Epigenetic Modifications in Prostate Cancer Metastasis and Microenvironment
Abstract
Simple Summary
Abstract
1. Introduction
2. Chromatin-Related Regulation
2.1. DNA Methylation
2.2. Histone Modification
2.3. Nucleosome Positioning
2.4. Higher-Order Chromatin Organization
3. RNA Modification
3.1. m6A
3.2. m6Am
3.3. m1A
3.4. m5C
3.5. ac4C
3.6. Ψ
3.7. m7G
4. Epigenetic Modifications in PCa
5. Epigenetic Modifications in Tumor Microenvironment (TME)
6. Epigenetic Modifications in Cancer Therapy
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Feinberg, A.P.; Levchenko, A. Epigenetics as a mediator of plasticity in cancer. Science 2023, 379, eaaw3835. [Google Scholar] [CrossRef]
- Kawasaki, F.; Beraldi, D.; Hardisty, R.E.; McInroy, G.R.; van Delft, P.; Balasubramanian, S. Genome-wide mapping of 5-hydroxymethyluracil in the eukaryote parasite Leishmania. Genome Biol. 2017, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Baginski, B.; Wirecki, T.K.; De Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Goel, S.; Bhatia, V.; Biswas, T.; Ateeq, B. Epigenetic reprogramming during prostate cancer progression: A perspective from development. Semin. Cancer Biol. 2022, 83, 136–151. [Google Scholar] [CrossRef] [PubMed]
- López, J.; Añazco-Guenkova, A.M.; Monteagudo-García, Ó.; Blanco, S. Epigenetic and Epitranscriptomic Control in Prostate Cancer. Genes 2022, 13, 378. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’His, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Skvortsova, K.; Stirzaker, C.; Taberlay, P. The DNA methylation landscape in cancer. Essays Biochem. 2019, 63, 797–811. [Google Scholar] [CrossRef]
- Maruyama, R.; Toyooka, S.; Toyooka, K.O.; Virmani, A.K.; Zöchbauer-Müller, S.; Farinas, A.J.; Minna, J.D.; McConnell, J.; Frenkel, E.P.; Gazdar, A.F. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin. Cancer Res. 2002, 8, 514–519. [Google Scholar]
- Nakayama, M.; Bennett, C.J.; Hicks, J.L.; Epstein, J.I.; Platz, E.A.; Nelson, W.G.; De Marzo, A.M. Hypermethylation of the human glutathione S-transferase-pi gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: A detailed study using laser-capture microdissection. Am. J. Pathol. 2003, 163, 923–933. [Google Scholar]
- Dryhurst, D.; Ausio, J. Histone H2A.Z deregulation in prostate cancer. Cause or effect? Cancer Metastasis Rev. 2014, 33, 429–439. [Google Scholar] [CrossRef]
- He, H.H.; Meyer, A.C.; Shin, H.; Bailey, S.T.; Wei, G.; Wang, Q.; Zhang, Y.; Xu, K.; Ni, M.; Lupien, M.; et al. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 2010, 42, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Hawley, J.R.; Zhou, S.; Arlidge, C.; Grillo, G.; Kron, K.J.; Hugh-White, R.; van der Kwast, T.H.; Fraser, M.; Boutros, P.C.; Bristow, R.G.; et al. Reorganization of the 3D Genome Pinpoints Noncoding Drivers of Primary Prostate Tumors. Cancer Res. 2021, 81, 5833–5848. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2016, 14, 23–31, Erratum in Nat. Methods 2017, 14, 323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Y.; Zeng, Y. Research progress of m(6)A methylation in prostate cancer. Asian J. Androl. 2023, 25, 166–170. [Google Scholar] [CrossRef]
- Ji, G.; Huang, C.; He, S.; Gong, Y.; Song, G.; Li, X.; Zhou, L. Comprehensive analysis of m6A regulators prognostic value in prostate cancer. Aging 2020, 12, 14863–14884. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Zhang, J.; Su, J.; Zuo, Z.; Zeng, L.; Liu, K.; Zheng, Y.; Huang, X.; Bai, R.; Zhuang, L.; et al. RNA m6A regulates transcription via DNA demethylation and chromatin accessibility. Nat. Genet. 2022, 54, 1427–1437. [Google Scholar] [CrossRef]
- Urabe, F.; Yamamoto, Y.; Kimura, T. miRNAs in prostate cancer: Intercellular and extracellular communications. Int. J. Urol. 2022, 29, 1429–1438. [Google Scholar] [CrossRef]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Wolffe, A.P.; Matzke, M.A. Epigenetics: Regulation Through Repression. Science 1999, 286, 481–486. [Google Scholar] [CrossRef]
- Massie, C.E.; Mills, I.G.; Lynch, A.G. The importance of DNA methylation in prostate cancer development. J. Steroid Biochem. Mol. Biol. 2017, 166, 1–15. [Google Scholar] [CrossRef]
- Luo, J.-H.; Ding, Y.; Chen, R.; Michalopoulos, G.; Nelson, J.; Tseng, G.; Yu, Y.P. Genome-Wide Methylation Analysis of Prostate Tissues Reveals Global Methylation Patterns of Prostate Cancer. Am. J. Pathol. 2013, 182, 2028–2036. [Google Scholar] [CrossRef]
- Yang, B.; Bhusari, S.; Kueck, J.; Weeratunga, P.; Wagner, J.; Leverson, G.; Huang, W.; Jarrard, D.F. Methylation Profiling Defines an Extensive Field Defect in Histologically Normal Prostate Tissues Associated with Prostate Cancer. Neoplasia 2013, 15, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.O. DNA methylation changes in prostate cancer: Current developments and future clinical implementation. Expert Rev. Mol. Diagn. 2009, 9, 243–257. [Google Scholar] [CrossRef]
- Sjöström, M.; Zhao, S.G.; Levy, S.; Zhang, M.; Ning, Y.; Shrestha, R.; Lundberg, A.; Herberts, C.; Foye, A.; Aggarwal, R.; et al. The 5-Hydroxymethylcytosine Landscape of Prostate Cancer. Cancer Res. 2022, 82, 3888–3902. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Rando, O.J.; Chang, H.Y. Genome-Wide Views of Chromatin Structure. Annu. Rev. Biochem. 2009, 78, 245–271. [Google Scholar] [CrossRef]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of Alternative Splicing by Histone Modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef]
- Huertas, D.; Sendra, R.; Muñoz, P. Chromatin dynamics coupled to DNA repair. Epigenetics 2009, 4, 31–42. [Google Scholar] [CrossRef]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.A.B.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Wissmann, M.; Yin, N.; Müller, J.M.; Schneider, R.; Peters, A.H.F.M.; Günther, T.; Buettner, R.; Schüle, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Gaughan, L.; Stockley, J.; Wang, N.; McCracken, S.R.; Treumann, A.; Armstrong, K.; Shaheen, F.; Watt, K.; McEwan, I.J.; Wang, C.; et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res. 2011, 39, 1266–1279. [Google Scholar] [CrossRef] [PubMed]
- Seligson, D.B.; Horvath, S.; Shi, T.; Yu, H.; Tze, S.; Grunstein, M.; Kurdistani, S.K. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005, 435, 1262–1266. [Google Scholar] [CrossRef]
- Bianco-Miotto, T.; Chiam, K.; Buchanan, G.; Jindal, S.; Day, T.K.; Thomas, M.; Pickering, M.A.; O’Loughlin, M.A.; Ryan, N.K.; Raymond, W.A.; et al. Global Levels of Specific Histone Modifications and an Epigenetic Gene Signature Predict Prostate Cancer Progression and Development. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2611–2622. [Google Scholar] [CrossRef]
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2006, 39, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Kn, H.; Chow, M.Z.; Baker, E.K.; Pal, S.; Bassal, S.; Brasacchio, D.; Wang, L.; Craig, J.M.; Jones, P.L.; Sif, S.; et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005, 37, 254–264. [Google Scholar]
- Attard, G.; Paeker, C.; Eeles, A.R.; Schröder, F.; Tomlins, A.S.; Tannock, I.; Drake, G.C.; Bono, S.J. Prostate cancer. Lancet 2016, 387, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gulzar, Z.G.; Salari, K.; Lapointe, J.; Brooks, J.D.; Pollack, J.R. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene 2012, 31, 4164–4170. [Google Scholar] [CrossRef]
- Reddy, K.L.; Feinberg, A.P. Higher order chromatin organization in cancer. Semin. Cancer Biol. 2013, 23, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Taberlay, P.C.; Achinger-Kawecka, J.; Lun, A.T.; Buske, F.A.; Sabir, K.; Gould, C.M.; Zotenko, E.; Bert, S.A.; Giles, K.A.; Bauer, D.C.; et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 2016, 26, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Staibano, S.; Mascolo, M.; Mancini, F.P.; Kisslinger, A.; Salvatore, G.; Di Benedetto, M.; Chieffi, P.; Altieri, V.; Prezioso, D.; Ilardi, G.; et al. Overexpression of chromatin assembly factor-1 (CAF-1) p60 is predictive of adverse behaviour of prostatic cancer. Histopathology 2009, 54, 580–589. [Google Scholar] [CrossRef]
- Ahmed, M.; Soares, F.; Xia, J.-H.; Yang, Y.; Li, J.; Guo, H.; Su, P.; Tian, Y.; Lee, H.J.; Wang, M.; et al. CRISPRi screens reveal a DNA methylation-mediated 3D genome dependent causal mechanism in prostate cancer. Nat. Commun. 2021, 12, 1781. [Google Scholar] [CrossRef] [PubMed]
- Wiener, D.; Schwartz, S. The epitranscriptome beyond m6A. Nat. Rev. Genet. 2021, 22, 119–131. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Somasekharan, S.P.; Saxena, N.; Zhang, F.; Beraldi, E.; Ni Huang, J.; Gentle, C.; Fazli, L.; Thi, M.; Sorensen, P.H.; Gleave, M. Regulation of AR mRNA translation in response to acute AR pathway inhibition. Nucleic Acids Res. 2022, 50, 1069–1091. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhong, J.; Zeng, J.; Duan, X.; Lu, J.; Sun, X.; Liu, Q.; Liang, Y.; Lin, Z.; Zhong, W.; et al. Characterization of the m6A-Associated Tumor Immune Microenvironment in Prostate Cancer to Aid Immunotherapy. Front. Immunol. 2021, 12, 735170. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.-J.; Chen, Q.; et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Liu, J.; Li, K.; Cai, J.; Zhang, M.; Zhang, X.; Xiong, X.; Meng, H.; Xu, X.; Huang, Z.; Peng, J.; et al. Landscape and Regulation of m6A and m6Am Methylome across Human and Mouse Tissues. Mol. Cell 2020, 77, 426–440.e6. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, M.; Li, K.; Bai, D.; Yi, C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019, 29, 80–82. [Google Scholar] [CrossRef]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Jin, M.-Z.; Zhang, Y.-G.; Jin, W.-L.; Wang, X.-P. A Pan-Cancer Analysis of the Oncogenic and Immunogenic Role of m6Am Methyltransferase PCIF1. Front. Oncol. 2021, 11, 753393. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Wang, H. N1-methyladenosine modification in cancer biology: Current status and future perspectives. Comput. Struct. Biotechnol. J. 2022, 20, 6578–6585. [Google Scholar] [CrossRef] [PubMed]
- Legrand, C.; Tuorto, F.; Hartmann, M.; Liebers, R.; Jacob, D.; Helm, M.; Lyko, F. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017, 27, 1589–1596. [Google Scholar] [CrossRef]
- Li, X.; Xiong, X.; Zhang, M.; Wang, K.; Chen, Y.; Zhou, J.; Mao, Y.; Lv, J.; Yi, D.; Chen, X.-W.; et al. Base-Resolution Mapping Reveals Distinct m1A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol. Cell 2017, 68, 993–1005.e9. [Google Scholar] [CrossRef]
- Macon, J.B.; Wolfenden, R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry 1968, 7, 3453–3458. [Google Scholar] [CrossRef] [PubMed]
- García-Vílchez, R.; Sevilla, A.; Blanco, S. Post-transcriptional regulation by cytosine-5 methylation of RNA. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Bujnicki, J.M.; Feder, M.; Ayres, C.L.; Redman, K.L. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004, 32, 2453–2463. [Google Scholar] [CrossRef]
- Nombela, P.; Miguel-Lopez, B.; Blanco, S. The role of m6A, m5C and Psi RNA modifications in cancer: Novel therapeutic opportunities. Mol. Cancer 2021, 20, 18. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, S.; Zhang, Y.; Liu, R.; Chen, M. Roles of m5C RNA Modification Patterns in Biochemical Recurrence and Tumor Microenvironment Characterization of Prostate Adenocarcinoma. Front. Immunol. 2022, 13, 869759. [Google Scholar] [CrossRef]
- Yu, G.; Bao, J.; Zhan, M.; Wang, J.; Li, X.; Gu, X.; Song, S.; Yang, Q.; Liu, Y.; Wang, Z.; et al. Comprehensive Analysis of m5C Methylation Regulatory Genes and Tumor Microenvironment in Prostate Cancer. Front. Immunol. 2022, 13, 914577. [Google Scholar] [CrossRef]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Arango, D.; Sturgill, D.; Alhusaini, N.; Dillman, A.A.; Sweet, T.J.; Hanson, G.; Hosogane, M.; Sinclair, W.R.; Nanan, K.K.; Mandler, M.D.; et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 2018, 175, 1872–1886.e24. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Liu, H.; Wu, Y.; Yao, M.; Zhang, B. Inhibition of N-Acetyltransferase 10 Suppresses the Progression of Prostate Cancer through Regulation of DNA Replication. Int. J. Mol. Sci. 2022, 23, 6573. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Guerrieri, A.N.; Zacchini, F.; Treré, D.; Montanaro, L. RNA Pseudouridylation in Physiology and Medicine: For Better and for Worse. Genes 2017, 8, 301. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef]
- Yu, Y.-T.; Meier, U.T. RNA-guided isomerization of uridine to pseudouridine—Pseudouridylation. RNA Biol. 2014, 11, 1483–1494. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef]
- McMahon, M.; Contreras, A.; Ruggero, D. Small RNAs with big implications: New insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip. Rev. RNA 2015, 6, 173–189. [Google Scholar] [CrossRef]
- Martens-Uzunova, E.S.; Jalava, E.S.; Dits, N.F.; van Leenders, G.J.L.H.; Møller, S.; Trapman, J.; Bangma, C.H.; Litman, T.; Visakorpi, T.; Jenster, G. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene 2012, 31, 978–991. [Google Scholar] [CrossRef]
- Crea, F.; Quagliata, L.; Michael, A.; Liu, H.H.; Frumento, P.; Azad, A.A.; Xue, H.; Pikor, L.; Watahiki, A.; Morant, R.; et al. Integrated analysis of the prostate cancer small-nucleolar transcriptome reveals SNORA55 as a driver of prostate cancer progression. Mol. Oncol. 2016, 10, 693–703. [Google Scholar] [CrossRef]
- Luo, Y.; Yao, Y.; Wu, P.; Zi, X.; Sun, N.; He, J. The potential role of N7-methylguanosine (m7G) in cancer. J. Hematol. Oncol. 2022, 15, 63. [Google Scholar] [CrossRef]
- Xin, S.; Deng, Y.; Mao, J.; Wang, T.; Liu, J.; Wang, S.; Song, X.; Song, W.; Liu, X. Characterization of 7-Methylguanosine Identified Biochemical Recurrence and Tumor Immune Microenvironment in Prostate Cancer. Front. Oncol. 2022, 12, 900203. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Li, F.; Takeda, D.Y.; Lenci, R.; Chonkar, A.; Chabot, M.S.; Cejas, P.; Vazquez, F.; Cook, J.; Shivdasani, R.A.; et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 2015, 47, 1346–1351. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Qiu, X.; Zhu, Y.; Takeda, D.Y.; Pan, W.; Baca, S.C.; Gusev, A.; Korthauer, K.; Severson, T.M.; Ha, G.; et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 2020, 52, 790–799. [Google Scholar] [CrossRef]
- Andreu-Vieyra, C.; Lai, J.; Berman, B.P.; Frenkel, B.; Jia, L.; Jones, P.A.; Coetzee, G.A. Dynamic Nucleosome-Depleted Regions at Androgen Receptor Enhancers in the Absence of Ligand in Prostate Cancer Cells. Mol. Cell. Biol. 2011, 31, 4648–4662. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.L.; Weinberg, R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013, 19, 1438–1449. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Tsai, Y.-P.; Wu, M.-Z.; Teng, S.-C.; Wu, K.-J. Epigenetic reprogramming and post-transcriptional regulation during the epithelial–mesenchymal transition. Trends Genet. 2012, 28, 454–463. [Google Scholar] [CrossRef]
- Ezponda, T.; Popovic, R.; Shah, M.Y.; Martinez-Garcia, E.; Zheng, Y.; Min, D.-J.; Will, C.; Neri, A.; Kelleher, N.L.; Yu, J.; et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial–mesenchymal transition and invasive properties of prostate cancer. Oncogene 2013, 32, 2882–2890. [Google Scholar] [CrossRef]
- Zavridou, M.; Strati, A.; Bournakis, E.; Smilkou, S.; Tserpeli, V.; Lianidou, E. Prognostic Significance of Gene Expression and DNA Methylation Markers in Circulating Tumor Cells and Paired Plasma Derived Exosomes in Metastatic Castration Resistant Prostate Cancer. Cancers 2021, 13, 780. [Google Scholar] [CrossRef]
- Friedlander, T.W.; Ngo, V.T.; Dong, H.; Premasekharan, G.; Weinberg, V.; Doty, S.; Zhao, Q.; Gilbert, E.G.; Ryan, C.J.; Chen, W.-T.; et al. Detection and characterization of invasive circulating tumor cells derived from men with metastatic castration-resistant prostate cancer. Int. J. Cancer 2014, 134, 2284–2293. [Google Scholar] [CrossRef]
- Cui, J.; Li, G.; Yin, J.; Li, L.; Tan, Y.; Wei, H.; Liu, B.; Deng, L.; Tang, J.; Chen, Y.; et al. GSTP1 and cancer: Expression, methylation, polymorphisms and signaling (Review). Int. J. Oncol. 2020, 56, 867–878. [Google Scholar] [CrossRef]
- Dubois, F.; Bergot, E.; Zalcman, G.; Levallet, G. RASSF1A, puppeteer of cellular homeostasis, fights tumorigenesis, and metastasis—An updated review. Cell Death Dis. 2019, 10, 928. [Google Scholar] [CrossRef] [PubMed]
- De Sarkar, N.; Patton, R.D.; Doebley, A.-L.; Hanratty, B.; Adil, M.; Kreitzman, A.J.; Sarthy, J.F.; Ko, M.; Brahma, S.; Meers, M.P.; et al. Nucleosome Patterns in Circulating Tumor DNA Reveal Transcriptional Regulation of Advanced Prostate Cancer Phenotypes. Cancer Discov. 2023, 13, 632–653. [Google Scholar] [CrossRef] [PubMed]
- Shiao, S.L.; Chu, G.C.-Y.; Chung, L.W. Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett. 2016, 380, 340–348. [Google Scholar] [CrossRef]
- Giannoni, E.; Bianchini, F.; Masieri, L.; Serni, S.; Torre, E.; Calorini, L.; Chiarugi, P. Reciprocal Activation of Prostate Cancer Cells and Cancer-Associated Fibroblasts Stimulates Epithelial-Mesenchymal Transition and Cancer Stemness. Cancer Res. 2010, 70, 6945–6956. [Google Scholar] [CrossRef]
- Zhao, D.; Cai, L.; Lu, X.; Liang, X.; Li, J.; Chen, P.; Ittmann, M.; Shang, X.; Jiang, S.; Li, H.; et al. Chromatin Regulator CHD1 Remodels the Immunosuppressive Tumor Microenvironment in PTEN-Deficient Prostate Cancer. Cancer Discov. 2020, 10, 1374–1387. [Google Scholar] [CrossRef]
- Taoka, R.; Tsukuda, F.; Ishikawa, M.; Haba, R.; Kakehi, Y. Association of prostatic inflammation with down-regulation of macrophage inhibitory cytokine-1 gene in symptomatic benign prostatic hyperplasia. J. Urol. 2004, 171, 2330–2335. [Google Scholar] [CrossRef]
- Bruzzese, F.; Hägglöf, C.; Leone, A.; Sjöberg, E.; Roca, M.S.; Kiflemariam, S.; Sjöblom, T.; Hammarsten, P.; Egevad, L.; Bergh, A.; et al. Local and Systemic Protumorigenic Effects of Cancer-Associated Fibroblast-Derived GDF15. Cancer Res. 2014, 74, 3408–3417. [Google Scholar] [CrossRef]
- Costa, V.L.; Henrique, R.; Danielsen, S.A.; Duarte-Pereira, S.; Eknaes, M.; Skotheim, R.I.; Rodrigues, Â.; Magalhães, J.S.; Oliveira, J.; Lothe, R.A.; et al. Three Epigenetic Biomarkers, GDF15, TMEFF2, and VIM, Accurately Predict Bladder Cancer from DNA-Based Analyses of Urine Samples. Clin. Cancer Res. 2010, 16, 5842–5851. [Google Scholar] [CrossRef]
- Ippolito, L.; Comito, G.; Parri, M.; Iozzo, M.; Duatti, A.; Virgilio, F.; Lorito, N.; Bacci, M.; Pardella, E.; Sandrini, G.; et al. Lactate Rewires Lipid Metabolism and Sustains a Metabolic–Epigenetic Axis in Prostate Cancer. Cancer Res. 2022, 82, 1267–1282. [Google Scholar] [CrossRef] [PubMed]
- Lugassy, C.; Vermeulen, P.B.; Ribatti, D.; Pezzella, F.; Barnhill, R.L. Vessel co-option and angiotropic extravascular migratory metastasis: A continuum of tumour growth and spread? Br. J. Cancer 2022, 126, 973–980. [Google Scholar] [CrossRef]
- Heidegger, I.; Fotakis, G.; Offermann, A.; Goveia, J.; Daum, S.; Salcher, S.; Noureen, A.; Timmer-Bosscha, H.; Schäfer, G.; Walenkamp, A.; et al. Comprehensive characterization of the prostate tumor microenvironment identifies CXCR4/CXCL12 crosstalk as a novel antiangiogenic therapeutic target in prostate cancer. Mol. Cancer 2022, 21, 132. [Google Scholar] [CrossRef]
- Janssens, R.; Struyf, S.; Proost, P. The unique structural and functional features of CXCL12. Cell. Mol. Immunol. 2018, 15, 299–311. [Google Scholar] [CrossRef]
- Yu, L.; Cecil, J.; Peng, S.-B.; Schrementi, J.; Kovacevic, S.; Paul, D.; Su, E.W.; Wang, J. Identification and expression of novel isoforms of human stromal cell-derived factor 1. Gene 2006, 374, 174–179. [Google Scholar] [CrossRef]
- Shirozu, M.; Nakano, T.; Inazawa, J.; Tashiro, K.; Tada, H.; Shinohara, T.; Honjo, T. Structure and Chromosomal Localization of the Human Stromal Cell-Derived Factor 1 (SDF1) Gene. Genomics 1995, 28, 495–500. [Google Scholar] [CrossRef]
- Altenburg, J.D.; Broxmeyer, H.E.; Jin, Q.; Cooper, S.; Basu, S.; Alkhatib, G. A naturally occurring splice variant of CXCL12/stromal cell-derived factor 1 is a potent human immunodeficiency virus type 1 inhibitor with weak chemotaxis and cell survival activities. J. Virol. 2007, 81, 8140–8148. [Google Scholar] [CrossRef]
- Alsayed, R.K.M.; Khan, A.Q.; Ahmad, F.; Ansari, A.W.; Alam, M.A.; Buddenkotte, J.; Steinhoff, M.; Uddin, S.; Ahmad, A. Epigenetic regulation of CXCR4 signaling in cancer pathogenesis and progression. Semin. Cancer Biol. 2022, 86, 697–708. [Google Scholar] [CrossRef]
- Shen, P.-F.; Chen, X.-Q.; Liao, Y.-C.; Chen, N.; Zhou, Q.; Wei, Q.; Li, X.; Wang, J.; Zeng, H. MicroRNA-494-3p targets CXCR4 to suppress the proliferation, invasion, and migration of prostate cancer. Prostate 2014, 74, 756–767. [Google Scholar] [CrossRef]
- He, C.; Lu, X.; Yang, F.; Qin, L.; Guo, Z.; Sun, Y.; Wu, J. LncRNA UCA1 acts as a sponge of miR-204 to up-regulate CXCR4 expression and promote prostate cancer progression. Biosci. Rep. 2019, 39, BSR20181465. [Google Scholar] [CrossRef]
- Aleyasin, S.A.; Shirkavand, A.; Boroujeni, Z.N. Examination of methylation changes of VIM, CXCR4, DOK7, and SPDEF genes in peripheral blood DNA in breast cancer patients. Indian J. Cancer 2018, 55, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Stuckel, A.J.; Zhang, W.; Zhang, X.; Zeng, S.; Dougherty, U.; Mustafi, R.; Zhang, Q.; Perreand, E.; Khare, T.; Joshi, T.; et al. Enhanced CXCR4 Expression Associates with Increased Gene Body 5-Hydroxymethylcytosine Modification but not Decreased Promoter Methylation in Colorectal Cancer. Cancers 2020, 12, 539. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Matsubayashi, H.; Fukushima, N.; Goggins, M. The Chemokine Receptor CXCR4 is Regulated by DNA Methylation in Pancreatic Cancer. Cancer Biol. Ther. 2005, 4, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Kim, J.; Yamano, T.; Takeuchi, H.; Huang, S.; Umetani, N.; Koyanagi, K.; Hoon, D.S. Epigenetic Up-regulation of C-C Chemokine Receptor 7 and C-X-C Chemokine Receptor 4 Expression in Melanoma Cells. Cancer Res. 2005, 65, 1800–1807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, Y.; Wu, Q.; Xie, L.; Barwick, B.; Fu, C.; Li, X.; Wu, D.; Xia, S.; Chen, J.; et al. Acetylation of KLF5 maintains EMT and tumorigenicity to cause chemoresistant bone metastasis in prostate cancer. Nat. Commun. 2021, 12, 1714. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Pandolfini, L.; Barbieri, I.; Bannister, A.J.; Hendrick, A.; Andrews, B.; Webster, N.; Murat, P.; Mach, P.; Brandi, R.; Robson, S.C.; et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol. Cell 2019, 74, 1278–1290.e9. [Google Scholar] [CrossRef]
- Yi, Y.-C.; Chen, X.-Y.; Zhang, J.; Zhu, J.-S. Novel insights into the interplay between m6A modification and noncoding RNAs in cancer. Mol. Cancer 2020, 19, 121. [Google Scholar] [CrossRef]
- Chen, T.; Hao, Y.-J.; Zhang, Y.; Li, M.-M.; Wang, M.; Han, W.; Wu, Y.; Lv, Y.; Hao, J.; Wang, L.; et al. m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 2015, 16, 289–301, Erratum in Cell Stem Cell 2015, 16, 338. [Google Scholar] [CrossRef]
- Li, Y.; Yang, F.; Gao, M.; Gong, R.; Jin, M.; Liu, T.; Sun, Y.; Fu, Y.; Huang, Q.; Zhang, W.; et al. miR-149-3p Regulates the Switch between Adipogenic and Osteogenic Differentiation of BMSCs by Targeting FTO. Mol. Ther.-Nucleic Acids 2019, 17, 590–600. [Google Scholar] [CrossRef]
- Du, M.; Zhang, Y.; Mao, Y.; Mou, J.; Zhao, J.; Xue, Q.; Wang, D.; Huang, J.; Gao, S.; Gao, Y. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem. Biophys. Res. Commun. 2017, 482, 582–589. [Google Scholar] [CrossRef]
- Gururajan, M.; Josson, S.; Chu, G.C.-Y.; Lu, C.-L.; Lu, Y.-T.; Haga, C.L.; Zhau, H.E.; Liu, C.; Lichterman, J.; Duan, P.; et al. miR-154* and miR-379 in the DLK1-DIO3 MicroRNA Mega-Cluster Regulate Epithelial to Mesenchymal Transition and Bone Metastasis of Prostate Cancer. Clin. Cancer Res. 2014, 20, 6559–6569. [Google Scholar] [CrossRef]
- Josson, S.; Gururajan, M.; Sung, S.Y.; Hu, P.; Shao, C.; Zhau, H.E.; Liu, C.; Lichterman, J.; Duan, P.; Li, Q.; et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 2015, 34, 2690–2699. [Google Scholar] [CrossRef]
- Ren, G.; Baritaki, S.; Marathe, H.; Feng, J.; Park, S.; Beach, S.; Bazeley, P.S.; Beshir, A.B.; Fenteany, G.; Mehra, R.; et al. Polycomb Protein EZH2 Regulates Tumor Invasion via the Transcriptional Repression of the Metastasis Suppressor RKIP in Breast and Prostate Cancer. Cancer Res. 2012, 72, 3091–3104. [Google Scholar] [CrossRef]
- Lu, J.; Huang, Y.; Zhang, X.; Xu, Y.; Nie, S. Noncoding RNAs involved in DNA methylation and histone methylation, and acetylation in diabetic vascular complications. Pharmacol. Res. 2021, 170, 105520. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Zhang, X. Interactions between cancer cells and bone microenvironment promote bone metastasis in prostate cancer. Cancer Commun. 2019, 39, 76. [Google Scholar] [CrossRef]
- Liu, L.; Kron, K.J.; Pethe, V.V.; Demetrashvili, N.; Nesbitt, M.E.; Trachtenberg, J.; Ozcelik, H.; Fleshner, N.E.; Briollais, L.; van der Kwast, T.H.; et al. Association of tissue promoter methylation levels of APC, TGFβ2, HOXD3 and RASSF1A with prostate cancer progression. Int. J. Cancer 2011, 129, 2454–2462. [Google Scholar] [CrossRef]
- Kim, J.K.; Jung, Y.; Wang, J.; Joseph, J.; Mishra, A.; Hill, E.E.; Krebsbach, P.H.; Pienta, K.J.; Shiozawa, Y.; Taichman, R.S. TBK1 Regulates Prostate Cancer Dormancy through mTOR Inhibition. Neoplasia 2013, 15, 1064–1074. [Google Scholar] [CrossRef]
- Jin, S.; Li, M.; Chang, H.; Wang, R.; Zhang, Z.; Zhang, J.; He, Y.; Ma, H. The m6A demethylase ALKBH5 promotes tumor progression by inhibiting RIG-I expression and interferon alpha production through the IKKepsilon/TBK1/IRF3 pathway in head and neck squamous cell carcinoma. Mol. Cancer 2022, 21, 97. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Wei, Y.; Zen, C.; Xiong, W.; Niu, Y.; Zhao, Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer 2020, 19, 171. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Ochi, H.; Sunamura, S.; Kosaka, N.; Mabuchi, Y.; Fukuda, T.; Yao, K.; Kanda, H.; Ae, K.; Okawa, A.; et al. Cancer-secreted hsa-miR-940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting ARHGAP1 and FAM134A. Proc. Natl. Acad. Sci. USA 2018, 115, 2204–2209. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wodzenski, D.; Gao, D.; Shiraishi, T.; Terada, N.; Li, Y.; Griend, D.J.V.; Luo, J.; Kong, C.; Getzenberg, R.H.; et al. Stress-Response Protein RBM3 Attenuates the Stem-like Properties of Prostate Cancer Cells by Interfering with CD44 Variant Splicing. Cancer Res. 2013, 73, 4123–4133. [Google Scholar] [CrossRef]
- Wellmann, S.; Truss, M.; Bruder, E.; Tornillo, L.; Zelmer, A.; Seeger, K.; Bührer, C. The RNA-Binding Protein RBM3 Is Required for Cell Proliferation and Protects Against Serum Deprivation-Induced Cell Death. Pediatr. Res. 2010, 67, 35–41. [Google Scholar] [CrossRef]
- Pilotte, J.; Cunningham, B.A.; Edelman, G.M.; Vanderklish, P.W. Developmentally regulated expression of the cold-inducible RNA-binding motif protein 3 in euthermic rat brain. Brain Res. 2009, 1258, 12–24. [Google Scholar] [CrossRef]
- Dresios, J.; Aschrafi, A.; Owens, G.C.; Vanderklish, P.W.; Edelman, G.M.; Mauro, V.P. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl. Acad. Sci. USA 2005, 102, 1865–1870. [Google Scholar] [CrossRef]
- Maruyama, K.; Sato, N.; Ohta, N. Conservation of structure and cold-regulation of RNA-binding proteins in cyanobacteria: Probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nucleic Acids Res. 1999, 27, 2029–2036. [Google Scholar] [CrossRef]
- Baron, R.; Kneissel, M. WNT signaling in bone homeostasis and disease: From human mutations to treatments. Nat. Med. 2013, 19, 179–192. [Google Scholar] [CrossRef]
- Ren, D.; Dai, Y.; Yang, Q.; Zhang, X.; Guo, W.; Ye, L.; Huang, S.; Chen, X.; Lai, Y.; Du, H.; et al. Wnt5a induces and maintains prostate cancer cells dormancy in bone. J. Exp. Med. 2019, 216, 428–449. [Google Scholar] [CrossRef]
- Zhang, K.; Guo, Y.; Wang, X.; Zhao, H.; Ji, Z.; Cheng, C.; Li, L.; Fang, Y.; Xu, D.; Zhu, H.; et al. WNT/beta-Catenin Directs Self-Renewal Symmetric Cell Division of hTERT (high) Prostate Cancer Stem Cells. Cancer Res. 2017, 77, 2534–2547. [Google Scholar] [CrossRef]
- Zhang, W.; Bado, I.L.; Hu, J.; Wan, Y.-W.; Wu, L.; Wang, H.; Gao, Y.; Jeong, H.-H.; Xu, Z.; Hao, X.; et al. The bone microenvironment invigorates metastatic seeds for further dissemination. Cell 2021, 184, 2471–2486.e20. [Google Scholar] [CrossRef]
- Zhang, S.; Lv, C.; Niu, Y.; Li, C.; Li, X.; Shang, Y.; Zhang, Y.; Zhang, Y.; Zhang, Y.; Zeng, Y. RBM3 suppresses stemness remodeling of prostate cancer in bone microenvironment by modulating N6-methyladenosine on CTNNB1 mRNA. Cell Death Dis. 2023, 14, 91. [Google Scholar] [CrossRef]
- Autin, P.; Blanquart, C.; Fradin, D. Epigenetic Drugs for Cancer and microRNAs: A Focus on Histone Deacetylase Inhibitors. Cancers 2019, 11, 1530. [Google Scholar] [CrossRef]
- Ku, S.Y.; Rosario, S.; Wang, Y.; Mu, P.; Seshadri, M.; Goodrich, Z.W.; Goodrich, M.M.; Labbé, D.P.; Gomez, E.C.; Wang, J.; et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355, 78–83. [Google Scholar] [CrossRef]
- Segel, M.; Lash, B.; Song, J.; Ladha, A.; Liu, C.C.; Jin, X.; Mekhedov, S.L.; Macrae, R.K.; Koonin, E.V.; Zhang, F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021, 373, 882–889. [Google Scholar] [CrossRef]

| Modification | Structure | Molecular | Events on PCa | Experimental Evidence | Reference (PMID) |
|---|---|---|---|---|---|
| m6A | 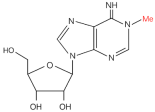 | Writer: METTL3/14 Reader: YTHDF1/2/3, YTHDC1/2 Eraser: ALKBH5, FTO | High expression: METTL3, RBM15, HNRNPA2B1 Low expression: ALKBH5, FTO | Transcriptome information and gene-level alteration data from The Cancer Genome Atlas datasets | 32710725 |
| m6Am | 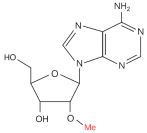 | Writer: PCIF1 Reader: — Eraser: FTO | PCIF1 gene deep deletion | TCGA database and GTEx database | 34888238 |
| m1A |  | Writer: TRMT6/61A/10C Reader: YTHDF1/2 Eraser: FTO, ALKBH1/3 | — | — | — |
| m5C |  | Writer: NSUN2, TRDMT1 Reader: YBX1, ALYREF Eraser: TET1/2/3 | High expression: NSUN2/5, DNMT3A/3B, ALYREF, TET1 Low expression: TET2 | TCGA and GEO databases | 35603206 |
| ac4C | 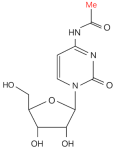 | Writer: NAT10, THUMPD1 Reader: — Eraser: — | High expression: NAT10 | PCa cell line assay | 35743017 |
| Ψ | 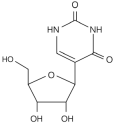 | Writer: TRUB1/2, PUS1/7 Reader: — Eraser: — | — | — | — |
| m7G | 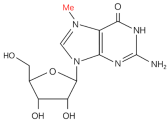 | Writer: METTL1/WDR4, RNMT/RAM, WBSCR22/TRMT112 Relative: EIF3D/4A1, LARP1 | High expression: METTL1 | TCGA and CPTAC databases | 34285249 |
| 5mC | 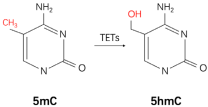 | Writer: DNMT1/3A/3B Reader: MeCP2, MBD1/2/4, UHRF1/2 Cytosine Dioxygenase: TETs | Low expression: TET2 Low level: 5hmC | Clinical samples immunohistochemistry | 26404510 |
| Histone modification | — | Core histones: H2A, H2B, H3, H4 | High expression: EZH2, LSD1, SET9 | Clinical samples by immunohistochemistry | 12374981 16079795 20959290 |
| Nucleosome positioning | — | Chromatin remodeling complexes: SWI/SNF, ISWI, CHD, and INO80 | CDH1 gene deletion | Clinical samples | 22179824 |
| Higher-order chromatin organization | — | Conformational molecules: CAF-1, CTCF, etc. | High expression: CAF-1 Low expression: CTCF | PCa cell line assay or clinical samples immunohistochemistry | 19309489 31736271 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Shen, T.; Zeng, Y. Epigenetic Modifications in Prostate Cancer Metastasis and Microenvironment. Cancers 2023, 15, 2243. https://doi.org/10.3390/cancers15082243
Zhang S, Shen T, Zeng Y. Epigenetic Modifications in Prostate Cancer Metastasis and Microenvironment. Cancers. 2023; 15(8):2243. https://doi.org/10.3390/cancers15082243
Chicago/Turabian StyleZhang, Shouyi, Tao Shen, and Yu Zeng. 2023. "Epigenetic Modifications in Prostate Cancer Metastasis and Microenvironment" Cancers 15, no. 8: 2243. https://doi.org/10.3390/cancers15082243
APA StyleZhang, S., Shen, T., & Zeng, Y. (2023). Epigenetic Modifications in Prostate Cancer Metastasis and Microenvironment. Cancers, 15(8), 2243. https://doi.org/10.3390/cancers15082243






