Increased SEC23A Expression Correlates with Poor Prognosis and Immune Infiltration in Stomach Adenocarcinoma †
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression Analyses of SEC23A by Bioinformatics
2.2. Tissue Samples and Real-Time Fluorescence Quantitative PCR (qRT–PCR) Analysis
2.3. Immunohistochemistry and Western Blot Assay
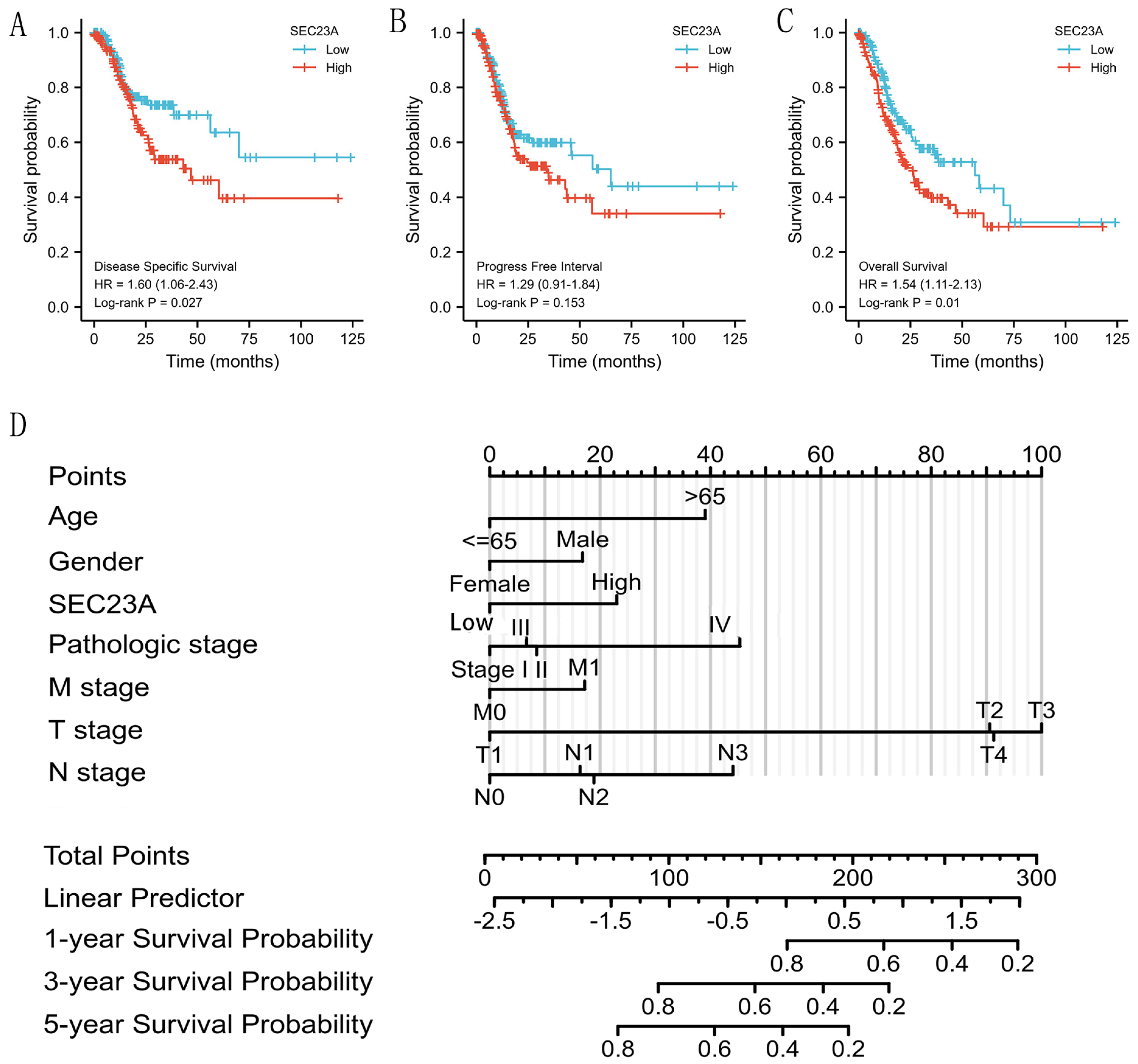
2.4. Analysis of SEC23A Expression with Clinicopathological Features and Survival
2.5. Analyses of Genes Co-Expressed with SEC23A and Gene Set Enrichment Analysis (GSEA)
2.6. Evaluation of Tumor-Infiltrating Immune Cells and Expression of Checkpoint-Related Genes
2.7. Statistical Methods
3. Results
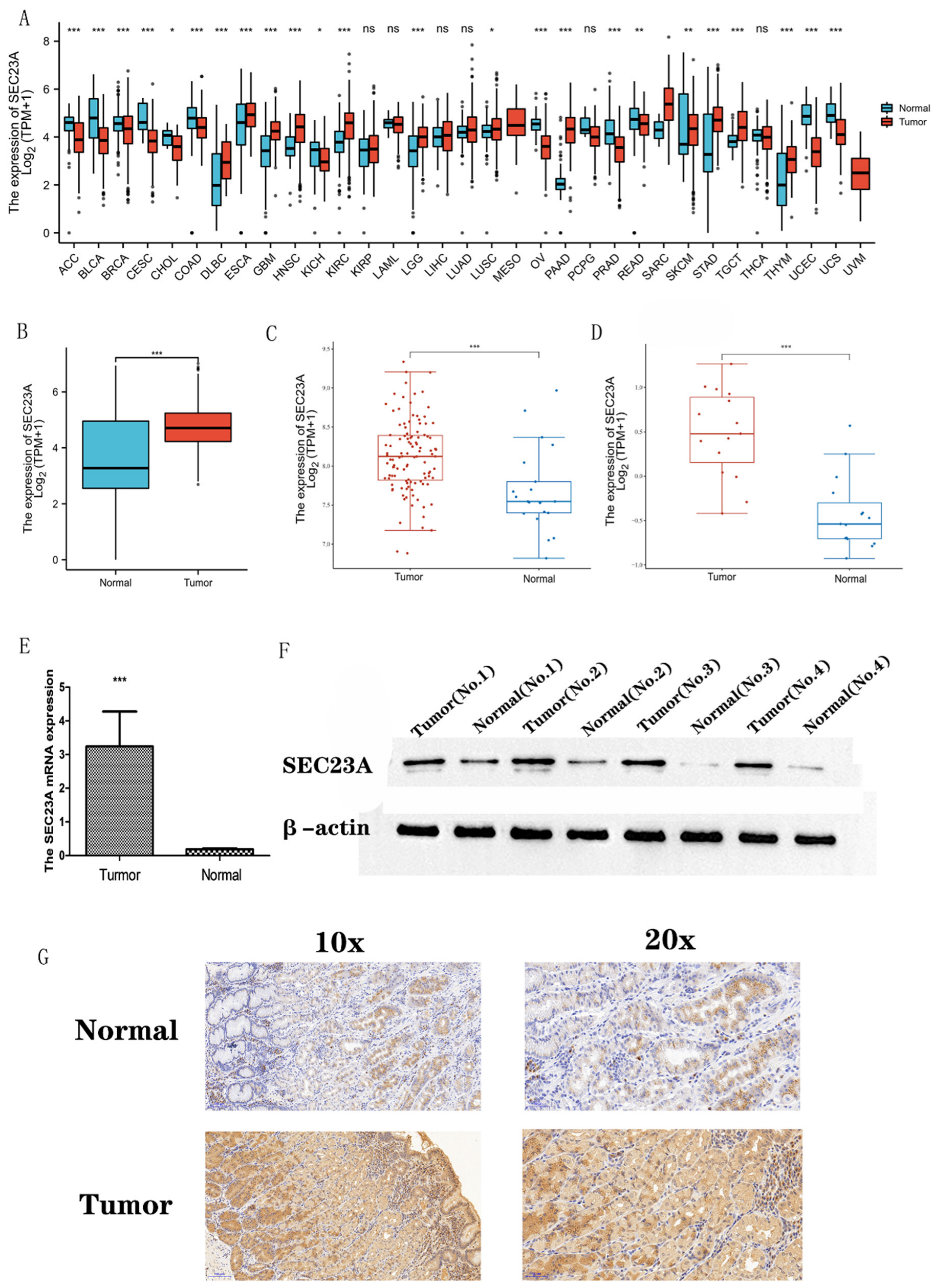
3.1. Transcriptional Levels of SEC23A in Pan-Cancer and STAD
3.2. sec23a Protein Was Upregulated in Clinical STAD Specimens
3.3. Association between SEC23A Expression and Clinicopathological Variables
3.4. High SEC23A Expression Predicted Poor Prognosis in GC Patients
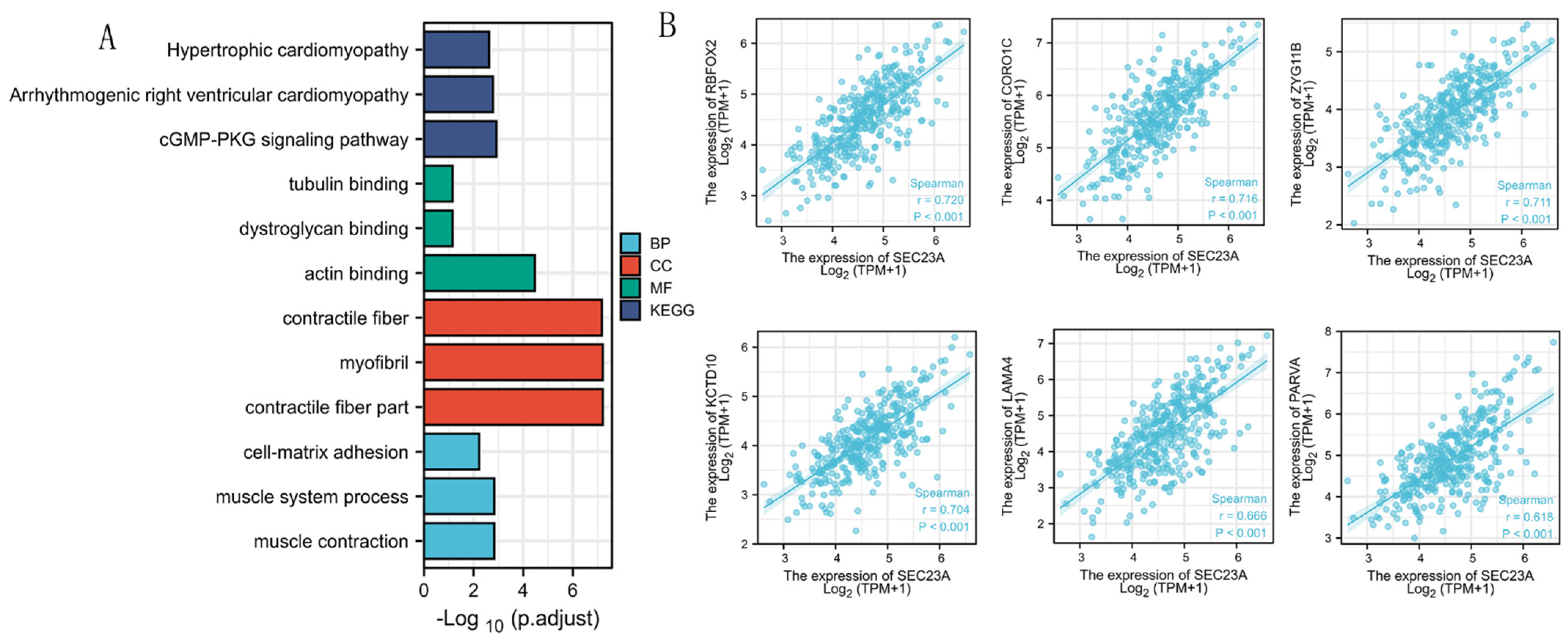
3.5. Analyses of Genes Co-Expressed with SEC23A in STAD
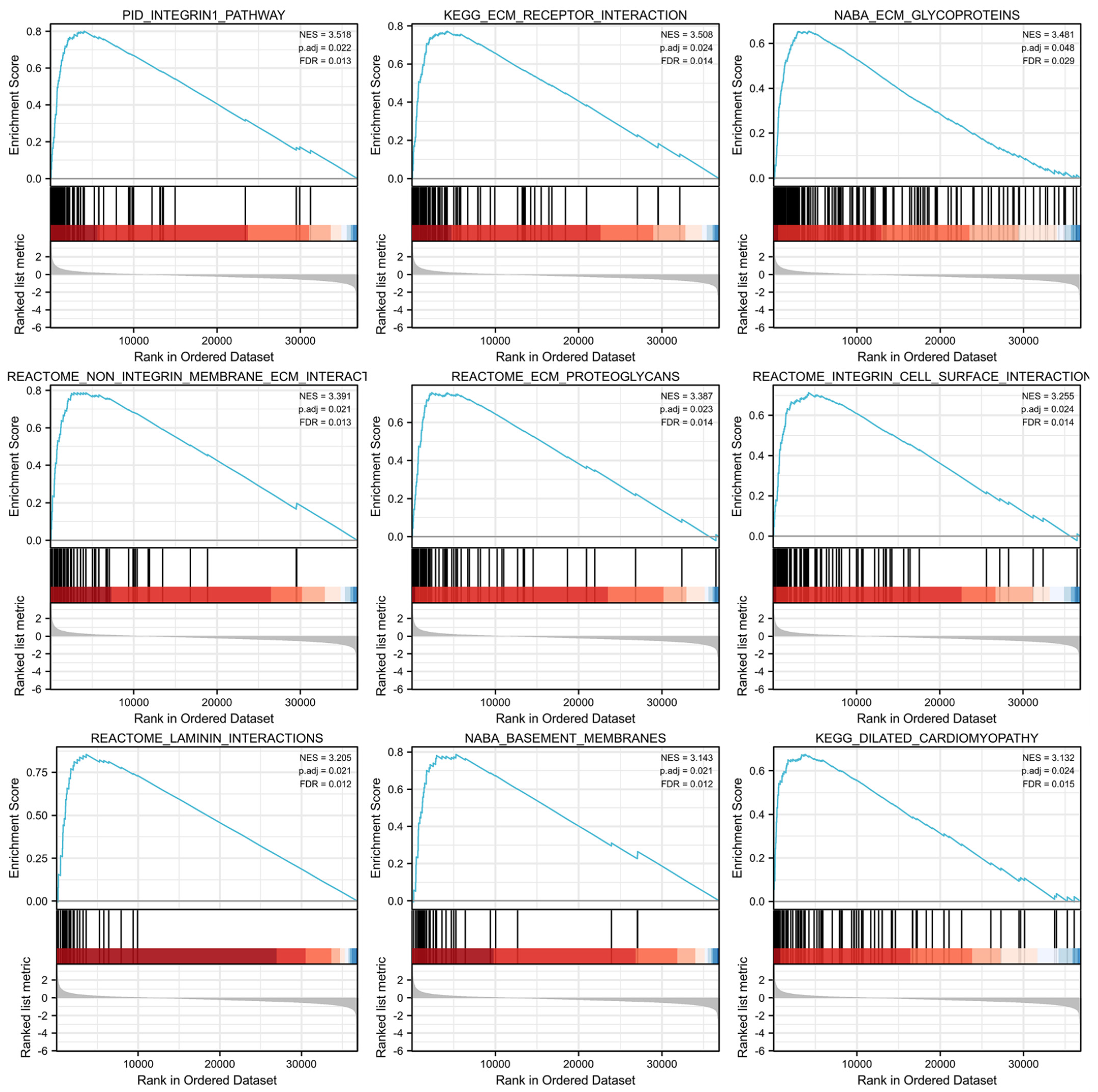
3.6. GSEA Identified SEC23A-Related Pathways
3.7. Expression of Tumor-Infiltrating Immune Cells and Checkpoint-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, H.; Zhang, H.; Wang, Y.; Wang, X.; Hou, H. Global Health Epidemiology Reference Group. Survival of gastric cancer in China from 2000 to 2022: A nationwide systematic review of hospital-based studies. J. Glob. Health 2022, 17, 11014. [Google Scholar] [CrossRef]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Paltridge, J.L.; Belle, L.; Khew-Goodall, Y. The secretome in cancer progression. Biochim. Biophys. Acta 2013, 1834, 2233–2341. [Google Scholar] [CrossRef] [PubMed]
- Barlowe, C. Twenty-five years after coat protein complex II. Mol. Biol. Cell 2020, 1, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Khoriaty, R.; Hesketh, G.G.; Bernard, A.; Weyand, A.C.; Mellacheruvu, D.; Zhu, G.; Hoenerhoff, M.J.; McGee, B.; Everett, L.; Adams, E.J.; et al. Functions of the COPII gene paralogs SEC23A and SEC23B are interchangeable In Vivo. Proc. Natl. Acad. Sci. USA 2018, 115, E7748–E7757. [Google Scholar] [CrossRef]
- Schwarz, K.; Iolascon, A.; Verissimo, F.; Trede, N.S.; Horsley, W.; Chen, W.; Paw, B.; Hopfner, K.-P.; Holzmann, K.; Russo, R.; et al. Mutations affecting the secretory COPII coat component SEC23B cause congenital dyserythropoietic anemia type II. Nat. Genet. 2009, 41, 936–940. [Google Scholar] [CrossRef]
- Boyadjiev, S.; Fromme, J.C.; Ben, J.; Chong, S.S.; Nauta, C.; Hur, D.; Zhang, G.; Hamamoto, S.; Schekman, R.; Ravazzola, M.; et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat. Genet. 2006, 38, 1192–1197. [Google Scholar] [CrossRef]
- Zeng, B.; Zhao, Q.; Sun, Z.; Liu, D.; Chen, H.; Li, X.; Wang, J.; Xing, H.R. SEC23A Is an Independent Prognostic Biomarker in Bladder Cancer Correlated with MAPK Signaling. Front. Genet. 2021, 12, 672832. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, D.; Bin Zeng, B.; Zhao, Q.; Li, X.; Chen, H.; Wang, J.; Xing, H.R. Sec23a inhibits the self-renewal of melanoma cancer stem cells via inactivation of ER-phagy. Cell Commun. Signal. 2022, 20, 22. [Google Scholar] [CrossRef]
- Sun, Z.; Zhou, S.; Tang, J.; Ye, T.; Li, J.; Liu, D.; Zhou, J.; Wang, J.; Xing, H.R. Sec23a mediates miR-200c augmented oligometastatic to polymetastatic progression. Ebiomedicine 2018, 37, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Szczyrba, J.; Nolte, E.; Wach, S.; Kremmer, E.; Stöhr, R.; Hartmann, A.; Wieland, W.; Wullich, B.; Grässer, F.A. Downregulation of Sec23A protein by miRNA-375 in prostate carcinoma. Mol. Cancer Res. 2011, 9, 791–800. [Google Scholar] [CrossRef]
- Su, Z.; Shu, K.; Li, G. Increased ANXA5 expression in stomach adenocarcinoma infers a poor prognosis and high level of immune infiltration. Cancer Biomark. 2022, 35, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Hippo, Y.; Taniguchi, H.; Tsutsumi, S.; Machida, N.; Chong, J.-M.; Fukayama, M.; Kodama, T.; Aburatani, H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002, 62, 233–420. [Google Scholar] [PubMed]
- Li, L.; Zhu, Z.; Zhao, Y.; Zhang, Q.; Wu, X.; Miao, B.; Cao, J.; Fei, S. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci. Rep. 2019, 9, 7827. [Google Scholar] [CrossRef]
- Varghese, F.; Bukhari, A.B.; Malhotra, R.; De, A. IHC Profiler: An open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE 2014, 9, e96801. [Google Scholar] [CrossRef] [PubMed]
- Peotter, J.; Kasberg, W.; Pustova, I.; Audhya, A. COPII-mediated trafficking at the ER/ERGIC interface. Traffic 2019, 20, 491–503. [Google Scholar] [CrossRef]
- Li, B.; Zeng, Y.; Jiang, L. COPII vesicles in plant autophagy and endomembrane trafficking. FEBS Lett. 2022, 596, 2314–2323. [Google Scholar] [CrossRef]
- Sun, Z.; Zeng, B.; Liu, D.; Zhao, Q.; Wang, J.; Xing, H.R. S100A8 transported by SEC23A inhibits metastatic colonization via autocrine activation of autophagy. Cell Death Dis. 2020, 11, 650. [Google Scholar] [CrossRef]
- Wang, Y.; Lieberman, R.; Pan, J.; Zhang, Q.; Du, M.; Zhang, P.; Nevalainen, M.; Kohli, M.; Shenoy, N.K.; Meng, H.; et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol. Cancer 2016, 15, 70. [Google Scholar] [CrossRef]
- Korpal, M.; Ell, B.J.; Buffa, F.; Ibrahim, T.; Blanco, M.A.; Celià-Terrassa, T.; Mercatali, L.; Khan, Z.; Goodarzi, H.; Hua, Y.; et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011, 17, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.; Nolte, E.; Wach, S.; Szczyrba, J.; Taubert, H.; Rau, T.T.; Hartmann, A.; Grässer, F.A.; Wullich, B. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol. Cancer Res. 2014, 12, 250–263. [Google Scholar] [CrossRef]
- Kubota, D.; Yoshida, A.; Tsuda, H.; Suehara, Y.; Okubo, T.; Saito, T.; Orita, H.; Sato, K.; Taguchi, T.; Yao, T.; et al. Gene expression network analysis of ETV1 reveals KCTD10 as a novel prognostic biomarker in gastrointestinal stromal tumor. PLoS ONE 2013, 8, e738962013. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, X.; Wu, Z.; Tan, S.; Zhu, T.; Ding, K. CORO1C expression is associated with poor survival rates in gastric cancer and promotes metastasis in vitro. FEBS Open Bio 2019, 9, 1097–1108. [Google Scholar] [CrossRef]
- Liao, M.; Peng, L. MiR-206 may suppress non-small lung cancer metastasis by targeting CORO1C. Cell. Mol. Biol. Lett. 2020, 25, 13–22. [Google Scholar] [CrossRef]
- Cai, H.; Kondo, M.; Sandhow, L.; Xiao, P.; Johansson, A.-S.; Sasaki, T.; Zawacka-Pankau, J.; Tryggvason, K.; Ungerstedt, J.S.; Walfridsson, J.; et al. Critical role of Lama4 for hematopoiesis regeneration and acute myeloid leukemia progression. Blood 2022, 139, 3040–3057. [Google Scholar] [CrossRef]
- Wang, M.; Li, C.; Liu, Y.; Wang, Z. Effect of LAMA4 on Prognosis and Its Correlation with Immune Infiltration in Gastric Cancer. BioMed Res. Int. 2021, 2021, 6428873. [Google Scholar] [CrossRef]
- Zheng, B.; Qu, J.; Ohuchida, K.; Feng, H.; Chong, S.J.F.; Yan, Z.; Piao, Y.; Liu, P.; Sheng, N.; Eguchi, D.; et al. LAMA4 upregulation is associated with high liver metastasis potential and poor survival outcome of Pancreatic Cancer. Theranostics 2020, 10, 10274–10289. [Google Scholar] [CrossRef]
- Yang, Z.-X.; Zhang, B.; Wei, J.; Jiang, G.-Q.; Wu, Y.-L.; Leng, B.-J.; Xing, C.-G. MiR-539 inhibits proliferation and migration of triple-negative breast cancer cells by down-regulating LAMA4 expression. Cancer Cell Int. 2018, 18, 16. [Google Scholar] [CrossRef]
- Murai, T.; Kawashima, H.; Naor, D. Editorial: Cell-Cell and Cell-Matrix Adhesion in Immunobiology and Cancer. Front. Immunol. 2020, 10, 3126. [Google Scholar] [CrossRef]
- Jia, H.; Yu, F.; Li, B.; Gao, Z. Actin-binding protein Anillin promotes the progression of gastric cancer in vitro and in mice. J. Clin. Lab. Anal. 2021, 35, e236352021. [Google Scholar] [CrossRef]
- Xiang, T.; Yuan, C.; Guo, X.; Wang, H.; Cai, Q.; Xiang, Y.; Luo, W.; Liu, G. The novel ZEB1-upregulated protein PRTG induced by Helicobacter pylori infection promotes gastric carcinogenesis through the cGMP/PKG signaling pathway. Cell Death Dis. 2021, 12, 150. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 2018, 120, 2782–2790. [Google Scholar] [CrossRef]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef]
- Li, C.; Teixeira, A.F.; Zhu, H.-J.; Dijke, P.T. Cancer associated-fibroblast-derived exosomes in cancer progression. Mol. Cancer 2021, 20, 154. [Google Scholar] [CrossRef]
- He, X.; Kiratipaiboon, C.; Porter, D.W.; Rojanasakul, L.W.; Dinu, C.Z.; Wang, K.; Yang, Y.; Rojanasakul, Y. Predicting Nanotube Fibrogenicity through Stem Cell-Mediated Fibroblast Focus and Spheroid Formation. Nano Lett. 2018, 10, 6500–6508. [Google Scholar] [CrossRef]
- Gupta, R. Epigenetic regulation and targeting of ECM for cancer therapy. Am. J. Physiol. Physiol. 2022, 322, C762–C768. [Google Scholar] [CrossRef]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- Kumar, A.; Watkins, R.; Vilgelm, A.E. Cell Therapy with TILs: Training and Taming T Cells to Fight Cancer. Front. Immunol. 2021, 12, 690499. [Google Scholar] [CrossRef]
- Chan, L.F.; Sadahiro, S.; Suzuki, T.; Okada, K.; Miyakita, H.; Yamamoto, S.; Kajiwara, H. Tissue-Infiltrating Lymphocytes as a Predictive Factor for Recurrence in Patients with Curatively Resected Colon Cancer: A Propensity Score Matching Analysis. Oncology 2020, 98, 680–688. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.-S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Laghi, L.; Negri, F.; Gaiani, F.; Cavalleri, T.; Grizzi, F.; Angelis, G.L.D.; Malesci, A. Prognostic and Predictive Cross-Roads of Microsatellite Instability and Immune Response to Colon Cancer. Int. J. Mol. Sci. 2020, 21, 9680. [Google Scholar] [CrossRef]
- Savas, P.; Virassamy, B.; Ye, C.; Salim, A.; Mintoff, C.P.; Caramia, F.; Salgado, R.; Byrne, D.J.; Teo, Z.L.; Dushyanthen, S.; et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018, 24, 986–993. [Google Scholar] [CrossRef]


| Characteristics | Total (N) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| Hazard Ratio (95% CI) | p Value | Hazard Ratio (95% CI) | p Value | ||
| T stage | 347 | ||||
| T1 | 18 | reference | |||
| T2 | 78 | 6.725 (0.913–49.524) | 0.061 | 4.332 (0.548–34.233) | 0.165 |
| T3 | 157 | 9.548 (1.326–68.748) | 0.025 | 4.984 (0.564–44.019) | 0.148 |
| T4 | 94 | 9.634 (1.323–70.151) | 0.025 | 4.197 (0.460–38.322) | 0.204 |
| N stage | 347 | ||||
| N0 | 102 | reference | |||
| N1 | 97 | 1.629 (1.001–2.649) | 0.049 | 1.299 (0.644–2.620) | 0.465 |
| N2 | 74 | 1.655 (0.979–2.797) | 0.060 | 1.366 (0.581–3.212) | 0.475 |
| N3 | 74 | 2.709 (1.669–4.396) | <0.001 | 1.985 (0.845–4.664) | 0.116 |
| M stage | 347 | ||||
| M0 | 322 | reference | |||
| M1 | 25 | 2.254 (1.295–3.924) | 0.004 | 1.216 (0.513–2.881) | 0.658 |
| Age | 347 | ||||
| ≤65 | 153 | reference | |||
| >65 | 194 | 1.620 (1.154–2.276) | 0.005 | 1.836 (1.264–2.667) | 0.001 |
| Gender | 347 | ||||
| Female | 123 | reference | |||
| Male | 224 | 1.267 (0.891–1.804) | 0.188 | ||
| Pathologic stage | 347 | ||||
| UICC I | 50 | reference | |||
| UICC II | 110 | 1.551 (0.782–3.078) | 0.209 | 1.113 (0.392–3.159) | 0.840 |
| UICC III | 149 | 2.381 (1.256–4.515) | 0.008 | 1.119 (0.284–4.415) | 0.873 |
| UICC IV | 38 | 3.991 (1.944–8.192) | <0.001 | 2.204 (0.539–9.010) | 0.271 |
| SEC23A | 347 | ||||
| Low | 184 | Reference | |||
| High | 183 | 1.542 (1.106–2.151) | 0.011 | 1.460 (1.016–2.098) | 0.041 |
| Ontology | ID | Description | GeneRatio | BgRatio | p Value | p.Adjust | q Value |
|---|---|---|---|---|---|---|---|
| BP | GO:0006936 | muscle contraction | 11/88 | 360/18,670 | 9.99 × 10−7 | 0.001 | 0.001 |
| BP | GO:0003012 | muscle system process | 12/88 | 465/18,670 | 1.83 × 10−6 | 0.001 | 0.001 |
| BP | GO:0007160 | cell-matrix adhesion | 8/88 | 225/18,670 | 1.10 × 10−5 | 0.006 | 0.005 |
| CC | GO:0044449 | contractile fiber part | 12/91 | 221/19,717 | 4.22 × 10−10 | 6.16 × 10−8 | 4.75 × 10−8 |
| CC | GO:0030016 | myofibril | 12/91 | 224/19,717 | 4.93 × 10−10 | 6.16 × 10−8 | 4.75 × 10−8 |
| CC | GO:0043292 | contractile fiber | 12/91 | 234/19,717 | 8.13 × 10−10 | 6.77 × 10−8 | 5.22 × 10−8 |
| MF | GO:0003779 | actin binding | 13/86 | 431/17,697 | 1.53 × 10−7 | 3.47 × 10−5 | 3.25 × 10−5 |
| MF | GO:0002162 | dystroglycan binding | 2/86 | 10/17,697 | 0.001 | 0.071 | 0.067 |
| MF | GO:0015631 | tubulin binding | 7/86 | 336/17,697 | 0.001 | 0.071 | 0.067 |
| KEGG | hsa04022 | cGMP-PKG signaling pathway | 7/37 | 167/8076 | 8.70 × 10−6 | 0.001 | 0.001 |
| KEGG | hsa05412 | Arrhythmogenic right ventricular cardiomyopathy | 5/37 | 77/8076 | 2.37 × 10−5 | 0.002 | 0.001 |
| KEGG | hsa05410 | Hypertrophic cardiomyopathy | 5/37 | 90/8076 | 5.06 × 10−5 | 0.002 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhaoran, S.; Linnebacher, C.S.; Linnebacher, M. Increased SEC23A Expression Correlates with Poor Prognosis and Immune Infiltration in Stomach Adenocarcinoma. Cancers 2023, 15, 2065. https://doi.org/10.3390/cancers15072065
Zhaoran S, Linnebacher CS, Linnebacher M. Increased SEC23A Expression Correlates with Poor Prognosis and Immune Infiltration in Stomach Adenocarcinoma. Cancers. 2023; 15(7):2065. https://doi.org/10.3390/cancers15072065
Chicago/Turabian StyleZhaoran, Su, Christina Susanne Linnebacher, and Michael Linnebacher. 2023. "Increased SEC23A Expression Correlates with Poor Prognosis and Immune Infiltration in Stomach Adenocarcinoma" Cancers 15, no. 7: 2065. https://doi.org/10.3390/cancers15072065
APA StyleZhaoran, S., Linnebacher, C. S., & Linnebacher, M. (2023). Increased SEC23A Expression Correlates with Poor Prognosis and Immune Infiltration in Stomach Adenocarcinoma. Cancers, 15(7), 2065. https://doi.org/10.3390/cancers15072065







