The Gut-Prostate Axis: A New Perspective of Prostate Cancer Biology through the Gut Microbiome
Abstract
Simple Summary
Abstract
1. Introduction
2. The Gut Microbiome
| Prostate Cancer Risk | Risk Factors | Changes in Gut Microbiota | References |
|---|---|---|---|
| High | Obesity | The ratio of Firmicutes to Bacteroidetes ↓ Bifidobacterium ↓ | [24,38] |
| High fat diet | Bacteroidetes↓ Firmicutes ↑ Proteobacteria ↑ | [9,26] | |
| Dairy product | Lactobacillus ↑ Bifidobacterium ↑ Bacteroidetes↓ | [28,29,30] | |
| Low | Mediterranean diet | Lachnospiaceae↑ | [39] |
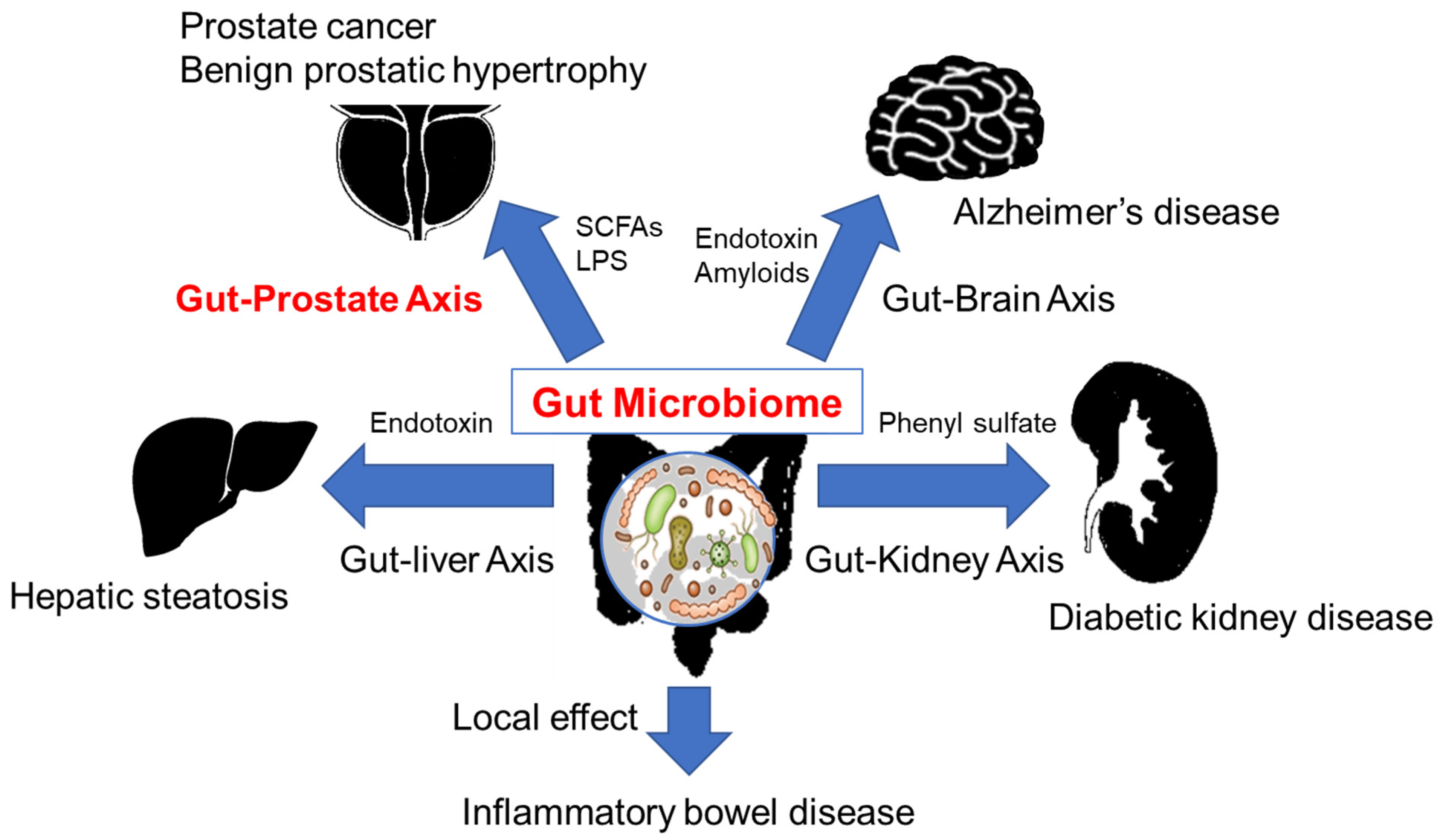
3. The Gut Microbiome and Diseases
4. The Gut Microbiome and Cancer
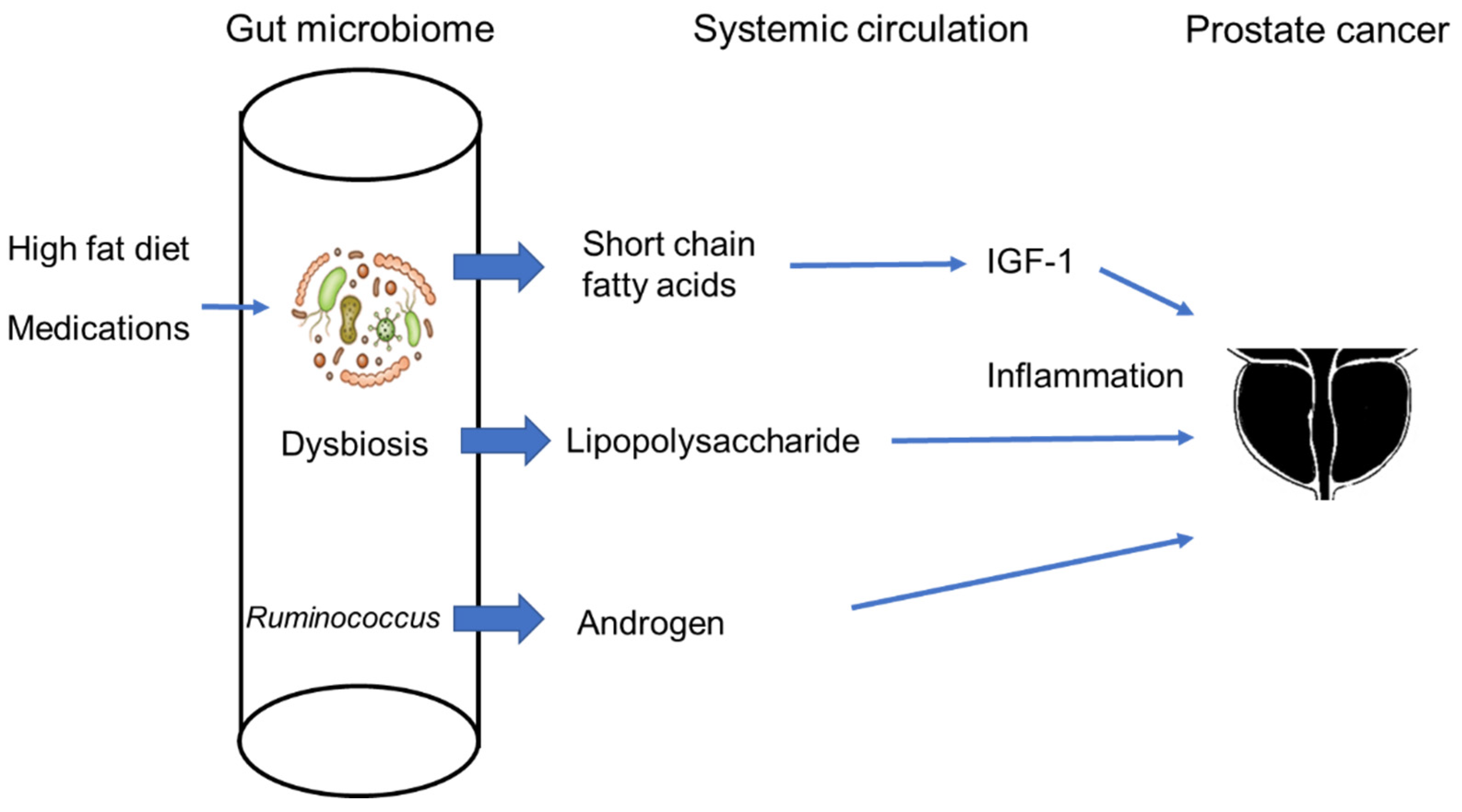
5. The Gut Microbiome and Prostate Cancer
6. The Gut Microbiome in Androgenesis
7. The Association with the Epigenetics
8. Limitations
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ito, K.; Oki, R.; Sekine, Y.; Arai, S.; Miyazawa, Y.; Shibata, Y.; Suzuki, K.; Kurosawa, I. Screening for prostate cancer: History, evidence, controversies and future perspectives toward individualized screening. Int. J. Urol. 2019, 26, 956–970. [Google Scholar] [CrossRef]
- Peto, J. Cancer epidemiology in the last century and the next decade. Nature 2001, 411, 390–395. [Google Scholar] [CrossRef]
- Discacciati, A.; Orsini, N.; Wolk, A. Body mass index and incidence of localized and advanced prostate cancer--a dose-response meta-analysis of prospective studies. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23, 1665–1671. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, K.; Nonomura, N. Influence of Diet and Nutrition on Prostate Cancer. Int. J. Mol. Sci. 2020, 21, 1447. [Google Scholar] [CrossRef]
- Hayashi, T.; Fujita, K.; Nojima, S.; Hayashi, Y.; Nakano, K.; Ishizuya, Y.; Wang, C.; Yamamoto, Y.; Kinouchi, T.; Matsuzaki, K.; et al. High-Fat Diet-Induced Inflammation Accelerates Prostate Cancer Growth via IL6 Signaling. Clin. Cancer Res. 2018, 24, 4309–4318. [Google Scholar] [CrossRef]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef]
- Fujita, K.; Matsushita, M.; Banno, E.; De Velasco, M.A.; Hatano, K.; Nonomura, N.; Uemura, H. Gut microbiome and prostate cancer. Int. J. Urol. 2022, 29, 793–798. [Google Scholar] [CrossRef]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldaferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients 2019, 11, 2393. [Google Scholar] [CrossRef]
- Davis, D.D.; Davis, D.D. The Giant Panda: A Morphological Study of Evolutionary Mechanisms; Chicago Natural History Museum: Chicago, IL, USA, 1964. [Google Scholar]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Li, R.; Fan, W.; Tian, G.; Zhu, H.; He, L.; Cai, J.; Huang, Q.; Cai, Q.; Li, B.; Bai, Y.; et al. The sequence and de novo assembly of the giant panda genome. Nature 2010, 463, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wu, Q.; Dai, J.; Zhang, S.; Wei, F. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 17714–17719. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, e00261-19. [Google Scholar] [CrossRef]
- Yuan, X.; Chen, R.; Zhang, Y.; Lin, X.; Yang, X. Gut microbiota: Effect of pubertal status. BMC Microbiol. 2020, 20, 334. [Google Scholar] [CrossRef] [PubMed]
- Harada, N.; Minami, Y.; Hanada, K.; Hanaoka, R.; Kobayashi, Y.; Izawa, T.; Sato, T.; Kato, S.; Inui, H.; Yamaji, R. Relationship between gut environment, feces-to-food ratio, and androgen deficiency-induced metabolic disorders. Gut Microbes 2020, 12, 1817719. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, S.; Suda, W.; Oshima, K.; Kim, S.W.; Hirose, Y.; Morita, H.; Hattori, M. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016, 23, 125–133. [Google Scholar] [CrossRef]
- He, Y.; Wu, W.; Zheng, H.M.; Li, P.; McDonald, D.; Sheng, H.F.; Chen, M.X.; Chen, Z.H.; Ji, G.Y.; Zheng, Z.D.X.; et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 2018, 24, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010, 18, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Kurahashi, N.; Inoue, M.; Iwasaki, M.; Sasazuki, S.; Tsugane, S. Dairy product, saturated fatty acid, and calcium intake and prostate cancer in a prospective cohort of Japanese men. Cancer Epidemiol. Biomarkers Prev. 2008, 17, 930–937. [Google Scholar] [CrossRef]
- Aslam, H.; Marx, W.; Rocks, T.; Loughman, A.; Chandrasekaran, V.; Ruusunen, A.; Dawson, S.L.; West, M.; Mullarkey, E.; Pasco, J.A.; et al. The effects of dairy and dairy derivatives on the gut microbiota: A systematic literature review. Gut Microbes 2020, 12. [Google Scholar] [CrossRef]
- Odamaki, T.; Kato, K.; Sugahara, H.; Xiao, J.Z.; Abe, F.; Benno, Y. Effect of probiotic yoghurt on animal-based diet-induced change in gut microbiota: An open, randomised, parallel-group study. Benef. Microbes 2016, 7, 473–484. [Google Scholar] [CrossRef]
- Odamaki, T.; Sugahara, H.; Yonezawa, S.; Yaeshima, T.; Iwatsuki, K.; Tanabe, S.; Tominaga, T.; Togashi, H.; Benno, Y.; Xiao, J. zhong Effect of the oral intake of yogurt containing Bifidobacterium longum BB536 on the cell numbers of enterotoxigenic Bacteroides fragilis in microbiota. Anaerobe 2012, 18, 14–18. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, Y.; Fang, W.; Cui, K.; Xu, D.; Liu, G.; Chi, S.; Tan, B.; Mai, K.; Ai, Q. Octanoate Alleviates Dietary Soybean Oil-Induced Intestinal Physical Barrier Damage, Oxidative Stress, Inflammatory Response and Microbial Dysbiosis in Large Yellow Croaker (Larimichthys Crocea). Front. Immunol. 2022, 13, 3314. [Google Scholar] [CrossRef]
- Nascimento, J.C.; Matheus, V.A.; Oliveira, R.B.; Tada, S.F.S.; Collares-Buzato, C.B. High-Fat Diet Induces Disruption of the Tight Junction-Mediated Paracellular Barrier in the Proximal Small Intestine Before the Onset of Type 2 Diabetes and Endotoxemia. Dig. Dis. Sci. 2021, 66, 3359–3374. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Reunanen, J.; Meijerink, M.; Pietila, T.E.; Kainulainen, V.; Klievink, J.; Huuskonen, L.; Aalvink, S.; Skurnik, M.; Boeren, S.; et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE 2017, 12, e0173004. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H.J. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Dao, M.C.; Everard, A.; Aron-Wisnewsky, J.; Sokolovska, N.; Prifti, E.; Verger, E.O.; Kayser, B.D.; Levenez, F.; Chilloux, J.; Hoyles, L.; et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut 2016, 65, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Roediger, W.E.W. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 1980, 21, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.N.; Yao, Y.; Ju, S.Y. Short chain fatty acids and fecal microbiota abundance in humans with obesity: A systematic review and meta-analysis. Nutrients 2019, 11, 2512. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- Muralidharan, J.; Moreno-Indias, I.; Bulló, M.; Lopez, J.V.; Corella, D.; Castañer, O.; Vidal, J.; Atzeni, A.; Fernandez-García, J.C.; Torres-Collado, L.; et al. Effect on gut microbiota of a 1-y lifestyle intervention with Mediterranean diet compared with energy-reduced Mediterranean diet and physical activity promotion: PREDIMED-Plus Study. Am. J. Clin. Nutr. 2021, 114, 1148–1158. [Google Scholar] [CrossRef]
- Kayama, H.; Takeda, K. Manipulation of epithelial integrity and mucosal immunity by host and microbiota-derived metabolites. Eur. J. Immunol. 2020, 50, 921–931. [Google Scholar] [CrossRef]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 204. [Google Scholar] [CrossRef]
- Ohkusa, T.; Sato, N.; Ogihara, T.; Morita, K.; Ogawa, M.; Okayasu, I. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J. Gastroenterol. Hepatol. 2002, 17, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Imdad, A.; Nicholson, M.R.; Tanner-Smith, E.E.; Zackular, J.P.; Gomez-Duarte, O.G.; Beaulieu, D.B.; Acra, S. Fecal transplantation for treatment of inflammatory bowel disease. Cochrane Database Syst. Rev. 2018, 11, CD012774. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Yin, Y.; Li, Z.; Zhang, W. Gut microbiota-derived components and metabolites in the progression of non-alcoholic fatty liver disease (NAFLD). Nutrients 2019, 11, 1712. [Google Scholar] [CrossRef]
- Kikuchi, K.; Saigusa, D.; Kanemitsu, Y.; Matsumoto, Y.; Thanai, P.; Suzuki, N.; Mise, K.; Yamaguchi, H.; Nakamura, T.; Asaji, K.; et al. Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat. Commun. 2019, 10, 1835. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.J.; Kikuchi, K.; Oikawa, D.; Watanabe, S.; Kanemitsu, Y.; Saigusa, D.; Kujirai, R.; Ikeda-Ohtsubo, W.; Ichijo, M.; Akiyama, Y.; et al. SGLT-1-specific inhibition ameliorates renal failure and alters the gut microbial community in mice with adenine-induced renal failure. Physiol. Rep. 2021, 9, e15092. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C.A.; Adegboyega, P.; van Rooijen, N.; Tagalicud, A.; Allman, M.; Wallace, M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J. Hepatol. 2007, 47, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Henao-Mejia, J.; Elinav, E.; Jin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J.; et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef]
- Liang, H.; Hussey, S.E.; Sanchez-Avila, A.; Tantiwong, P.; Musi, N. Effect of Lipopolysaccharide on Inflammation and Insulin Action in Human Muscle. PLoS ONE 2013, 8, e63983. [Google Scholar] [CrossRef]
- Kim, H.s.; Kim, S.; Shin, S.J.; Park, Y.H.; Nam, Y.; Kim, C.w.; Lee, K.w.; Kim, S.-M.; Jung, I.D.; Yang, H.D.; et al. Gram-negative bacteria and their lipopolysaccharides in Alzheimer’s disease: Pathologic roles and therapeutic implications. Transl. Neurodegener. 2021, 10, 49. [Google Scholar] [CrossRef]
- Seo, D.O.; O’Donnell, D.; Jain, N.; Ulrich, J.D.; Herz, J.; Li, Y.; Lemieux, M.; Cheng, J.; Hu, H.; Serrano, J.R.; et al. ApoE isoform- and microbiota-dependent progression of neurodegeneration in a mouse model of tauopathy. Science 2023, 379, eadd1236. [Google Scholar] [CrossRef]
- Mizutani, S.; Yamada, T.; Yachida, S. Significance of the gut microbiome in multistep colorectal carcinogenesis. Cancer Sci. 2020, 111, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE 2-Mediated Suppression of Antitumor Immunity. Cancer Discov. 2017, 7, 522–538. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Tan, Q.; Fu, Q.; Zhou, Y.; Hu, Y.; Tang, S.; Zhou, Y.; Zhang, J.; Qiu, J.; Lv, Q. Gastrointestinal microbiome and breast cancer: Correlations, mechanisms and potential clinical implications. Breast Cancer 2017, 24, 220–228. [Google Scholar] [CrossRef]
- Parida, S.; Sharma, D. The Microbiome-Estrogen Connection and Breast Cancer Risk. Cells 2019, 8, 1642. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ma, Y.; Raoult, D.; Kroemer, G.; Gajewski, T.F. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018, 359, 1366–1370. [Google Scholar] [CrossRef]
- Derosa, L.; Hellmann, M.D.; Spaziano, M.; Halpenny, D.; Fidelle, M.; Rizvi, H.; Long, N.; Plodkowski, A.J.; Arbour, K.C.; Chaft, J.E.; et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1437–1444. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Fidelle, M.; Iebba, V.; Alla, L.; Pasolli, E.; Segata, N.; Desnoyer, A.; Pietrantonio, F.; Ferrere, G.; et al. Gut Bacteria Composition Drives Primary Resistance to Cancer Immunotherapy in Renal Cell Carcinoma Patients. Eur. Urol. 2020, 78, 195–206. [Google Scholar] [CrossRef]
- Golombos, D.M.; Ayangbesan, A.; O’Malley, P.; Lewicki, P.; Barlow, L.M.; Barbieri, C.E.; Chan, C.; DuLong, C.; Abu-Ali, G.; Huttenhower, C.; et al. The Role of Gut Microbiome in the Pathogenesis of Prostate Cancer: A Prospective, Pilot Study. Urology 2018, 111, 122–128. [Google Scholar] [CrossRef]
- Sfanos, K.S.; Markowski, M.C.; Peiffer, L.B.; Ernst, S.E.; White, J.R.; Pienta, K.J.; Antonarakis, E.S.; Ross, A.E. Compositional differences in gastrointestinal microbiota in prostate cancer patients treated with androgen axis-targeted therapies. Prostate Cancer Prostatic Dis. 2018, 21, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Liss, M.A.; White, J.R.; Goros, M.; Gelfond, J.; Leach, R.; Johnson-Pais, T.; Lai, Z.; Rourke, E.; Basler, J.; Ankerst, D.; et al. Metabolic Biosynthesis Pathways Identified from Fecal Microbiome Associated with Prostate Cancer. Eur. Urol. 2018, 74, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Fujita, K.; Motooka, D.; Hatano, K.; Fukae, S.; Kawamura, N.; Tomiyama, E.; Hayashi, Y.; Banno, E.; Takao, T.; et al. The gut microbiota associated with high-Gleason prostate cancer. Cancer Sci. 2021, 112, 3125–3135. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, L.B.; White, J.R.; Jones, C.B.; Slottke, R.E.; Ernst, S.E.; Moran, A.E.; Graff, J.N.; Sfanos, K.S. Composition of gastrointestinal microbiota in association with treatment response in individuals with metastatic castrate resistant prostate cancer progressing on enzalutamide and initiating treatment with anti-PD-1 (pembrolizumab). Neoplasia (United States) 2022, 32, 100822. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, K.; Hayashi, T.; Kayama, H.; Motooka, D.; Hase, H.; Jingushi, K.; Yamamichi, G.; Yumiba, S.; Tomiyama, E.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Prostate Cancer Growth via IGF1 Signaling. Cancer Res. 2021, 81, 4014–4026. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Belcheva, A.; Irrazabal, T.; Robertson, S.J.; Streutker, C.; Maughan, H.; Rubino, S.; Moriyama, E.H.; Copeland, J.K.; Kumar, S.; Green, B.; et al. Gut microbial metabolism drives transformation of msh2-deficient colon epithelial cells. Cell 2014, 158, 288–299. [Google Scholar] [CrossRef]
- Yan, J.; Herzog, J.W.; Tsang, K.; Brennan, C.A.; Bower, M.A.; Garrett, W.S.; Sartor, B.R.; Aliprantis, A.O.; Charles, J.F. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. USA 2016, 113, E7554–E7563. [Google Scholar] [CrossRef]
- Ratajczak, W.; Mizerski, A.; Rył, A.; Słojewski, M.; Sipak, O.; Piasecka, M.; Laszczyńska, M. Alterations in fecal short chain fatty acids (SCFAs) and branched short-chain fatty acids (BCFAs) in men with benign prostatic hyperplasia (BPH) and metabolic syndrome (MetS). Aging (Albany. NY). 2021, 13, 10934–10954. [Google Scholar] [CrossRef]
- Takezawa, K.; Fujita, K.; Matsushita, M.; Motooka, D.; Hatano, K.; Banno, E.; Shimizu, N.; Takao, T.; Takada, S.; Okada, K.; et al. The Firmicutes/Bacteroidetes ratio of the human gut microbiota is associated with prostate enlargement. Prostate 2021, 81, 1287–1293. [Google Scholar] [CrossRef]
- Chughtai, B.; Forde, J.C.; Thomas, D.D.M.; Laor, L.; Hossack, T.; Woo, H.H.; Te, A.E.; Kaplan, S.A. Benign prostatic hyperplasia. Nat. Rev. Dis. Prim. 2016, 2, 16031. [Google Scholar] [CrossRef]
- Russo, G.I.; Broggi, G.; Cocci, A.; Capogrosso, P.; Falcone, M.; Sokolakis, I.; Gül, M.; Caltabiano, R.; Di Mauro, M. Relationship between dietary patterns with benign prostatic hyperplasia and erectile dysfunction: A collaborative review. Nutrients 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Sebastiano, C. Dietary patterns and prostatic diseases. Front. Biosci. 2012, E4, 195. [Google Scholar] [CrossRef]
- Li, Y.Z.; Shi, B.K.; Li, J.Y.; Zhu, X.W.; Liu, J.; Liu, Y.L. Role of p-ERK1/2 in Benign Prostatic Hyperplasia during Hyperinsulinemia. Urol. J. 2021, 18, 225–229. [Google Scholar] [CrossRef]
- Kristal, A.R.; Arnold, K.B.; Schenk, J.M.; Neuhouser, M.L.; Goodman, P.; Penson, D.F.; Thompson, I.M. Dietary patterns, supplement use, and the risk of symptomatic benign prostatic hyperplasia: Results from the Prostate Cancer Prevention Trial. Am. J. Epidemiol. 2008, 167, 925–934. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, K.; Hatano, K.; Hayashi, T.; Kayama, H.; Motooka, D.; Hase, H.; Yamamoto, A.; Uemura, T.; Yamamichi, G.; et al. High-fat diet promotes prostate cancer growth through histamine signaling. Int. J. Cancer 2022, 151, 623–636. [Google Scholar] [CrossRef]
- Ohtani, N.; Hara, E. Gut-liver axis-mediated mechanism of liver cancer: A special focus on the role of gut microbiota. Cancer Sci. 2021, 112, 4433–4443. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Wu, K.; Long, Z.; Zhou, X.; Zhong, C.; Wang, S.; Lai, H.; Guo, Y.; Lv, D.; Lu, J.; et al. Gut dysbiosis promotes prostate cancer progression and docetaxel resistance via activating NF-κB-IL6-STAT3 axis. Microbiome 2022, 10, 94. [Google Scholar] [CrossRef]
- Fujita, K.; Nonomura, N. Role of Androgen Receptor in Prostate Cancer: A Review. World J. Mens. Health 2019, 37, 288–295. [Google Scholar] [CrossRef]
- Shin, J.H.; Park, Y.H.; Sim, M.; Kim, S.A.; Joung, H.; Shin, D.M. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res. Microbiol. 2019, 170, 192–201. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, K.; Motooka, D.; Hatano, K.; Nishimoto, M.; Bannno, E.; Wang, Q.; Hata, J.; Takezawa, K.; Fukuhara, S.; et al. Firmicutes in gut microbiota correlate with blood testosterone levels in elderly men. World J. Mens. Health 2022, 40, 517–525. [Google Scholar] [CrossRef]
- Lindheim, L.; Bashir, M.; Münzker, J.; Trummer, C.; Zachhuber, V.; Leber, B.; Horvath, A.; Pieber, T.R.; Gorkiewicz, G.; Stadlbauer, V.; et al. Alterations in Gut Microbiome Composition and Barrier Function Are Associated with Reproductive and Metabolic Defects in Women with Polycystic Ovary Syndrome (PCOS): A Pilot Study. PLoS ONE 2017, 12, e0168390. [Google Scholar] [CrossRef] [PubMed]
- Giampaolino, P.; Foreste, V.; Di Filippo, C.; Gallo, A.; Mercorio, A.; Serafino, P.; Improda, F.P.; Verrazzo, P.; Zara, G.; Buonfantino, C.; et al. Microbiome and PCOS: State-of-Art and Future Aspects. Int. J. Mol. Sci. 2021, 22, 2048. [Google Scholar] [CrossRef] [PubMed]
- Cross, T.W.L.; Kasahara, K.; Rey, F.E. Sexual dimorphism of cardiometabolic dysfunction: Gut microbiome in the play? Mol. Metab. 2018, 15, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Pernigoni, N.; Zagato, E.; Calcinotto, A.; Troiani, M.; Mestre, R.P.; Calì, B.; Attanasio, G.; Troisi, J.; Minini, M.; Mosole, S.; et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science 2021, 374, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Daisley, B.A.; Chanyi, R.M.; Abdur-Rashid, K.; Al, K.F.; Gibbons, S.; Chmiel, J.A.; Wilcox, H.; Reid, G.; Anderson, A.; Dewar, M.; et al. Abiraterone acetate preferentially enriches for the gut commensal Akkermansia muciniphila in castrate-resistant prostate cancer patients. Nat. Commun. 2020, 11, 4822. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M.; Sato, H.; Kanesaka, M.; Imamura, Y.; Sakamoto, S.; Ichikawa, T.; Kaneda, A. Epigenetic modifications in prostate cancer. Int. J. Urol. 2021, 28, 140–149. [Google Scholar] [CrossRef]
- Dawson, M.A. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science 2017, 355, 1147–1152. [Google Scholar] [CrossRef]
- Lee, W.H.; Morton, R.A.; Epstein, J.I.; Brooks, J.D.; Campbell, P.A.; Bova, G.S.; Hsieh, W.S.; Isaacs, W.B.; Nelson, W.G. Cytidine methylation of regulatory sequences near the π-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl. Acad. Sci. USA 1994, 91, 11733–11737. [Google Scholar] [CrossRef]
- Jasek, K.; Kubatka, P.; Samec, M.; Liskova, A.; Smejkal, K.; Vybohova, D.; Bugos, O.; Biskupska-Bodova, K.; Bielik, T.; Zubor, P.; et al. DNA methylation status in cancer disease: Modulations by plant-derived natural compounds and dietary interventions. Biomolecules 2019, 9, 289. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Mandhare, A.; Tryphena, K.P.; Srivastava, S.; Shaikh, M.F.; Singh, S.B.; Khatri, D.K. Epigenetics in depression and gut-brain axis: A molecular crosstalk. Front. Aging Neurosci. 2022, 14. [Google Scholar] [CrossRef]
- Rossi, M.; Amaretti, A.; Raimondi, S. Folate production by probiotic bacteria. Nutrients 2011, 3, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Sivaprakasam, S.; Prasad, P.D.; Singh, N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol. Ther. 2016, 164, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Fentiman, I.S.; Tirelli, U.; Monfardini, S.; Schneider, M.; Festen, J.; Cognetti, F.; Aapro, M.S. Cancer in the elderly: Why so badly treated? Lancet 1990, 335, 1020–1022. [Google Scholar] [CrossRef] [PubMed]
- Bui, V.N.; Ogawa, H.; Ngo, L.H.; Baatartsogt, T.; Abao, L.N.B.; Tamaki, S.; Saito, K.; Watanabe, Y.; Runstadler, J.; Imai, K. H5N1 highly pathogenic avian influenza virus isolated from conjunctiva of a whooper swan with neurological signs. Arch. Virol. 2013, 158, 451–455. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujita, K.; Matsushita, M.; De Velasco, M.A.; Hatano, K.; Minami, T.; Nonomura, N.; Uemura, H. The Gut-Prostate Axis: A New Perspective of Prostate Cancer Biology through the Gut Microbiome. Cancers 2023, 15, 1375. https://doi.org/10.3390/cancers15051375
Fujita K, Matsushita M, De Velasco MA, Hatano K, Minami T, Nonomura N, Uemura H. The Gut-Prostate Axis: A New Perspective of Prostate Cancer Biology through the Gut Microbiome. Cancers. 2023; 15(5):1375. https://doi.org/10.3390/cancers15051375
Chicago/Turabian StyleFujita, Kazutoshi, Makoto Matsushita, Marco A. De Velasco, Koji Hatano, Takafumi Minami, Norio Nonomura, and Hirotsugu Uemura. 2023. "The Gut-Prostate Axis: A New Perspective of Prostate Cancer Biology through the Gut Microbiome" Cancers 15, no. 5: 1375. https://doi.org/10.3390/cancers15051375
APA StyleFujita, K., Matsushita, M., De Velasco, M. A., Hatano, K., Minami, T., Nonomura, N., & Uemura, H. (2023). The Gut-Prostate Axis: A New Perspective of Prostate Cancer Biology through the Gut Microbiome. Cancers, 15(5), 1375. https://doi.org/10.3390/cancers15051375







