Critical Evaluation of a microRNA-Based Risk Classifier Predicting Cancer-Specific Survival in Renal Cell Carcinoma with Tumor Thrombus of the Inferior Vena Cava
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tumor Tissue Samples and Patients
2.2. RNA Extraction and qRT-PCR
2.3. Statistics and Computational Analysis
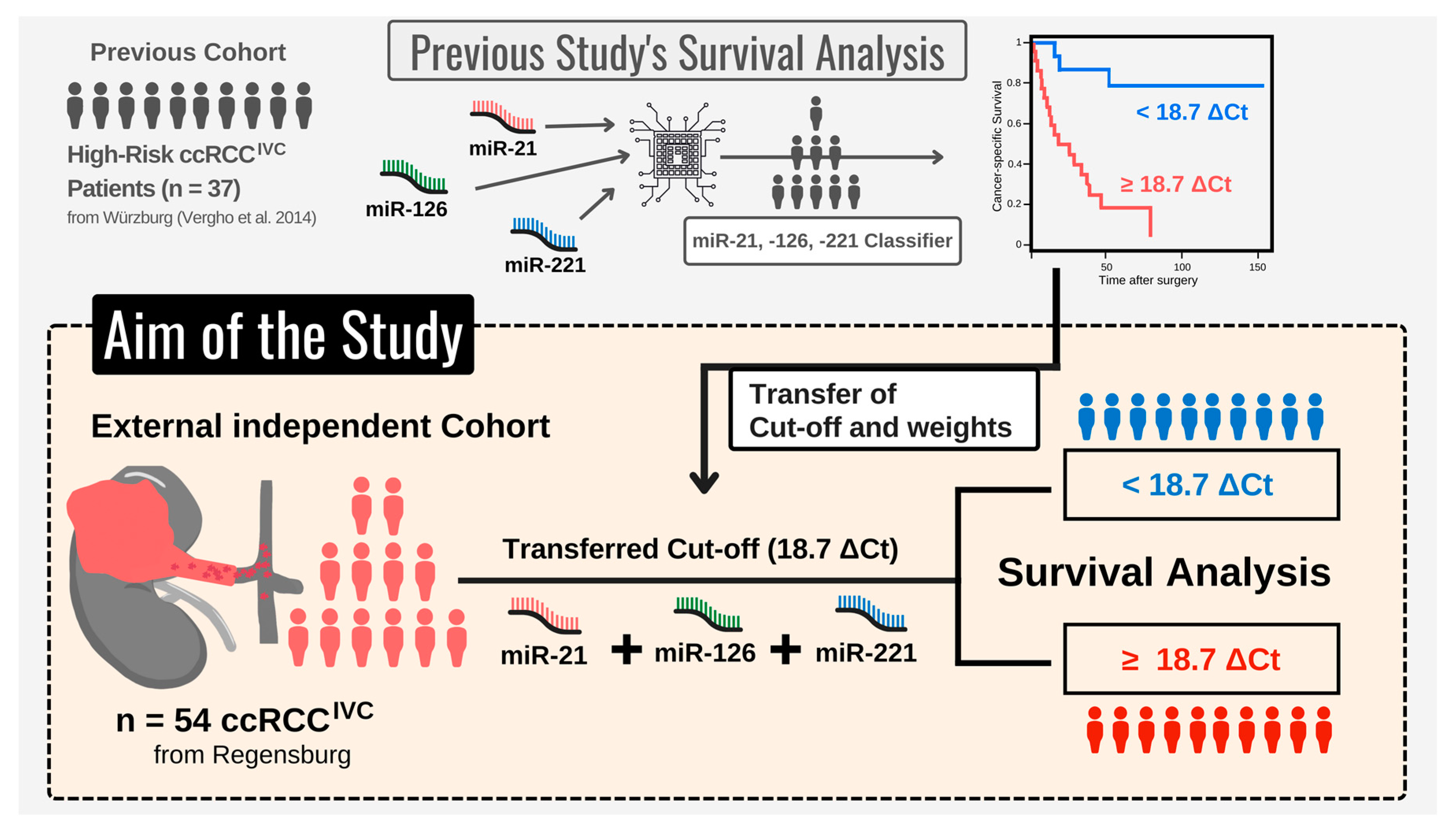
2.3.1. Initial microRNA-Based Risk Classifier Calculation
- Performing single-variable and multivariable Cox regression analysis, Vergho et al. evaluated the impact of clinicopathological parameters and various miRs on CSS.
- To select the best fitting Cox model, the relative goodness-of-fit was measured based on the Akaike information criterion (AIC) for different variable combinations. The combination of miR-21, -126, and -221 displayed the best prediction properties.
- Finally, the miR-based risk classifier was calculated based on the publication by Lossos et al. [22]. Hereby, the z-factor calculated by the multivariable Cox model (R package ‘survival’) for miR-21, -126, and -221 was multiplied with the relative expression levels (ΔCt) of the respective miR. This approach resulted in the following formula: (4.592 × ΔCt miR-21) + (−3.892 × ΔCt miR-126) + (−1.938 × ΔCt miR-221).
- Subsequently, a receiver operating characteristic (ROC) curve was plotted (R package pROC), showing the true positive rate (TPR) against the false positive rate (FPR) at various threshold settings. The risk score cut-off/threshold of 18.7 ΔCt was determined from ROC using the Youden’s J statistics [23] (integrated in pROC package [24], default settings).
2.3.2. Clinical Validation of microRNA-Based Risk Classifier
3. Results
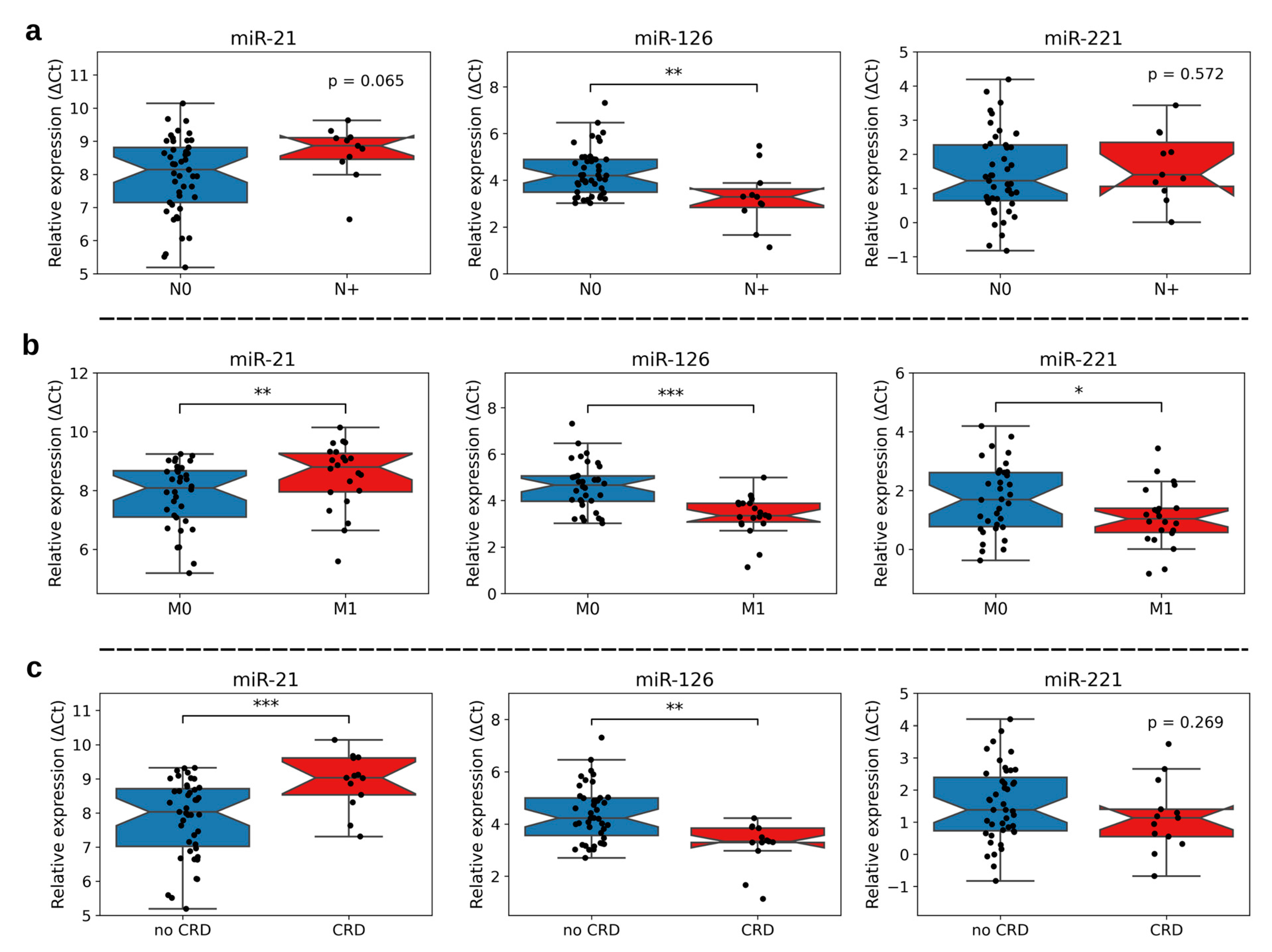
3.1. Association of miR-21, -126, and -221 Expression with Clinicopathological Characteristics
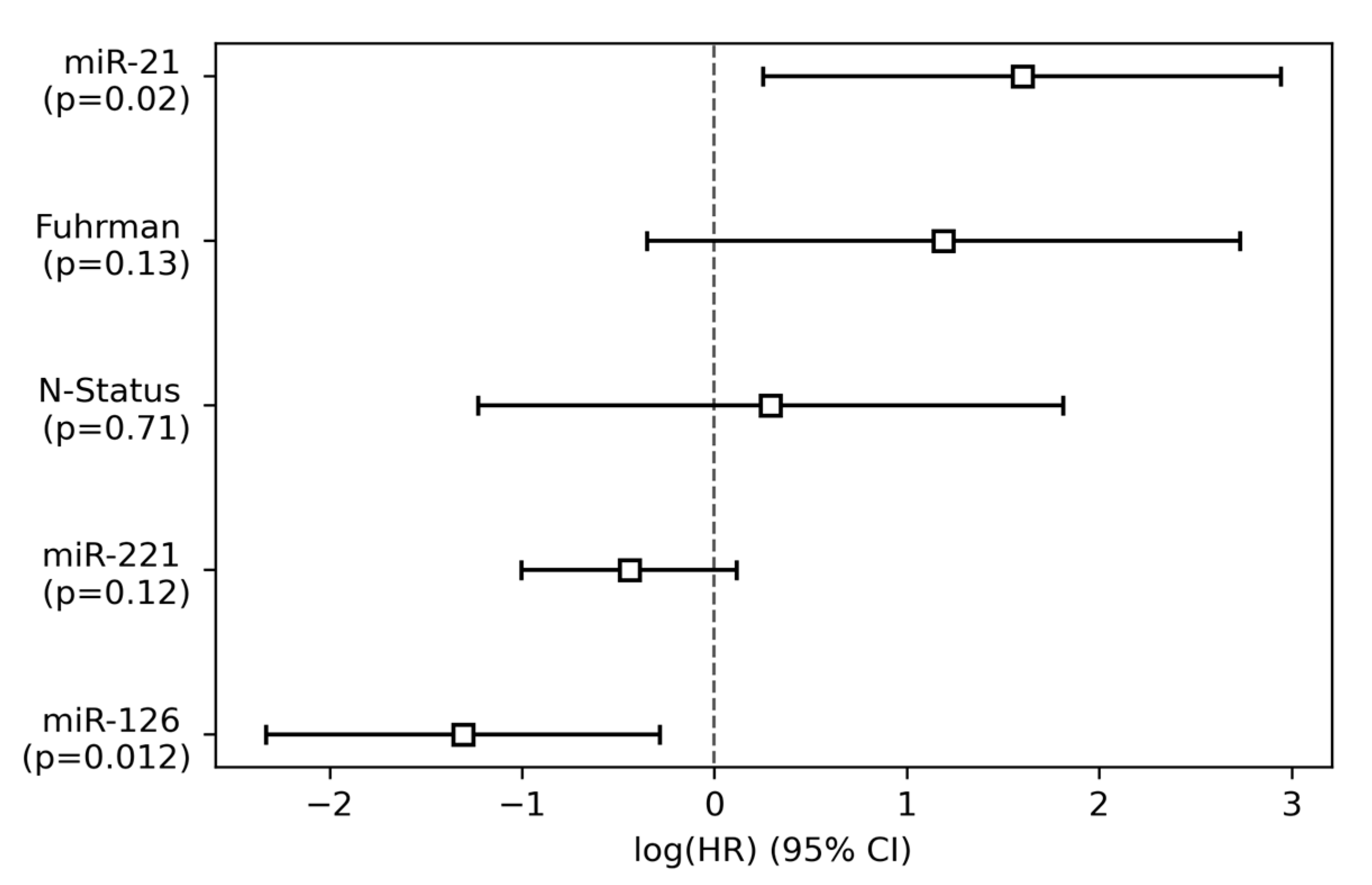
3.2. Single-Variable and Multivariable Cox Regression Analysis
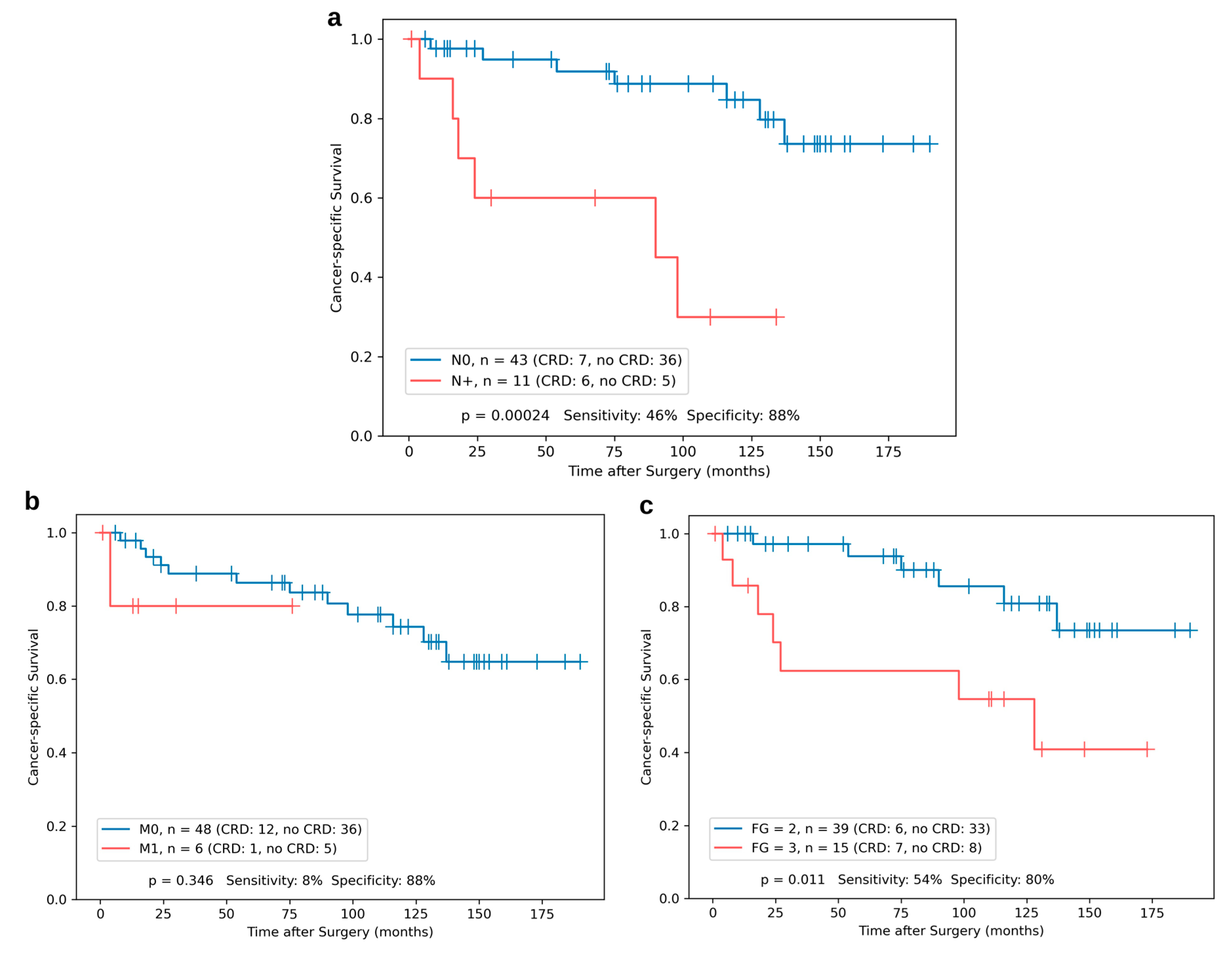
3.3. Kaplan–Meier Analyses for Single miR Expression and the Risk Classifier
3.4. Testing the Composition and Robustness of the Risk Classifier
4. Discussion
4.1. Evaluating an miR-Based Risk Classifier for RCC with Infiltration of the Vena Cava
4.2. Functional Roles of miR-21, miR-126, and miR-221 in Cancer and Thrombosis
4.3. Limitations and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ljungberg, B.; Stenling, R.; Osterdahl, B.; Farrelly, E.; Aberg, T.; Roos, G. Vein Invasion in Renal Cell Carcinoma: Impact on Metastatic Behavior and Survival. J. Urol. 1995, 154, 1681–1684. [Google Scholar] [CrossRef] [PubMed]
- Hoehn, W.; Hermanek, P. Invasion of Veins in Renal Cell Carcinoma -Frequency, Correlation and Prognosis. Eur. Urol. 1983, 9, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Novick, A.C.; Cosgrove, D.M. Surgical Approach for Removal of Renal Cell Carcinoma Extending into the Vena Cava and the Right Atrium. J. Urol. 1980, 123, 947–950. [Google Scholar] [CrossRef]
- Haferkamp, A.; Bastian, P.J.; Jakobi, H.; Pritsch, M.; Pfitzenmaier, J.; Albers, P.; Hallscheidt, P.; Müller, S.C.; Hohenfellner, M. Renal Cell Carcinoma With Tumor Thrombus Extension Into the Vena Cava: Prospective Long-Term Followup. J. Urol. 2007, 177, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Blute, M.L.; Leibovich, B.C.; Lohse, C.M.; Cheville, J.C.; Zincke, H. The Mayo Clinic Experience with Surgical Management, Complications and Outcome for Patients with Renal Cell Carcinoma and Venous Tumour Thrombus. BJU Int. 2004, 94, 33–41. [Google Scholar] [CrossRef]
- Vergho, D.C.; Loeser, A.; Kocot, A.; Spahn, M.; Riedmiller, H. Tumor Thrombus of Inferior Vena Cava in Patients with Renal Cell Carcinoma—Clinical and Oncological Outcome of 50 Patients after Surgery. BMC Res. Notes 2012, 5, 264. [Google Scholar] [CrossRef]
- Haddad, A.Q.; Wood, C.G.; Abel, E.J.; Krabbe, L.-M.; Darwish, O.M.; Thompson, R.H.; Heckman, J.E.; Merril, M.M.; Gayed, B.A.; Sagalowsky, A.I.; et al. Oncologic Outcomes Following Surgical Resection of Renal Cell Carcinoma with Inferior Vena Caval Thrombus Extending Above the Hepatic Veins: A Contemporary Multicenter Cohort. J. Urol. 2014, 192, 1050–1056. [Google Scholar] [CrossRef]
- Klatte, T.; Pantuck, A.J.; Riggs, S.B.; Kleid, M.D.; Shuch, B.; Zomorodian, N.; Kabbinavar, F.F.; Belldegrun, A.S. Prognostic Factors for Renal Cell Carcinoma With Tumor Thrombus Extension. J. Urol. 2007, 178, 1189–1195. [Google Scholar] [CrossRef]
- Berczi, A.; Flasko, T.; Szerafin, T.; Thomas, B.; Bacso, Z.; Berczi, C. Surgical Management and Outcome of Renal Cell Carcinoma with Inferior Vena Cava Tumor Thrombus. Urol. Int. 2017, 99, 267–271. [Google Scholar] [CrossRef]
- Vergho, D.C.; Kneitz, S.; Kalogirou, C.; Burger, M.; Krebs, M.; Rosenwald, A.; Spahn, M.; Löser, A.; Kocot, A.; Riedmiller, H.; et al. Impact of MiR-21, MiR-126 and MiR-221 as Prognostic Factors of Clear Cell Renal Cell Carcinoma with Tumor Thrombus of the Inferior Vena Cava. PLoS ONE 2014, 9, e109877. [Google Scholar] [CrossRef]
- Spachmann, P.J.; Breyer, J.; Kalogirou, C.; Lausenmeyer, E.-M.; Weber, F.; Prohaska, S.; Denzinger, S.; Burger, M.; Kübler, H.; Otto, W.; et al. Impact of E-Cadherin and β-Catenin as Prognostic Factor in Renal Cell Carcinoma with Tumor Thrombus of the Vena Cava. Urol. Int. 2019, 102, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. MicroRNAs. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a Role in Cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Kong, Y.W.; Ferland-McCollough, D.; Jackson, T.J.; Bushell, M. MicroRNAs in Cancer Management. Lancet Oncol. 2012, 13, e249–e258. [Google Scholar] [CrossRef] [PubMed]
- Zaman, M.S.; Shahryari, V.; Deng, G.; Thamminana, S.; Saini, S.; Majid, S.; Chang, I.; Hirata, H.; Ueno, K.; Yamamura, S.; et al. Up-Regulation of MicroRNA-21 Correlates with Lower Kidney Cancer Survival. PLoS ONE 2012, 7, e31060. [Google Scholar] [CrossRef]
- Vergho, D.; Kneitz, S.; Rosenwald, A.; Scherer, C.; Spahn, M.; Burger, M.; Riedmiller, H.; Kneitz, B. Combination of Expression Levels of MiR-21 and MiR-126 Is Associated with Cancer-Specific Survival in Clear-Cell Renal Cell Carcinoma. BMC Cancer 2014, 14, 25. [Google Scholar] [CrossRef]
- Spadaccino, F.; Gigante, M.; Netti, G.S.; Rocchetti, M.T.; Franzin, R.; Gesualdo, L.; Castellano, G.; Stallone, G.; Ranieri, E. The Ambivalent Role of MiRNAs in Carcinogenesis: Involvement in Renal Cell Carcinoma and Their Clinical Applications. Pharmaceuticals 2021, 14, 322. [Google Scholar] [CrossRef] [PubMed]
- Davidson-Pilon, C. Lifelines: Survival Analysis in Python. JOSS 2019, 4, 1317. [Google Scholar] [CrossRef]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- SciPy 1.0 Contributors; Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Lossos, I.S.; Czerwinski, D.K.; Alizadeh, A.A.; Wechser, M.A.; Tibshirani, R.; Botstein, D.; Levy, R. Prediction of Survival in Diffuse Large-B-Cell Lymphoma Based on the Expression of Six Genes. N. Engl. J. Med. 2004, 350, 1828–1837. [Google Scholar] [CrossRef]
- Youden, W.J. Index for Rating Diagnostic Tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Cho, M.C.; Kim, J.K.; Moon, K.C.; Kim, H.H.; Kwak, C. Prognostic Factor for Korean Patients with Renal Cell Carcinoma and Venous Tumor Thrombus Extension: Application of the New 2009 TNM Staging System. Int. Braz. J. Urol. 2013, 39, 353–363. [Google Scholar] [CrossRef]
- Abel, E.J.; Margulis, V.; Bauman, T.M.; Karam, J.A.; Christensen, W.P.; Krabbe, L.-M.; Haddad, A.; Golla, V.; Wood, C.G. Risk Factors for Recurrence after Surgery in Non-Metastatic RCC with Thrombus: A Contemporary Multicentre Analysis. BJU Int. 2016, 117, E87–E94. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Nakashima, J.; Ohori, M.; Tanaka, A.; Hashimoto, T.; Gondo, T.; Hatano, T.; Tachibana, M. Clinical Variables for Predicting Metastatic Renal Cell Carcinoma Patients Who Might Not Benefit from Cytoreductive Nephrectomy: Neutrophil-to-Lymphocyte Ratio and Performance Status. Int. J. Clin. Oncol. 2014, 19, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Kalogirou, C.; Mulfinger, P.; Sokolakis, I.; Krebs, M.; Kübler, H.; Riedmiller, H.; Vergho, D. Preoperative C-Reactive Protein Values as a Potential Component in Outcome Prediction Models of Metastasized Renal Cell Carcinoma Patients Receiving Cytoreductive Nephrectomy. Urol. Int. 2017, 99, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Xu, B.; Fan, Y.; Yu, W.; Zhang, Q.; Jin, J. Preoperative Gamma-Glutamyltransferase Is Associated with Cancer-Specific Survival and Recurrence-Free Survival of Nonmetastatic Renal Cell Carcinoma with Venous Tumor Thrombus. BioMed Res. Int. 2017, 2017, 3142926. [Google Scholar] [CrossRef]
- Shang, B.; Guo, L.; Shen, R.; Cao, C.; Xie, R.; Jiang, W.; Wen, L.; Bi, X.; Shi, H.; Zheng, S.; et al. Prognostic Significance of NLR About NETosis and Lymphocytes Perturbations in Localized Renal Cell Carcinoma With Tumor Thrombus. Front. Oncol. 2021, 11, 771545. [Google Scholar] [CrossRef]
- Nagamoto, S.; Urakami, S.; Oka, S.; Ogawa, K.; Kono, K.; Sakaguchi, K.; Kinowaki, K.; Yamada, D.; Kume, H. Impact of the Neutrophil-to-Lymphocyte Ratio as a Surgical Prognostic Factor in Renal Cell Carcinoma with Inferior-Vena-Cava Tumor Thrombus. Asian J. Surg. 2023, 46, 192–200. [Google Scholar] [CrossRef]
- Zapała, Ł.; Kunc, M.; Sharma, S.; Biernat, W.; Radziszewski, P. Low Lymphocyte-to-Monocyte Ratio Is the Potential Indicator of Worse Overall Survival in Patients with Renal Cell Carcinoma and Venous Tumor Thrombus. Diagnostics 2021, 11, 2159. [Google Scholar] [CrossRef]
- Argentiero, A.; Solimando, A.G.; Krebs, M.; Leone, P.; Susca, N.; Brunetti, O.; Racanelli, V.; Vacca, A.; Silvestris, N. Anti-Angiogenesis and Immunotherapy: Novel Paradigms to Envision Tailored Approaches in Renal Cell-Carcinoma. JCM 2020, 9, 1594. [Google Scholar] [CrossRef] [PubMed]
- Bedke, J.; Albiges, L.; Capitanio, U.; Giles, R.H.; Hora, M.; Lam, T.B.; Ljungberg, B.; Marconi, L.; Klatte, T.; Volpe, A.; et al. 2021 Updated European Association of Urology Guidelines on the Use of Adjuvant Pembrolizumab for Renal Cell Carcinoma. Eur. Urol. 2022, 81, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Ravaud, A.; Motzer, R.J.; Pandha, H.S.; George, D.J.; Pantuck, A.J.; Patel, A.; Chang, Y.-H.; Escudier, B.; Donskov, F.; Magheli, A.; et al. Adjuvant Sunitinib in High-Risk Renal-Cell Carcinoma after Nephrectomy. N. Engl. J. Med. 2016, 375, 2246–2254. [Google Scholar] [CrossRef]
- Haas, N.B.; Manola, J.; Uzzo, R.G.; Flaherty, K.T.; Wood, C.G.; Kane, C.; Jewett, M.; Dutcher, J.P.; Atkins, M.B.; Pins, M.; et al. Adjuvant Sunitinib or Sorafenib for High-Risk, Non-Metastatic Renal-Cell Carcinoma (ECOG-ACRIN E2805): A Double-Blind, Placebo-Controlled, Randomised, Phase 3 Trial. Lancet 2016, 387, 2008–2016. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Tomczak, P.; Park, S.H.; Venugopal, B.; Ferguson, T.; Chang, Y.-H.; Hajek, J.; Symeonides, S.N.; Lee, J.L.; Sarwar, N.; et al. Adjuvant Pembrolizumab after Nephrectomy in Renal-Cell Carcinoma. N. Engl. J. Med. 2021, 385, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Kalogirou, C.; Schäfer, D.; Krebs, M.; Kurz, F.; Schneider, A.; Riedmiller, H.; Kneitz, B.; Vergho, D. Metformin-Derived Growth Inhibition in Renal Cell Carcinoma Depends on MiR-21-Mediated PTEN Expression. Urol. Int. 2016, 96, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of MiR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. —Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Li, X.; Xin, S.; He, Z.; Che, X.; Wang, J.; Xiao, X.; Chen, J.; Song, X. MicroRNA-21 (MiR-21) Post-Transcriptionally Downregulates Tumor Suppressor PDCD4 and Promotes Cell Transformation, Proliferation, and Metastasis in Renal Cell Carcinoma. Cell Physiol Biochem. 2014, 33, 1631–1642. [Google Scholar] [CrossRef]
- Zhang, G.-M.; Luo, L.; Ding, X.-M.; Dong, D.-H.; Li, B.; Ma, X.-C.; Sun, L.-J. MicroRNA-126 Inhibits Tumor Cell Invasion and Metastasis by Downregulating ROCK1 in Renal Cell Carcinoma. Mol. Med. Rep. 2016, 13, 5029–5036. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara, M.; Bhawal, U.K.; Ueda, N.; Shimomoto, T.; Yamamoto, K.; Kirita, T.; Kuniyasu, H. Downregulation of MiR-126 Induces Angiogenesis and Lymphangiogenesis by Activation of VEGF-A in Oral Cancer. Br. J. Cancer 2012, 107, 700–706. [Google Scholar] [CrossRef]
- Garofalo, M.; Quintavalle, C.; Romano, G.M.; Croce, C.; Condorelli, G. MiR221/222 in Cancer: Their Role in Tumor Progression and Response to Therapy. CMM 2012, 12, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xu, J.-W.; Wang, L.; Liu, H.; Yan, Y.; Hu, S.-Y. MicroRNA-221-3p Is up-Regulated and Serves as a Potential Biomarker in Pancreatic Cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, 482–487. [Google Scholar] [CrossRef]
- Kneitz, B.; Krebs, M.; Kalogirou, C.; Schubert, M.; Joniau, S.; van Poppel, H.; Lerut, E.; Kneitz, S.; Scholz, C.J.; Ströbel, P.; et al. Survival in Patients with High-Risk Prostate Cancer Is Predicted by MiR-221, Which Regulates Proliferation, Apoptosis, and Invasion of Prostate Cancer Cells by Inhibiting IRF2 and SOCS3. Cancer Res. 2014, 74, 2591–2603. [Google Scholar] [CrossRef] [PubMed]
- Khella, H.W.Z.; Butz, H.; Ding, Q.; Rotondo, F.; Evans, K.R.; Kupchak, P.; Dharsee, M.; Latif, A.; Pasic, M.D.; Lianidou, E.; et al. MiR-221/222 Are Involved in Response to Sunitinib Treatment in Metastatic Renal Cell Carcinoma. Mol. Ther. 2015, 23, 1748–1758. [Google Scholar] [CrossRef]
- Krebs, M.; Solimando, A.G.; Kalogirou, C.; Marquardt, A.; Frank, T.; Sokolakis, I.; Hatzichristodoulou, G.; Kneitz, S.; Bargou, R.; Kübler, H.; et al. MiR-221-3p Regulates VEGFR2 Expression in High-Risk Prostate Cancer and Represents an Escape Mechanism from Sunitinib In Vitro. JCM 2020, 9, 670. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Vera, Y.M.; Marchat, L.A.; Gallardo-Rincón, D.; Ruiz-García, E.; Astudillo-De La Vega, H.; Echavarría-Zepeda, R.; López-Camarillo, C. AngiomiRs: MicroRNAs Driving Angiogenesis in Cancer (Review). Int. J. Mol. Med. 2019, 43, 657–670. [Google Scholar] [CrossRef]
- Wang, S.; Olson, E.N. AngiomiRs—Key Regulators of Angiogenesis. Curr. Opin. Genet. Dev. 2009, 19, 205–211. [Google Scholar] [CrossRef]
- Marquardt, A.; Solimando, A.G.; Kerscher, A.; Bittrich, M.; Kalogirou, C.; Kübler, H.; Rosenwald, A.; Bargou, R.; Kollmannsberger, P.; Schilling, B.; et al. Subgroup-Independent Mapping of Renal Cell Carcinoma—Machine Learning Reveals Prognostic Mitochondrial Gene Signature Beyond Histopathologic Boundaries. Front. Oncol. 2021, 11, 621278. [Google Scholar] [CrossRef]
- Jankowska, K.I.; Sauna, Z.E.; Atreya, C.D. Role of MicroRNAs in Hemophilia and Thrombosis in Humans. IJMS 2020, 21, 3598. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Kang, J.; Liu, F. Plasma Levels of MicroRNA-221 (MiR-221) Are Increased in Patients with Acute Pulmonary Embolism. Med. Sci. Monit. 2018, 24, 8621–8626. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sundquist, K.; Svensson, P.J.; Rastkhani, H.; Palmér, K.; Memon, A.A.; Sundquist, J.; Zöller, B. Association of Recurrent Venous Thromboembolism and Circulating MicroRNAs. Clin. Epigenet. 2019, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Warsow, G.; Hübschmann, D.; Kleinheinz, K.; Nientiedt, C.; Heller, M.; Van Coile, L.; Tolstov, Y.; Trennheuser, L.; Wieczorek, K.; Pecqueux, C.; et al. Genomic Features of Renal Cell Carcinoma with Venous Tumor Thrombus. Sci. Rep. 2018, 8, 7477. [Google Scholar] [CrossRef] [PubMed]

| microRNA | Assay-ID | Catalog Number | Manufacturer |
|---|---|---|---|
| hsa-miR-21-5p | 000397 | 4427975 | Applied Biosystems |
| hsa-miR-126-3p | 002228 | 4427975 | Applied Biosystems |
| hsa-miR-221-3p | 000524 | 4427975 | Applied Biosystems |
| RNU6B | 001093 | 4427975 | Applied Biosystems |
| TaqMan™ Universal PCR Master Mix, no AmpErase™ UNG | 4324020 | Applied Biosystems | |
| TaqMan™ MicroRNA Reverse Transcription Kit | 4366596 | Applied Biosystems | |
| Characteristics | |
|---|---|
| Median Follow-up (n = 54) | 94 (1–190) months |
| Median Age at surgery | 67 (41–89) years |
| Sex Female Male | 22 (39.3%) 34 (60.7%) |
| Tumor Stage: pT3b | 56 (100%) |
| Fuhrman Grade G2 G3 | 41 (73.2%) 15 (26.8%) |
| Nodal Status N0 N+ | 45 (80.4%) 11 (19.6%) |
| Distant Metastasis (synchronous and metachronous) M0 M1 (synchronous: n = 7, metachronous: n = 15) | 34 (60.7%) 22 (39.3%) |
| Median Tumor Size | 70 (18–225) mm |
| Overall survival yes no | 27 (48.2%) 29 (51.8%) |
| Cancer-related death yes no | 13 (23.2%) 43 (76.8%) |
| Cancer-Related Death | ||||
|---|---|---|---|---|
| (a) Single-Variable Analysis | (b) Multivariable Analysis | |||
| Parameters | HR (95% CI) | p Value | HR (95% CI) | p Value |
| miR-21 | 3.79 (1.55–9.26) | 0.003 ** | 4.94 (1.29–18.98) | 0.02 * |
| miR-126 | 0.19 (0.09–0.42) | 0.00003 *** | 0.27 (0.097–0.75) | 0.01 * |
| miR-221 | 0.74 (0.46–1.19) | 0.22 | 0.64 (0.37–1.12) | 0.12 |
| Age at surgery | 0.98 (0.92–1.03) | 0.42 | ||
| Sex | 2.10 (0.58–7.65) | 0.26 | ||
| Tumor size | 1.01 (1.00–1.03) | 0.07 | ||
| Fuhrman grade | 3.79 (1.27–11.33) | 0.02 * | 3.28 (0.70–15.32) | 0.13 |
| Nodal status | 6.70 (2.09–21.47) | 0.001 ** | 1.34 (0.29–6.12) | 0.71 |
| Distant metastasis (synchronous and metachronous) | ∞ | NA | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotlyar, M.J.; Krebs, M.; Solimando, A.G.; Marquardt, A.; Burger, M.; Kübler, H.; Bargou, R.; Kneitz, S.; Otto, W.; Breyer, J.; et al. Critical Evaluation of a microRNA-Based Risk Classifier Predicting Cancer-Specific Survival in Renal Cell Carcinoma with Tumor Thrombus of the Inferior Vena Cava. Cancers 2023, 15, 1981. https://doi.org/10.3390/cancers15071981
Kotlyar MJ, Krebs M, Solimando AG, Marquardt A, Burger M, Kübler H, Bargou R, Kneitz S, Otto W, Breyer J, et al. Critical Evaluation of a microRNA-Based Risk Classifier Predicting Cancer-Specific Survival in Renal Cell Carcinoma with Tumor Thrombus of the Inferior Vena Cava. Cancers. 2023; 15(7):1981. https://doi.org/10.3390/cancers15071981
Chicago/Turabian StyleKotlyar, Mischa J., Markus Krebs, Antonio Giovanni Solimando, André Marquardt, Maximilian Burger, Hubert Kübler, Ralf Bargou, Susanne Kneitz, Wolfgang Otto, Johannes Breyer, and et al. 2023. "Critical Evaluation of a microRNA-Based Risk Classifier Predicting Cancer-Specific Survival in Renal Cell Carcinoma with Tumor Thrombus of the Inferior Vena Cava" Cancers 15, no. 7: 1981. https://doi.org/10.3390/cancers15071981
APA StyleKotlyar, M. J., Krebs, M., Solimando, A. G., Marquardt, A., Burger, M., Kübler, H., Bargou, R., Kneitz, S., Otto, W., Breyer, J., Vergho, D. C., Kneitz, B., & Kalogirou, C. (2023). Critical Evaluation of a microRNA-Based Risk Classifier Predicting Cancer-Specific Survival in Renal Cell Carcinoma with Tumor Thrombus of the Inferior Vena Cava. Cancers, 15(7), 1981. https://doi.org/10.3390/cancers15071981






