Semi-Supervised Learning to Automate Tumor Bud Detection in Cytokeratin-Stained Whole-Slide Images of Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Related Work
1.2. Our Contribution
2. Materials and Methods
2.1. Materials
2.1.1. Model Development Data
2.1.2. Validation Data
2.2. Model Development
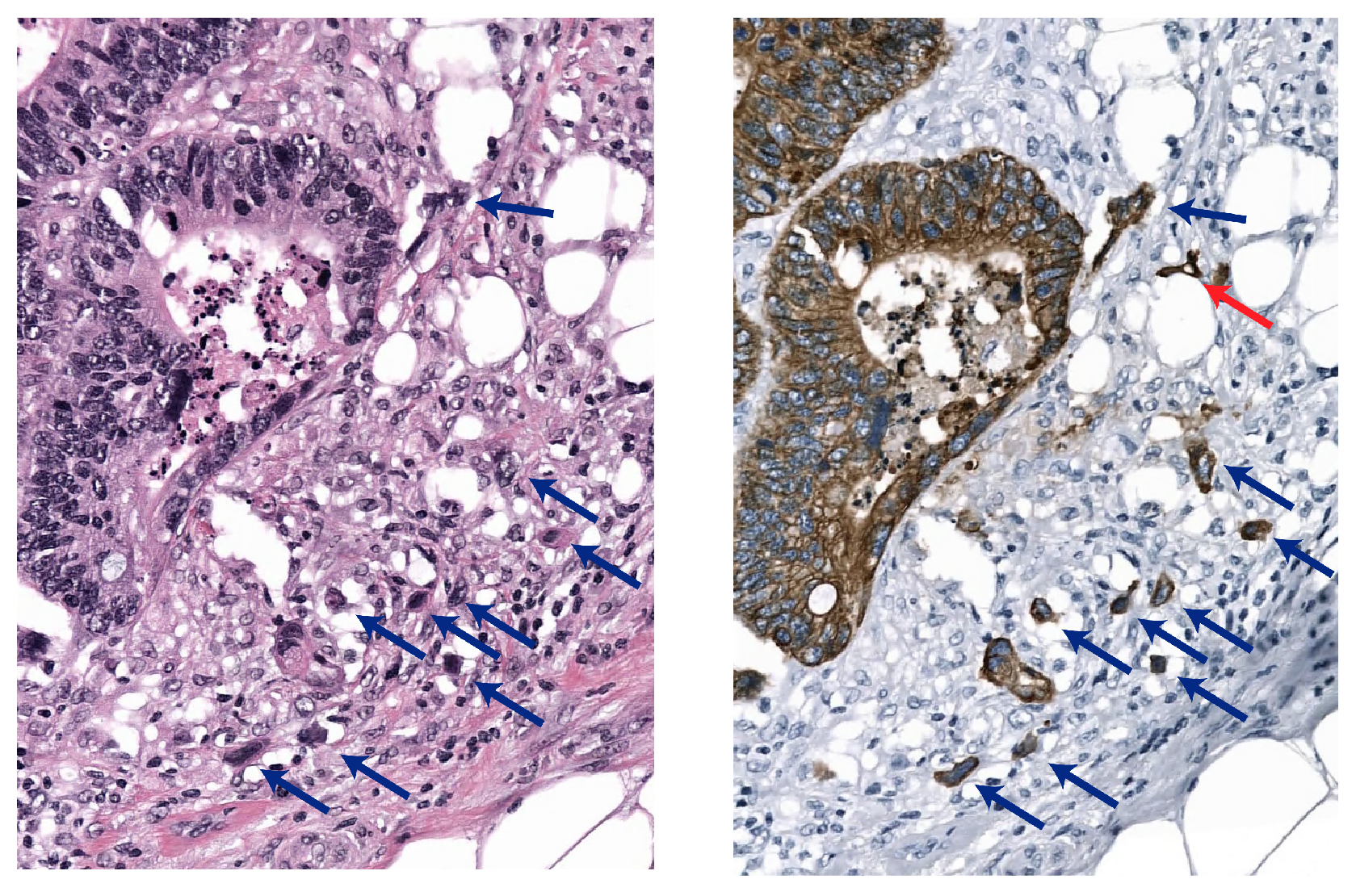
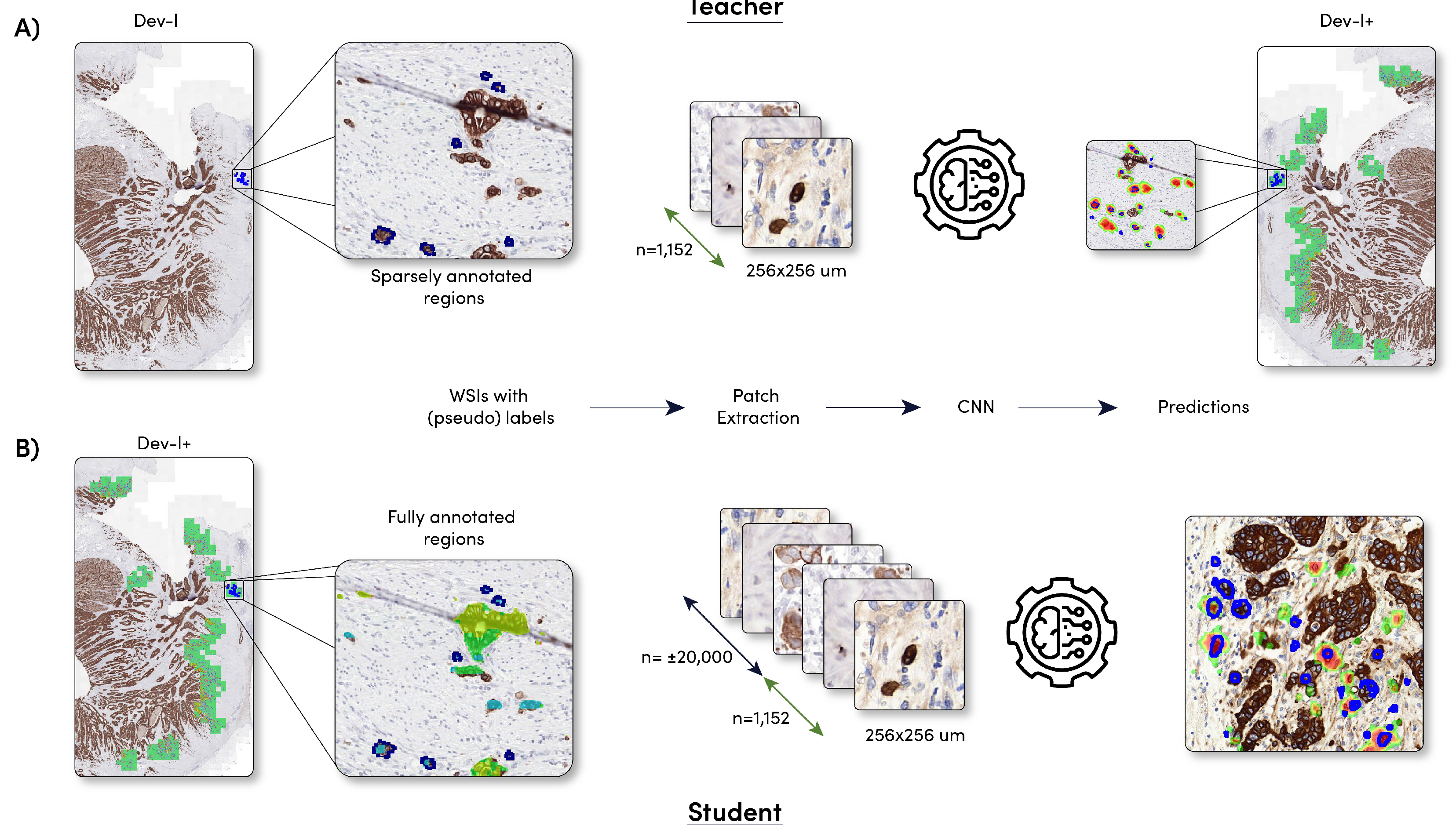
2.2.1. Sparsely Annotated Data and Object Detection
2.2.2. Student Development
2.2.3. Training Parameters
2.2.4. Automated Hotspot Selection
| Algorithm 1 Create tumor budding density map. |
| Ensure: The network was applied to the entire slide |
|
2.2.5. Tumor Bud Distribution
2.2.6. Statistical Analysis
3. Results
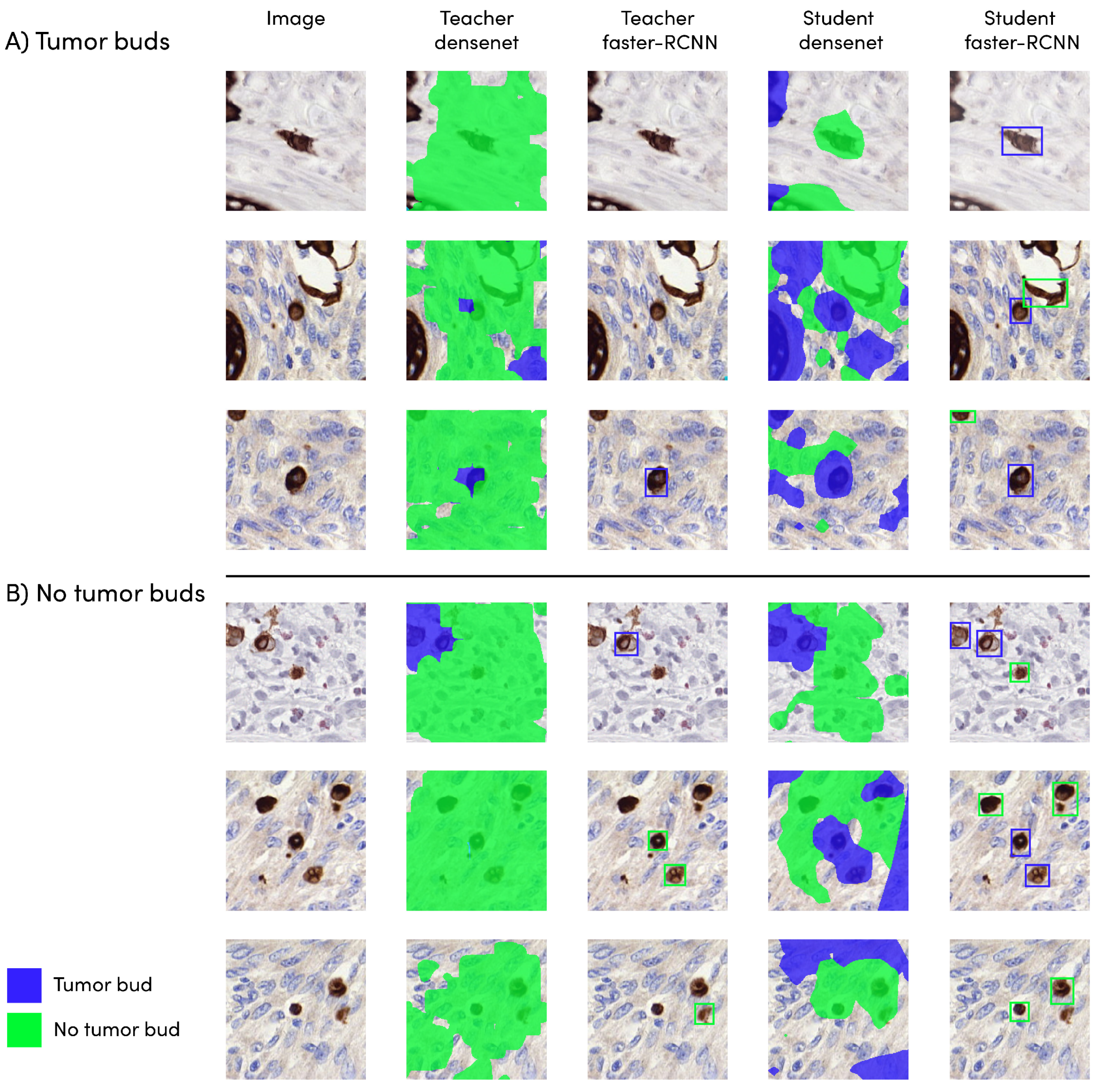
3.1. Detection Model Performance
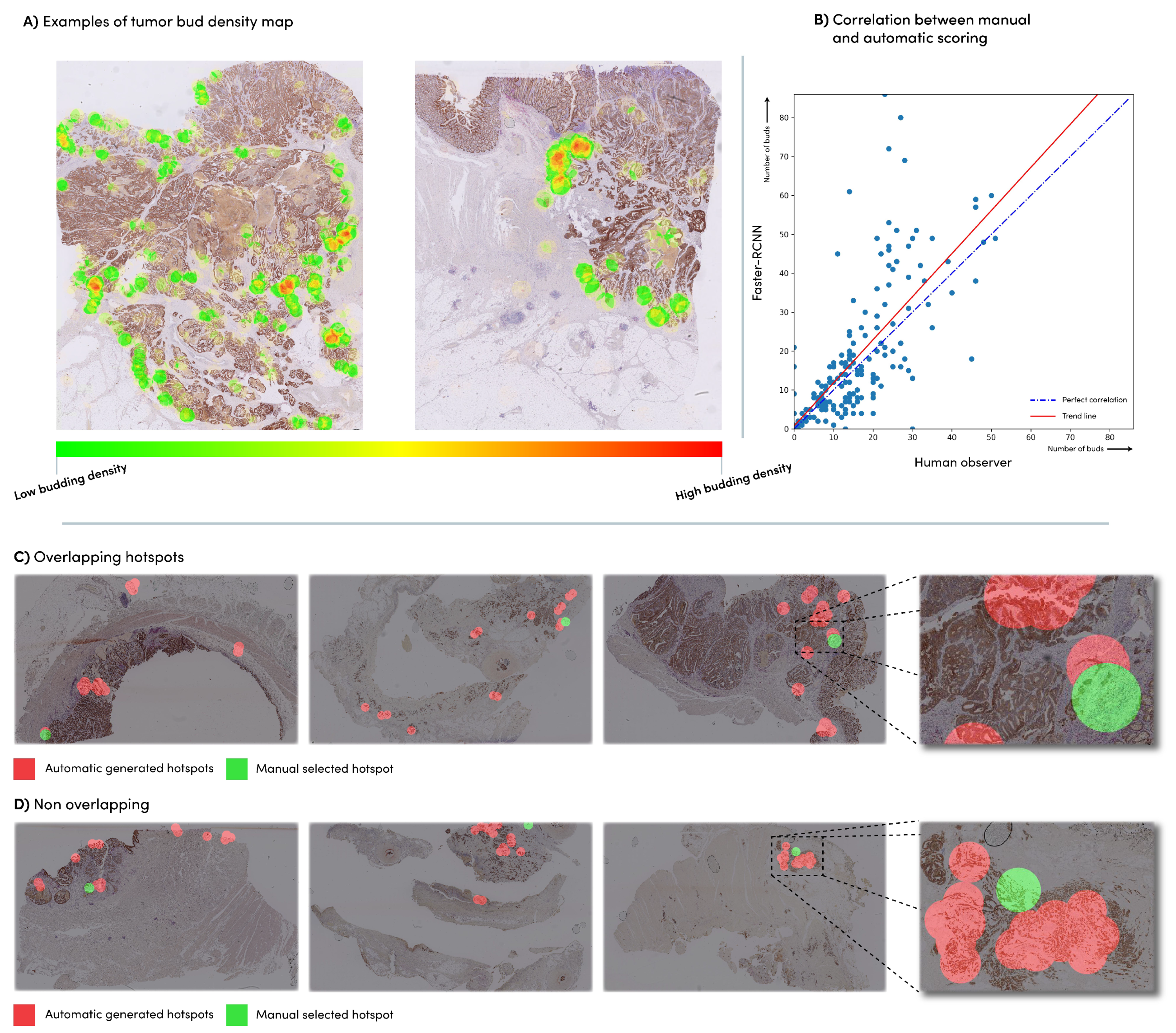
3.2. Automatic vs. Manual Tumor Bud Count
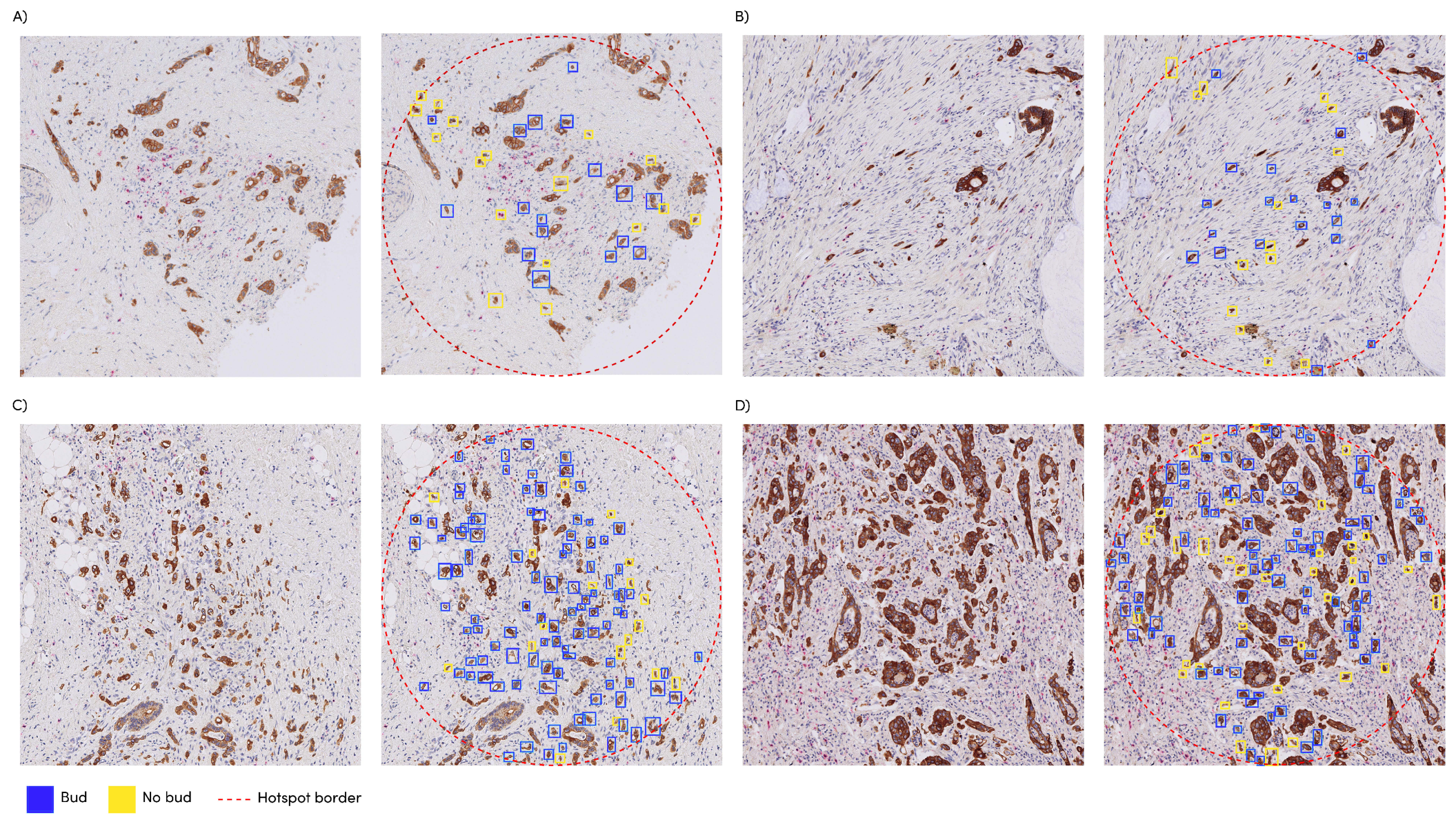
3.3. Automatic vs. Manual Hotspot Detection
3.4. Survival Analysis
4. Discussion
5. Conclusions
6. Code Availability
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berg, K.B.; Schaeffer, D.F. Tumor budding as a standardized parameter in gastrointestinal carcinomas: More than just the colon. Mod. Pathol. 2018, 31, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Lugli, A.; Zlobec, I.; Berger, M.D.; Kirsch, R.; Nagtegaal, I.D. Tumour budding in solid cancers. Nat. Rev. Clin. Oncol. 2021, 18, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Lugli, A.; Kirsch, R.; Ajioka, Y.; Bosman, F.; Cathomas, G.; Dawson, H.; El Zimaity, H.; Fléjou, J.F.; Hansen, T.P.; Hartmann, A.; et al. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod. Pathol. 2017, 30, 1299–1311. [Google Scholar] [CrossRef] [PubMed]
- Fisher, N.C.; Loughrey, M.B.; Coleman, H.G.; Gelbard, M.D.; Bankhead, P.; Dunne, P.D. Development of a semi-automated method for tumor budding assessment in colorectal cancer and comparison with manual methods. Histopathology 2022, 80, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Haddad, T.S.; Lugli, A.; Aherne, S.; Barresi, V.; Terris, B.; Bokhorst, J.M.; Brockmoeller, S.F.; Cuatrecasas, M.; Simmer, F.; El-Zimaity, H.; et al. Improving tumor budding reporting in colorectal cancer: A Delphi consensus study. Virchows Arch. 2021, 479, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Bokhorst, J.; Blank, A.; Lugli, A.; Zlobec, I.; Dawson, H.; Vieth, M.; Rijstenberg, L.; Brockmoeller, S.; Urbanowicz, M.; Flejou, J.; et al. Assessment of individual tumor buds using keratin immunohistochemistry: Moderate interobserver agreement suggests a role for machine learning. Mod. Pathol. 2020, 33, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Studer, L.; Blank, A.; Bokhorst, J.M.; Nagtegaal, I.D.; Zlobec, I.; Lugli, A.; Fischer, A.; Dawson, H. Taking tumour budding to the next frontier; a post International Tumour Budding Consensus Conference (ITBCC) 2016 review. Histopathology 2021, 78, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Fauzi, M.F.; Chen, W.; Knight, D.; Hampel, H.; Frankel, W.L.; Gurcan, M.N. Tumor budding detection system in whole slide pathology images. J. Med. Syst. 2020, 44, 38. [Google Scholar] [CrossRef] [PubMed]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.M.; Ciompi, F.; Van Der Laak, J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef] [PubMed]
- Bergler, M.; Benz, M.; Rauber, D.; Hartmann, D.; Kötter, M.; Eckstein, M.; Schneider-Stock, R.; Hartmann, A.; Merkel, S.; Bruns, V.; et al. Automatic detection of tumor buds in pan-cytokeratin stained colorectal cancer sections by a hybrid image analysis approach. In Proceedings of the European Congress on Digital Pathology, Warwick, UK, 10–13 April 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 83–90. [Google Scholar]
- Weis, C.A.; Kather, J.N.; Melchers, S.; Al-Ahmdi, H.; Pollheimer, M.J.; Langner, C.; Gaiser, T. Automatic evaluation of tumor budding in immunohistochemically stained colorectal carcinomas and correlation to clinical outcome. Diagn. Pathol. 2018, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Niazi, M.K.K.; Moore, K.; Berenhaut, K.S.; Hartman, D.J.; Pantanowitz, L.; Gurcan, M.N. Hotspot detection in pancreatic neuroendocrine images using local depth. In Proceedings of the Medical Imaging 2020: Digital Pathology, Houston, TX, USA, 15–20 February 2020; SPIE: Bellingham, WA, USA, 2020; Volume 11320, pp. 41–46. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. Ssd: Single shot multibox detector. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 21–37. [Google Scholar]
- Mahmood, T.; Arsalan, M.; Owais, M.; Lee, M.B.; Park, K.R. Artificial intelligence-based mitosis detection in breast cancer histopathology images using faster R-CNN and deep CNNs. J. Clin. Med. 2020, 9, 749. [Google Scholar] [CrossRef] [PubMed]
- Kawazoe, Y.; Shimamoto, K.; Yamaguchi, R.; Shintani-Domoto, Y.; Uozaki, H.; Fukayama, M.; Ohe, K. Faster R-CNN-based glomerular detection in multistained human whole slide images. J. Imaging 2018, 4, 91. [Google Scholar] [CrossRef]
- Harrison, P.; Park, K. Tumor Detection In Breast Histopathological Images Using Faster R-CNN. In Proceedings of the 2021 International Symposium on Medical Robotics (ISMR), Atlanta, GA, USA, 17–19 November 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–7. [Google Scholar]
- Shibly, K.H.; Dey, S.K.; Islam, M.T.U.; Rahman, M.M. COVID faster R–CNN: A novel framework to Diagnose Novel Coronavirus Disease (COVID-19) in X-ray images. Inform. Med. Unlocked 2020, 20, 100405. [Google Scholar] [CrossRef] [PubMed]
- Ruifrok, A.C.; Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar] [PubMed]
- Srivastava, S.; Divekar, A.V.; Anilkumar, C.; Naik, I.; Kulkarni, V.; Pattabiraman, V. Comparative analysis of deep learning image detection algorithms. J. Big Data 2021, 8, 66. [Google Scholar] [CrossRef]
- Bokhorst, J.M.; Rijstenberg, L.; Goudkade, D.; Nagtegaal, I.; van der Laak, J.; Ciompi, F. Automatic detection of tumor budding in colorectal carcinoma with deep learning. In Proceedings of the Computational Pathology and Ophthalmic Medical Image Analysis: First International Workshop, COMPAY 2018, and 5th International Workshop, OMIA 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 130–138. [Google Scholar]
- Pai, R.K.; Hartman, D.; Schaeffer, D.F.; Rosty, C.; Shivji, S.; Kirsch, R.; Pai, R.K. Development and initial validation of a deep learning algorithm to quantify histological features in colorectal carcinoma including tumour budding/poorly differentiated clusters. Histopathology 2021, 79, 391–405. [Google Scholar] [CrossRef] [PubMed]
| Dataset | Number of WSIs | Origin (# of Slides) | Annotations |
|---|---|---|---|
| dev-l | 51 | Bern (3), Dublin (1), Nijmegen (47) | 480 tumor bud candidates and 321 non-tumor-bud candidates |
| dev-v | 23 | Bern (1), Dublin (0), Nijmegen (21) | 200 tumor bud candidates and 151 non-tumor-bud candidates |
| dev-t | 10 | Bern (2), Dublin (1), Nijmegen (8) | 330 tumor bud candidates and 283 non-tumor-bud candidates |
| eval | 240 | Bern (240) | Manual hotspot locations and number of tumor buds within this hotspot |
| n | % | ||
|---|---|---|---|
| Sex | Male | 15 | 62.5 |
| Female | 9 | 37.5 | |
| Age, years | <65 | 14 | 58.3 |
| ≥65 | 10 | 41.6 | |
| Invasion depth | T1T2 | 7 | 29.1 |
| T3T4 | 17 | 70.9 | |
| Nodal status | 0 and 1 | 18 | 75.0 |
| 2 | 6 | 25.0 | |
| Death | Yes | 14 | 58.3 |
| No | 10 | 41.6 |
| Model | Sensitivity | |
|---|---|---|
| Teacher | DenseNet | 0.83 |
| Faster R-CNN | 0.47 | |
| Student | DenseNet | 0.87 |
| Faster R-CNN | 0.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bokhorst, J.-M.; Nagtegaal, I.D.; Zlobec, I.; Dawson, H.; Sheahan, K.; Simmer, F.; Kirsch, R.; Vieth, M.; Lugli, A.; van der Laak, J.; et al. Semi-Supervised Learning to Automate Tumor Bud Detection in Cytokeratin-Stained Whole-Slide Images of Colorectal Cancer. Cancers 2023, 15, 2079. https://doi.org/10.3390/cancers15072079
Bokhorst J-M, Nagtegaal ID, Zlobec I, Dawson H, Sheahan K, Simmer F, Kirsch R, Vieth M, Lugli A, van der Laak J, et al. Semi-Supervised Learning to Automate Tumor Bud Detection in Cytokeratin-Stained Whole-Slide Images of Colorectal Cancer. Cancers. 2023; 15(7):2079. https://doi.org/10.3390/cancers15072079
Chicago/Turabian StyleBokhorst, John-Melle, Iris D. Nagtegaal, Inti Zlobec, Heather Dawson, Kieran Sheahan, Femke Simmer, Richard Kirsch, Michael Vieth, Alessandro Lugli, Jeroen van der Laak, and et al. 2023. "Semi-Supervised Learning to Automate Tumor Bud Detection in Cytokeratin-Stained Whole-Slide Images of Colorectal Cancer" Cancers 15, no. 7: 2079. https://doi.org/10.3390/cancers15072079
APA StyleBokhorst, J.-M., Nagtegaal, I. D., Zlobec, I., Dawson, H., Sheahan, K., Simmer, F., Kirsch, R., Vieth, M., Lugli, A., van der Laak, J., & Ciompi, F. (2023). Semi-Supervised Learning to Automate Tumor Bud Detection in Cytokeratin-Stained Whole-Slide Images of Colorectal Cancer. Cancers, 15(7), 2079. https://doi.org/10.3390/cancers15072079






