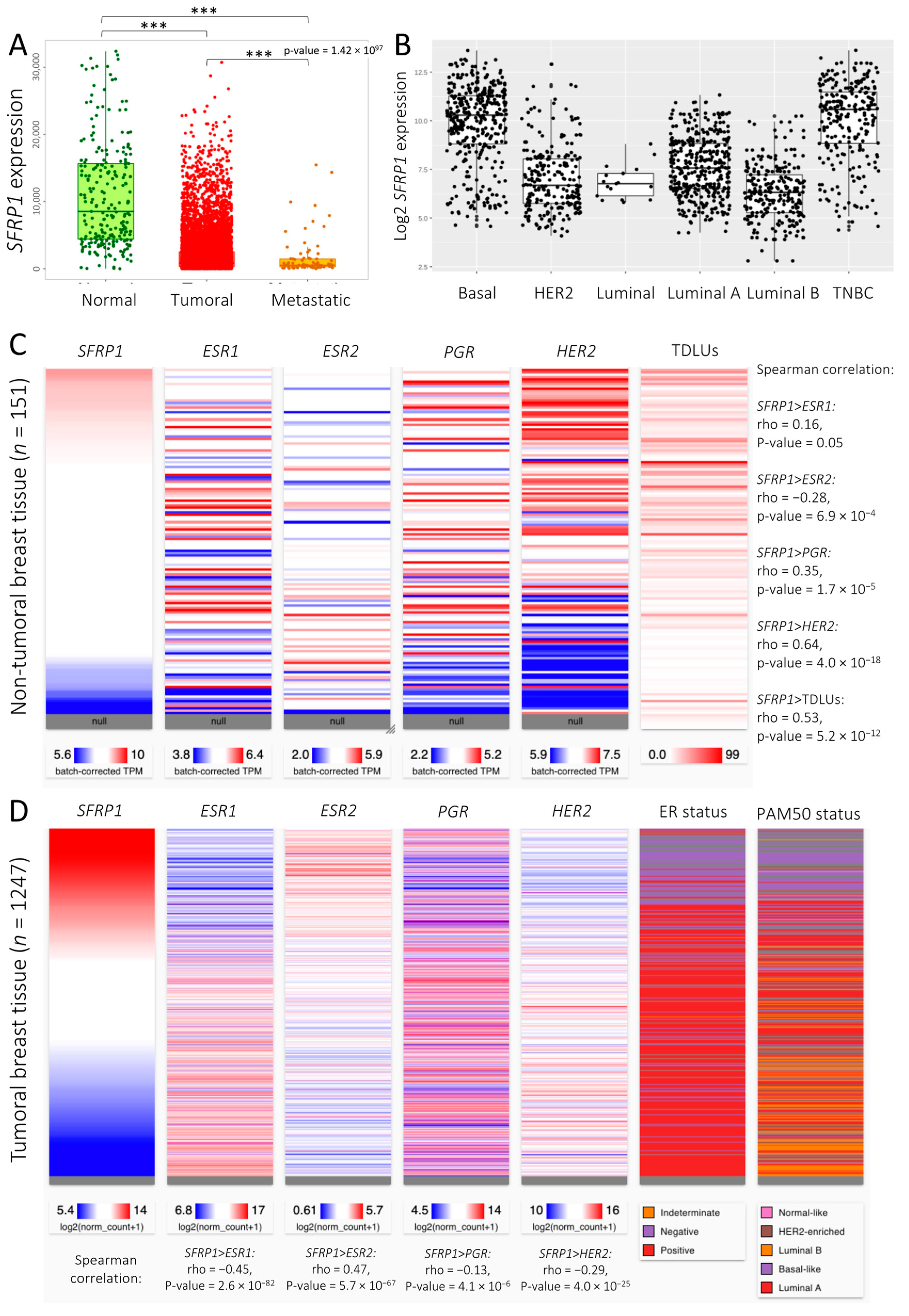
3.1. SFRP1 Expression and Co-Expression Pattern across Breast Cancer Tissues
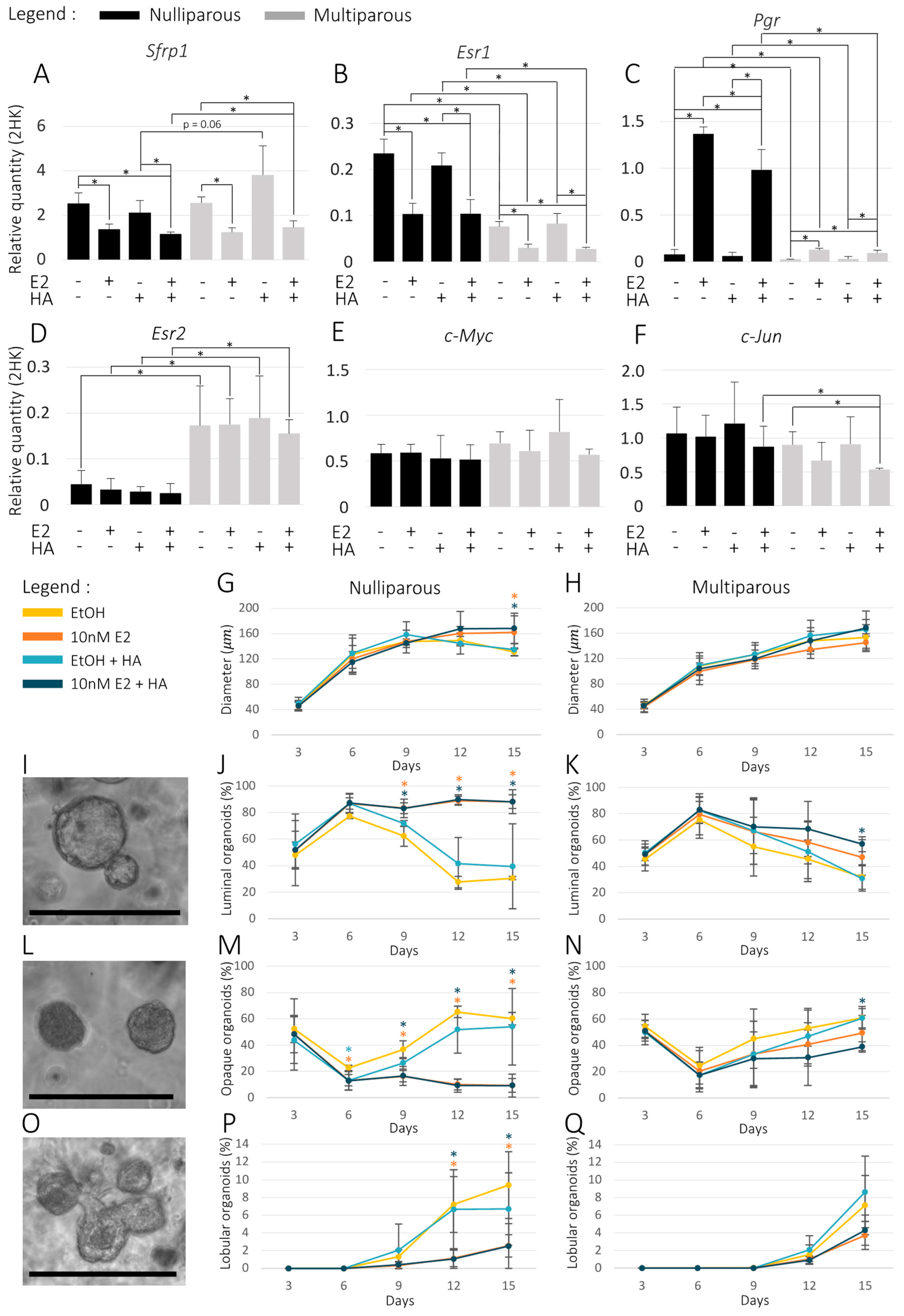
3.2. Sfrp1 Expression in Mice Organoids and Its Association with Organoid Phenotype
3.3. The Role of SFRP1 in Early Breast Carcinogenesis
3.3.1. Non-Tumoral Breast Epithelial Cell
3.3.2. Atypical Ductal Hyperplasia
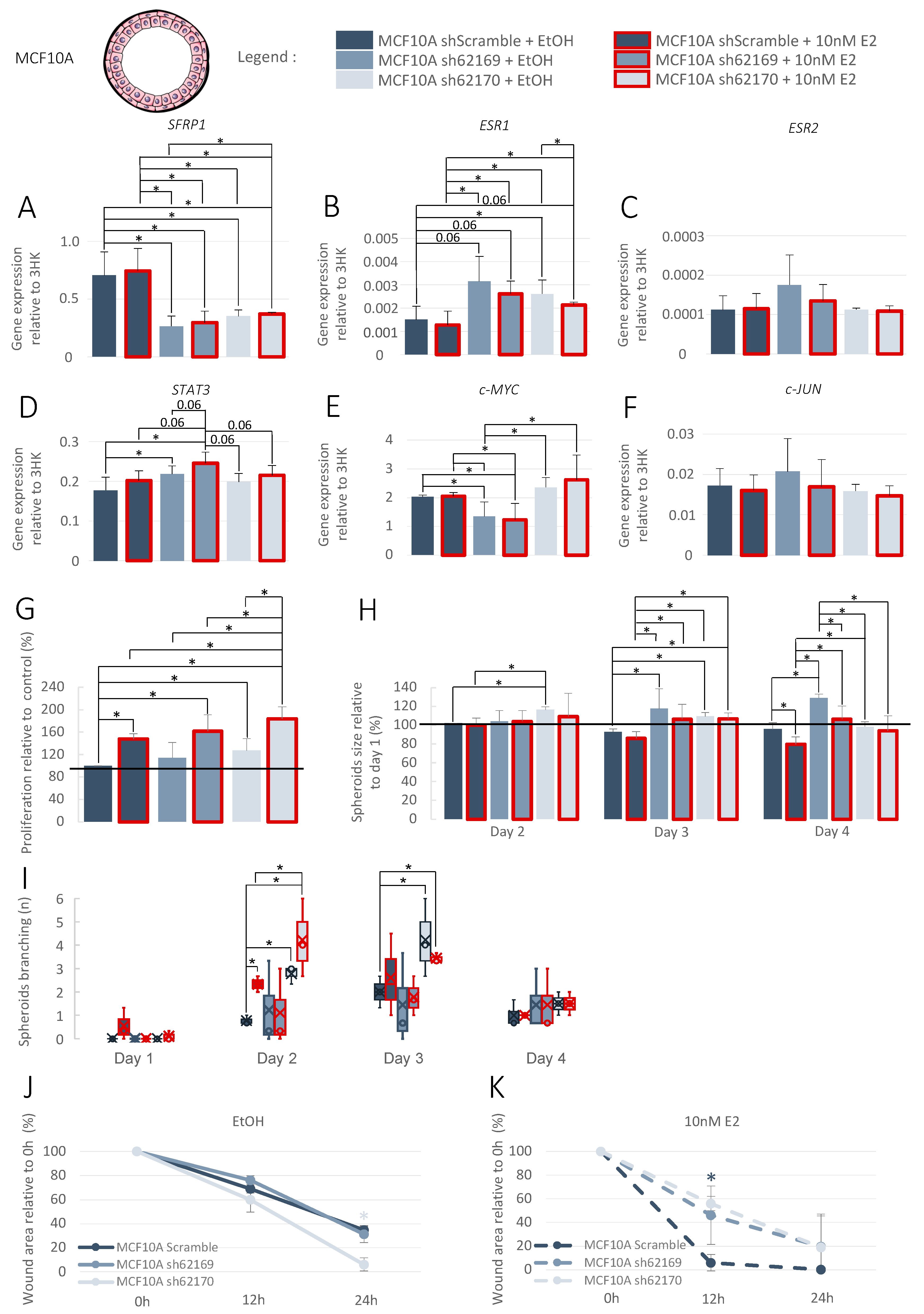
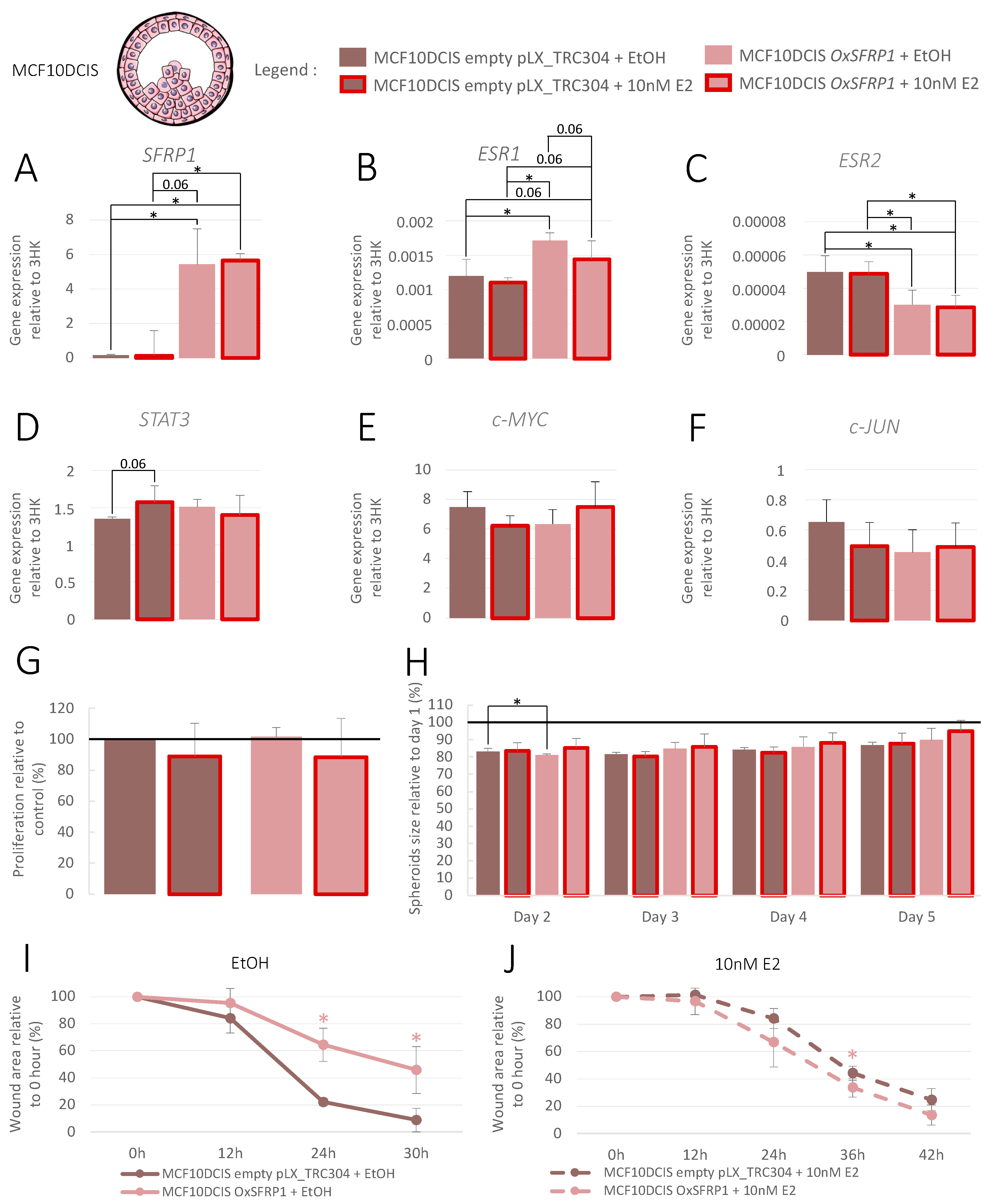
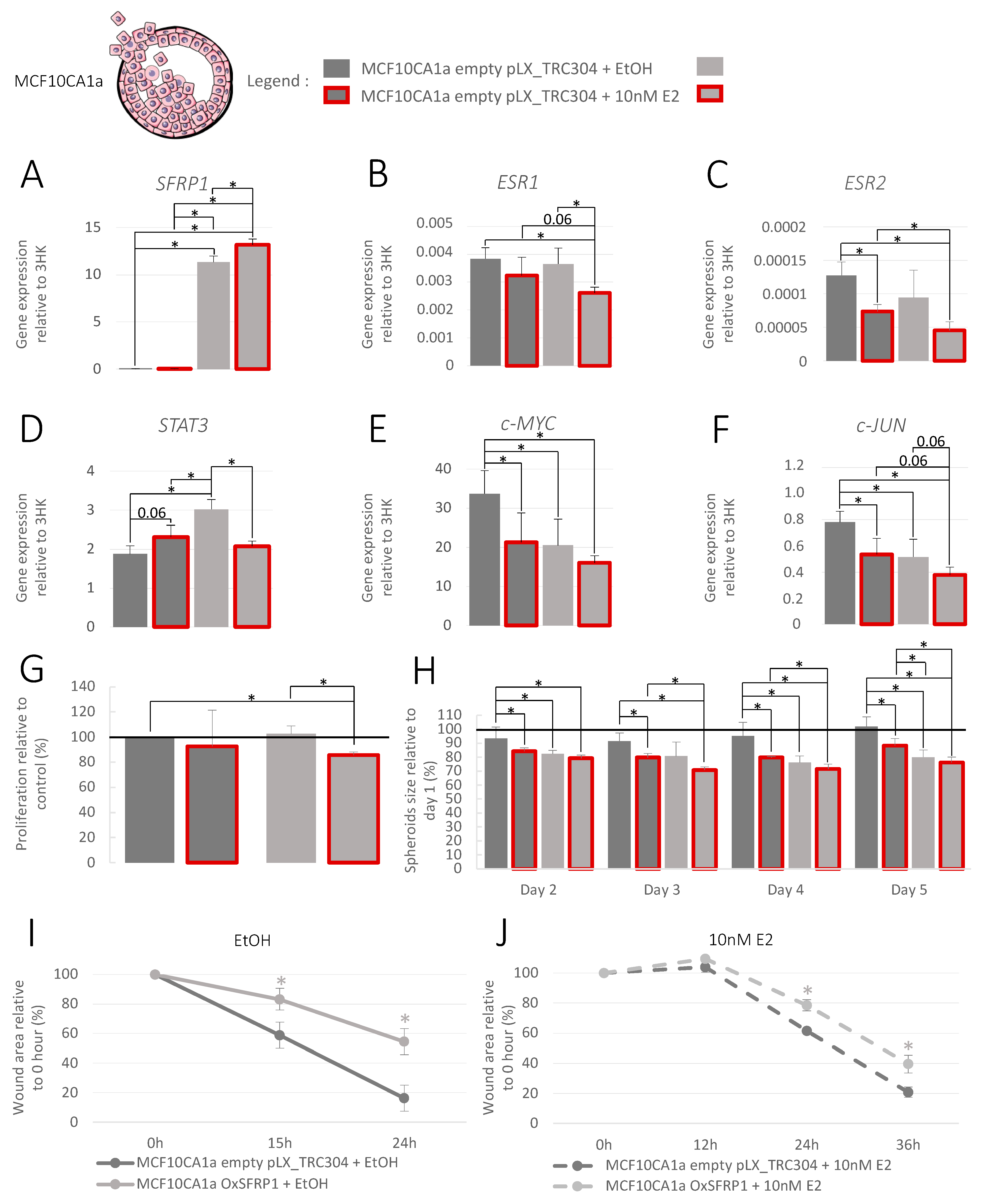
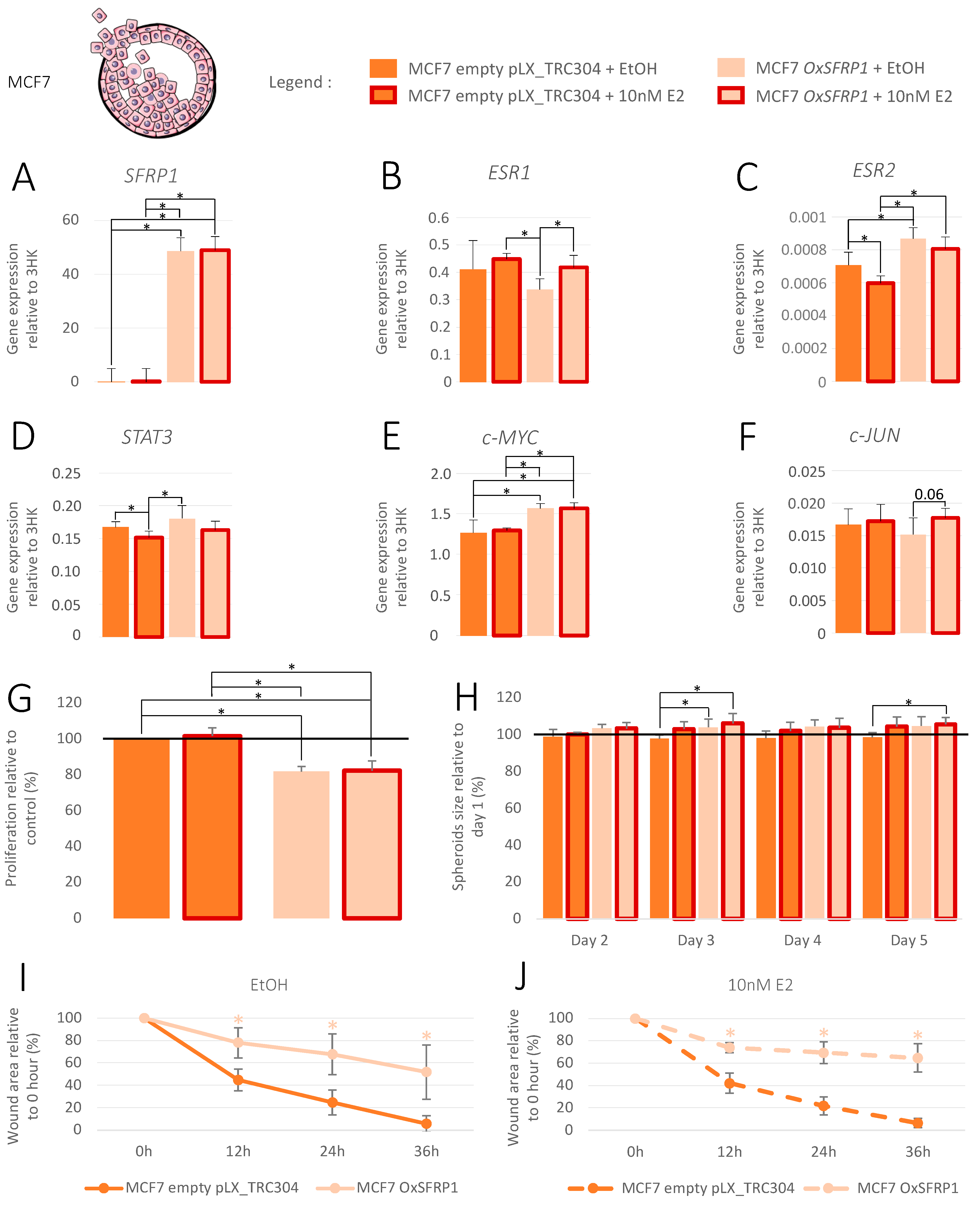
3.4. Increase in SFRP1 Expression in Pre-Invasive Cell Line
3.5. Increase in SFRP1 Expression in Triple-Negative Invasive Cell Line
3.6. Increase in SFRP1 Expression in Luminal A Invasive Cell Line
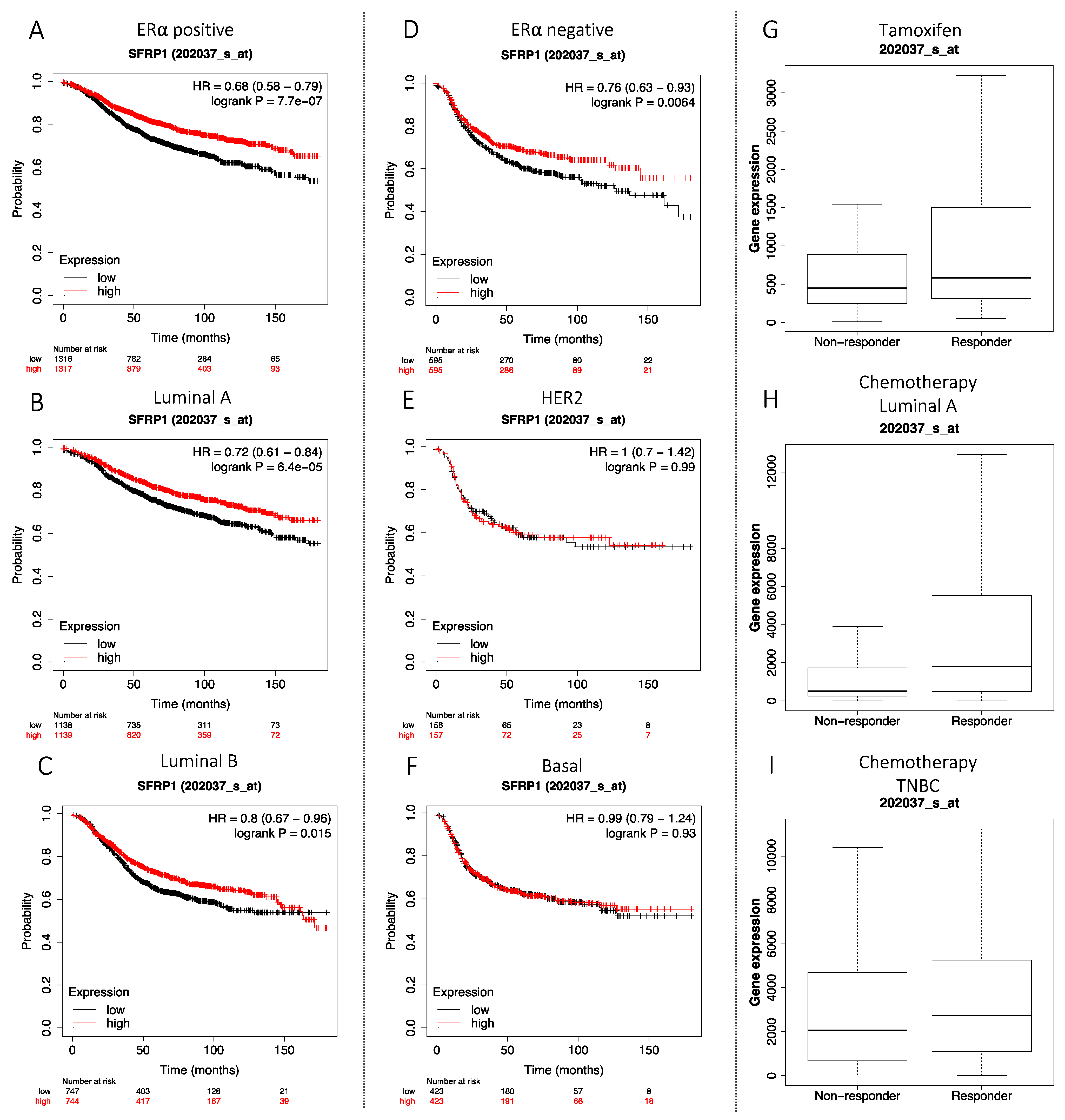
3.7. Association between SFRP1 Expression and Breast Cancer Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D. Strategies for Subtypes—Dealing with the Diversity of Breast Cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B. Personalizing the Treatment of Women with Early Breast Cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular Portraits of Human Breast Tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.M.; Yeo, S.K.; Holm, T.M.; Shaughnessy, E.; Guan, J.-L. Heterogeneity within Molecular Subtypes of Breast Cancer. Am. J. Physiol.-Cell Physiol. 2021, 321, C343–C354. [Google Scholar] [CrossRef]
- Keeling, J.W.; Özer, E.; King, G.; Walker, F. Oestrogen Receptor Alpha in Female Fetal, Infant, and Child Mammary Tissue. J. Pathol. 2000, 191, 449–451. [Google Scholar]
- Bartow, S.A. Use of the Autopsy to Study Ontogeny and Expression of the Estrogen Receptor Gene in Human Breast. J. Mammary Gland. Biol. Neoplasia 1998, 3, 37–48. [Google Scholar] [CrossRef]
- Naccarato, A.G.; Viacava, P.; Vignati, S.; Fanelli, G.; Bonadio, A.G.; Montruccoli, G.; Bevilacqua, G. Bio-Morphological Events in the Development of the Human Female Mammary Gland from Fetal Age to Puberty. Virchows Arch. 2000, 436, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Markopoulos, C.; Berger, U.; Wilson, P.; Gazet, J.-C.; Coombes, R.C. Oestrogen Receptor Content of Normal Breast Cells and Breast Carcinomas throughout the Menstrual Cycle. Br. Med. J. 1988, 296, 1349–1351. [Google Scholar] [CrossRef]
- Shoker, B.S.; Jarvis, C.; Sibson, D.R.; Walker, C.; Sloane, J.P. Oestrogen Receptor Expression in the Normal and Pre-Cancerous Breast. J. Pathol. 1999, 188, 237–244. [Google Scholar] [CrossRef]
- Walker, R.; Martin, C. The Aged Breast. J. Pathol. 2007, 211, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Hovey, R.C.; Trott, J.F.; Vonderhaar, B.K. Establishing a Framework for the Functional Mammary Gland: From Endocrinology to Morphology. J. Mammary Gland Biol. Neoplasia 2002, 7, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.; Ao, X.; Grill, C.; Russo, I.H. Pattern of Distribution of Cells Positive for Estrogen Receptor α and Progesterone Receptor in Relation to Proliferating Cells in the Mammary Gland. Breast Cancer Res. Treat. 1999, 53, 217–227. [Google Scholar] [CrossRef]
- Gaudet, M.M.; Press, M.F.; Haile, R.W.; Lynch, C.F.; Glaser, S.L.; Schildkraut, J.; Gammon, M.D.; Douglas Thompson, W.; Bernstein, J.L. Risk Factors by Molecular Subtypes of Breast Cancer across a Population-Based Study of Women 56 Years or Younger. Breast Cancer Res. Treat. 2011, 130, 587–597. [Google Scholar] [CrossRef]
- Ntirenganya, F.; Twagirumukiza, J.D.; Bucyibaruta, G.; Rugwizangoga, B.; Rulisa, S. Premenopausal Breast Cancer Risk Factors and Associations with Molecular Subtypes: A Case-Control Study. Int. J. Breast Cancer 2021, 2021, 5560559. [Google Scholar] [CrossRef] [PubMed]
- Collaborative Group on Hormonal Factors in Breast Cancer. Breast cancer and breastfeeding: Collaborative reanalysis of individual data from 47 epidemiological studies in 30 countries, including 50302 women with breast cancer and 96973 women without the disease. Lancet 2002, 360, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Ellingjord-Dale, M.; Vos, L.; Tretli, S.; Hofvind, S.; dos-Santos-Silva, I.; Ursin, G. Parity, Hormones and Breast Cancer Subtypes—Results from a Large Nested Case-Control Study in a National Screening Program. Breast Cancer Res. 2017, 19, 10. [Google Scholar] [CrossRef]
- Turkoz, F.P.; Solak, M.; Petekkaya, I.; Keskin, O.; Kertmen, N.; Sarici, F.; Arik, Z.; Babacan, T.; Ozisik, Y.; Altundag, K. Association between Common Risk Factors and Molecular Subtypes in Breast Cancer Patients. Breast 2013, 22, 344–350. [Google Scholar] [CrossRef]
- Barnard, M.E.; Boeke, C.E.; Tamimi, R.M. Established Breast Cancer Risk Factors and Risk of Intrinsic Tumor Subtypes. Biochim. Biophys. Acta BBA—Rev. Cancer 2015, 1856, 73–85. [Google Scholar] [CrossRef]
- Kothari, C.; Ouellette, G.; Labrie, Y.; Jacob, S.; Diorio, C.; Durocher, F. Identification of a Gene Signature for Different Stages of Breast Cancer Development That Could Be Used for Early Diagnosis and Specific Therapy. Oncotarget 2018, 9, 37407. [Google Scholar] [CrossRef]
- Clemenceau, A.; Diorio, C.; Durocher, F. Role of Secreted Frizzled-Related Protein 1 in Early Mammary Gland Tumorigenesis and Its Regulation in Breast Microenvironment. Cells 2020, 9, 208. [Google Scholar] [CrossRef]
- Gauger, K.J.; Shimono, A.; Crisi, G.M.; Schneider, S. Loss of Sfrp1 Promotes Ductal Branching in the Murine Mammary Gland. BMC Dev. Biol. 2012, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Clemenceau, A.; Hanna, M.; Ennour-Idrissi, K.; Burguin, A.; Diorio, C.; Durocher, F. Secreted Frizzled-Related Protein 1 as a Biomarker against Incomplete Age-Related Lobular Involution and Microcalcifications’ Development. Cancers 2020, 12, 2693. [Google Scholar] [CrossRef] [PubMed]
- Alsheh Ali, M.; Czene, K.; Hall, P.; Humphreys, K. Association of Microcalcification Clusters with Short-Term Invasive Breast Cancer Risk and Breast Cancer Risk Factors. Sci. Rep. 2019, 9, 14604. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot.Com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Park, S.-J.; Yoon, B.-H.; Kim, S.-K.; Kim, S.-Y. GENT2: An Updated Gene Expression Database for Normal and Tumor Tissues. BMC Med. Genom. 2019, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Lánczky, A.; Menyhárt, O.; Győrffy, B. Validation of MiRNA Prognostic Power in Hepatocellular Carcinoma Using Expression Data of Independent Datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef]
- Burstein, H.J.; Curigliano, G.; Thürlimann, B.; Weber, W.P.; Poortmans, P.; Regan, M.M.; Senn, H.J.; Winer, E.P.; Gnant, M.; Aebi, S.; et al. Customizing Local and Systemic Therapies for Women with Early Breast Cancer: The St. Gallen International Consensus Guidelines for Treatment of Early Breast Cancer 2021. Ann. Oncol. 2021, 32, 1216–1235. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The Prognostic Landscape of Genes and Infiltrating Immune Cells across Human Cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef]
- Lacouture, A.; Jobin, C.; Weidmann, C.; Berthiaume, L.; Bastien, D.; Laverdière, I.; Pelletier, M.; Audet-Walsh, É. A FACS-Free Purification Method to Study Estrogen Signaling, Organoid Formation, and Metabolic Reprogramming in Mammary Epithelial Cells. Front. Endocrinol. 2021, 12, 672466. [Google Scholar] [CrossRef]
- Klaus, A.; Birchmeier, W. Wnt Signalling and Its Impact on Development and Cancer. Nat. Rev. Cancer 2008, 8, 387–398. [Google Scholar] [CrossRef]
- Ma, J.; Qin, L.; Li, X. Role of STAT3 Signaling Pathway in Breast Cancer. Cell Commun. Signal. 2020, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Fallah, Y.; Brundage, J.; Allegakoen, P.; Shajahan-Haq, A.N. MYC-Driven Pathways in Breast Cancer Subtypes. Biomolecules 2017, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Sinha, I.; Fan, R.; Haldosen, L.-A.; Yan, F.; Zhao, C.; Dahlman-Wright, K. C-Jun/AP-1 Overexpression Reprograms ERα Signaling Related to Tamoxifen Response in ERα-Positive Breast Cancer. Oncogene 2018, 37, 2586–2600. [Google Scholar] [CrossRef] [PubMed]
- Furth, P.A.; Bar-Peled, U.; Li, M. Apoptosis and Mammary Gland Involution: Reviewing the Process. Apoptosis 1997, 2, 19–24. [Google Scholar] [CrossRef]
- Li, M.; Liu, X.; Robinson, G.; Bar-Peled, U.; Wagner, K.-U.; Young, W.S.; Hennighausen, L.; Furth, P.A. Mammary-Derived Signals Activate Programmed Cell Death during the First Stage of Mammary Gland Involution. Proc. Natl. Acad. Sci. USA 1997, 94, 3425–3430. [Google Scholar] [CrossRef]
- Jerry, D.J.; Dickinson, E.S.; Roberts, A.L.; Said, T.K. Regulation of Apoptosis during Mammary Involution by the P53 Tumor Suppressor Gene. J. Dairy Sci. 2002, 85, 1103–1110. [Google Scholar] [CrossRef]
- Baxter, F.O.; Neoh, K.; Tevendale, M.C. The Beginning of the End: Death Signaling in Early Involution. J. Mammary Gland. Biol. Neoplasia 2007, 12, 3–13. [Google Scholar] [CrossRef]
- Fekete, J.T.; Győrffy, B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3104 breast cancer patients. Int. J. Cancer 2019, 145, 3140–3151. [Google Scholar] [CrossRef]
- Veeck, J.; Niederacher, D.; An, H.; Klopocki, E.; Wiesmann, F.; Betz, B.; Galm, O.; Camara, O.; Dürst, M.; Kristiansen, G.; et al. Aberrant Methylation of the Wnt Antagonist SFRP1 in Breast Cancer Is Associated with Unfavourable Prognosis. Oncogene 2006, 25, 3479–3488. [Google Scholar] [CrossRef]
- Dahl, E.; Wiesmann, F.; Woenckhaus, M.; Stoehr, R.; Wild, P.J.; Veeck, J.; Knüchel, R.; Klopocki, E.; Sauter, G.; Simon, R.; et al. Frequent Loss of SFRP1 Expression in Multiple Human Solid Tumours: Association with Aberrant Promoter Methylation in Renal Cell Carcinoma. Oncogene 2007, 26, 5680–5691. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, N.; Bhattacharya, N.; Alam, N.; Roy, A.; Roychoudhury, S.; Panda, C.K. Subtype-Specific Alterations of the Wnt Signaling Pathway in Breast Cancer: Clinical and Prognostic Significance. Cancer Sci. 2012, 103, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Bernemann, C.; Hülsewig, C.; Ruckert, C.; Schäfer, S.; Blümel, L.; Hempel, G.; Götte, M.; Greve, B.; Barth, P.J.; Kiesel, L.; et al. Influence of Secreted Frizzled Receptor Protein 1 (SFRP1) on Neoadjuvant Chemotherapy in Triple Negative Breast Cancer Does Not Rely on WNT Signaling. Mol. Cancer 2014, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Huth, L.; Rose, M.; Kloubert, V.; Winkens, W.; Schlensog, M.; Hartmann, A.; Knüchel, R.; Dahl, E. BDNF Is Associated with SFRP1 Expression in Luminal and Basal-Like Breast Cancer Cell Lines and Primary Breast Cancer Tissues: A Novel Role in Tumor Suppression? PLoS ONE 2014, 9, e102558. [Google Scholar] [CrossRef]
- Gregory, K.J.; Schneider, S.S. Estrogen-Mediated Signaling Is Differentially Affected by the Expression Levels of Sfrp1 in Mammary Epithelial Cells: Estrogen Signaling and Sfrp1 Expression. Cell Biol. Int. 2015, 39, 873–879. [Google Scholar] [CrossRef]
- Li, Z.; Heng, J.; Yan, J.; Guo, X.; Tang, L.; Chen, M.; Peng, L.; Wu, Y.; Wang, S.; Xiao, Z.; et al. Integrated Analysis of Gene Expression and Methylation Profiles of 48 Candidate Genes in Breast Cancer Patients. Breast Cancer Res. Treat. 2016, 160, 371–383. [Google Scholar] [CrossRef]
- Zhu, Q.-N.; Renaud, H.; Guo, Y. Bioinformatics-Based Identification of MiR-542-5p as a Predictive Biomarker in Breast Cancer Therapy. Hereditas 2018, 155. [Google Scholar] [CrossRef]
- Gregory, K.J.; Roberts, A.L.; Conlon, E.M.; Mayfield, J.A.; Hagen, M.J.; Crisi, G.M.; Bentley, B.A.; Kane, J.J.; Makari-Judson, G.; Mason, H.S.; et al. Gene Expression Signature of Atypical Breast Hyperplasia and Regulation by SFRP1. Breast Cancer Res. 2019, 21, 76. [Google Scholar] [CrossRef]
- Poorolajal, J.; Heidarimoghis, F.; Karami, M.; Cheraghi, Z.; Gohari-Ensaf, F.; Shahbazi, F.; Zareie, B.; Ameri, P.; Sahraei, F. Factors for the Primary Prevention of Breast Cancer: A Meta-Analysis of Prospective Cohort Studies. J. Res. Health Sci. 2021, 21, e00520. [Google Scholar] [CrossRef]
- Russo, J.; Hu, Y.F.; Silva, I.D.C.G.; Russo, I.H. Cancer Risk Related to Mammary Gland Structure and Development. Microsc. Res. Tech. 2001, 52, 204–223. [Google Scholar] [CrossRef]
- dos Santos, C.O.; Dolzhenko, E.; Hodges, E.; Smith, A.D.; Hannon, G.J. An Epigenetic Memory of Pregnancy in the Mouse Mammary Gland. Cell Rep. 2015, 11, 1102–1109. [Google Scholar] [CrossRef]
- Feigman, M.J.; Moss, M.A.; Chen, C.; Cyrill, S.L.; Ciccone, M.F.; Trousdell, M.C.; Yang, S.-T.; Frey, W.D.; Wilkinson, J.E.; dos Santos, C.O. Pregnancy Reprograms the Epigenome of Mammary Epithelial Cells and Blocks the Development of Premalignant Lesions. Nat. Commun. 2020, 11, 2649. [Google Scholar] [CrossRef] [PubMed]
- Kanadys, W.; Barańska, A.; Malm, M.; Błaszczuk, A.; Polz-Dacewicz, M.; Janiszewska, M.; Jędrych, M. Use of Oral Contraceptives as a Potential Risk Factor for Breast Cancer: A Systematic Review and Meta-Analysis of Case-Control Studies Up to 2010. Int. J. Environ. Res. Public Health 2021, 18, 4638. [Google Scholar] [CrossRef] [PubMed]
- Veltmaat, J.M.; Ramsdell, A.F.; Sterneck, E. Positional Variations in Mammary Gland Development and Cancer. J. Mammary Gland Biol. Neoplasia 2013, 18, 179–188. [Google Scholar] [CrossRef]
- Garfinkel, L.; Craig, L.; Seidman, H. An appraisal of left and right breast cancer. J. Natl. Cancer Inst. 1959, 23, 617–631. [Google Scholar]
- Clancy, L.J.; Leitch, A.G. hypercapnia during oxygen therapy. Lancet 1977, 310, 763–764. [Google Scholar] [CrossRef]
- Senie, R.T.; Rosen, P.P.; Lesser, M.L.; Snyder, R.E.; Schottenfeld, D.; Duthie, K. Epidemiology of Breast Carcinoma II: Factors Related to the Predominance of Left-Sided Disease. Cancer 1980, 46, 1705–1713. [Google Scholar] [CrossRef]
- Tulinius, H.; Bjarnason, O.; Sigvaldason, H.; Bjarnadottir, G.; Olafsdottir, G. Tumours in Iceland 10. Malignant Tumours of the Female Breast. APMIS 1988, 96, 229–238. [Google Scholar] [CrossRef]
- Ekbom, A.; Adami, H.-O.; Trichopoulos, D.; Lambe, M.; Hsieh, C.; Pontén, J. Epidemiologic Correlates of Breast Cancer Laterality (Sweden). Cancer Causes Control 1994, 5, 510–516. [Google Scholar] [CrossRef]
- Weiss, H.A.; Devesa, S.S.; Brinton, L.A. Laterality of Breast Cancer in the United States. Cancer Causes Control 1996, 7, 539–543. [Google Scholar] [CrossRef]
- Roychoudhuri, R.; Putcha, V.; Møller, H. Cancer and Laterality: A Study of The Five Major Paired Organs (UK). Cancer Causes Control 2006, 17, 655–662. [Google Scholar] [CrossRef]
- Fatima, N.; Zaman, M.U.; Maqbool, A.; Khan, S.H.; Riaz, N. Lower Incidence but More Aggressive Behavior of Right Sided Breast Cancer in Pakistani Women: Does Right Deserve More Respect? Asian Pac. J. Cancer Prev. 2013, 14, 43–45. [Google Scholar] [CrossRef]
- Sun, X.; Nichols, H.B.; Tse, C.-K.; Bell, M.B.; Robinson, W.R.; Sherman, M.E.; Olshan, A.F.; Troester, M.A. Association of Parity and Time since Last Birth with Breast Cancer Prognosis by Intrinsic Subtype. Cancer Epidemiol. Biomark. Prev. 2016, 25, 60–67. [Google Scholar] [CrossRef]
- Butt, S.; Borgquist, S.; Garne, J.P.; Landberg, G.; Tengrup, I.; Olsson, Å.; Manjer, J. Parity in Relation to Survival Following Breast Cancer. Eur. J. Surg. Oncol. EJSO 2009, 35, 702–708. [Google Scholar] [CrossRef]

| Cell Lines | Type of Lesion | Molecular Subtype | Stade/Grade | Age (Years) | Culture Medium |
|---|---|---|---|---|---|
| MCF10A | Normal | NA | NA | 36 |
|
| MCF10AT1 | ADH | NA | NA | ||
| MCF10DCIS.com | DCIS | NA | NA |
| |
| MCF10CA1 | IDC | Basal-like | NA |
| |
| MCF7 | IDC | Luminal A | Stage 4 | 69 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clemenceau, A.; Lacouture, A.; Bherer, J.; Ouellette, G.; Michaud, A.; Audet-Walsh, É.; Diorio, C.; Durocher, F. Role of Secreted Frizzled-Related Protein 1 in Early Breast Carcinogenesis and Breast Cancer Aggressiveness. Cancers 2023, 15, 2251. https://doi.org/10.3390/cancers15082251
Clemenceau A, Lacouture A, Bherer J, Ouellette G, Michaud A, Audet-Walsh É, Diorio C, Durocher F. Role of Secreted Frizzled-Related Protein 1 in Early Breast Carcinogenesis and Breast Cancer Aggressiveness. Cancers. 2023; 15(8):2251. https://doi.org/10.3390/cancers15082251
Chicago/Turabian StyleClemenceau, Alisson, Aurélie Lacouture, Juliette Bherer, Geneviève Ouellette, Annick Michaud, Étienne Audet-Walsh, Caroline Diorio, and Francine Durocher. 2023. "Role of Secreted Frizzled-Related Protein 1 in Early Breast Carcinogenesis and Breast Cancer Aggressiveness" Cancers 15, no. 8: 2251. https://doi.org/10.3390/cancers15082251
APA StyleClemenceau, A., Lacouture, A., Bherer, J., Ouellette, G., Michaud, A., Audet-Walsh, É., Diorio, C., & Durocher, F. (2023). Role of Secreted Frizzled-Related Protein 1 in Early Breast Carcinogenesis and Breast Cancer Aggressiveness. Cancers, 15(8), 2251. https://doi.org/10.3390/cancers15082251






