Optical Biopsy of Dysplasia in Barrett’s Oesophagus Assisted by Artificial Intelligence
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
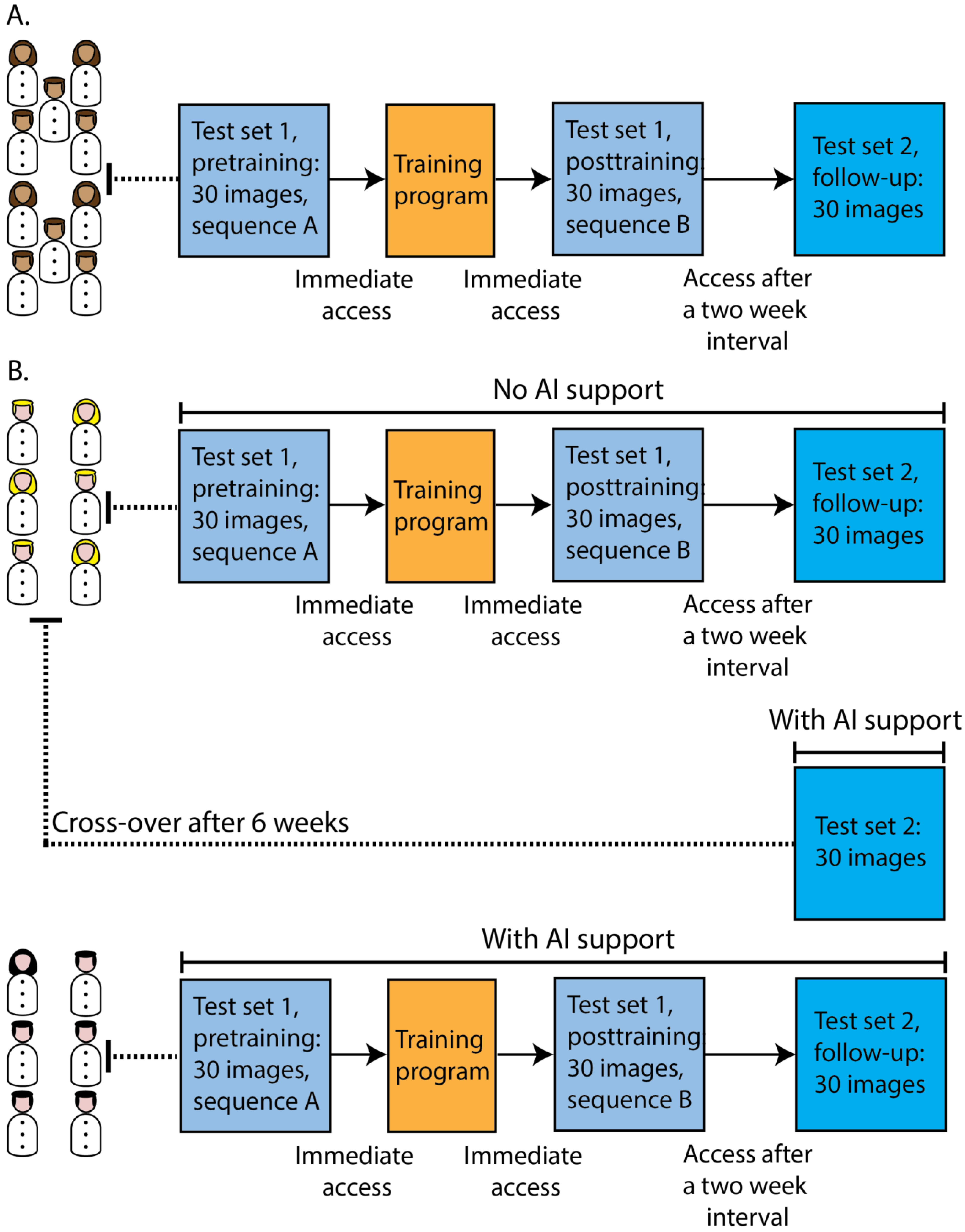
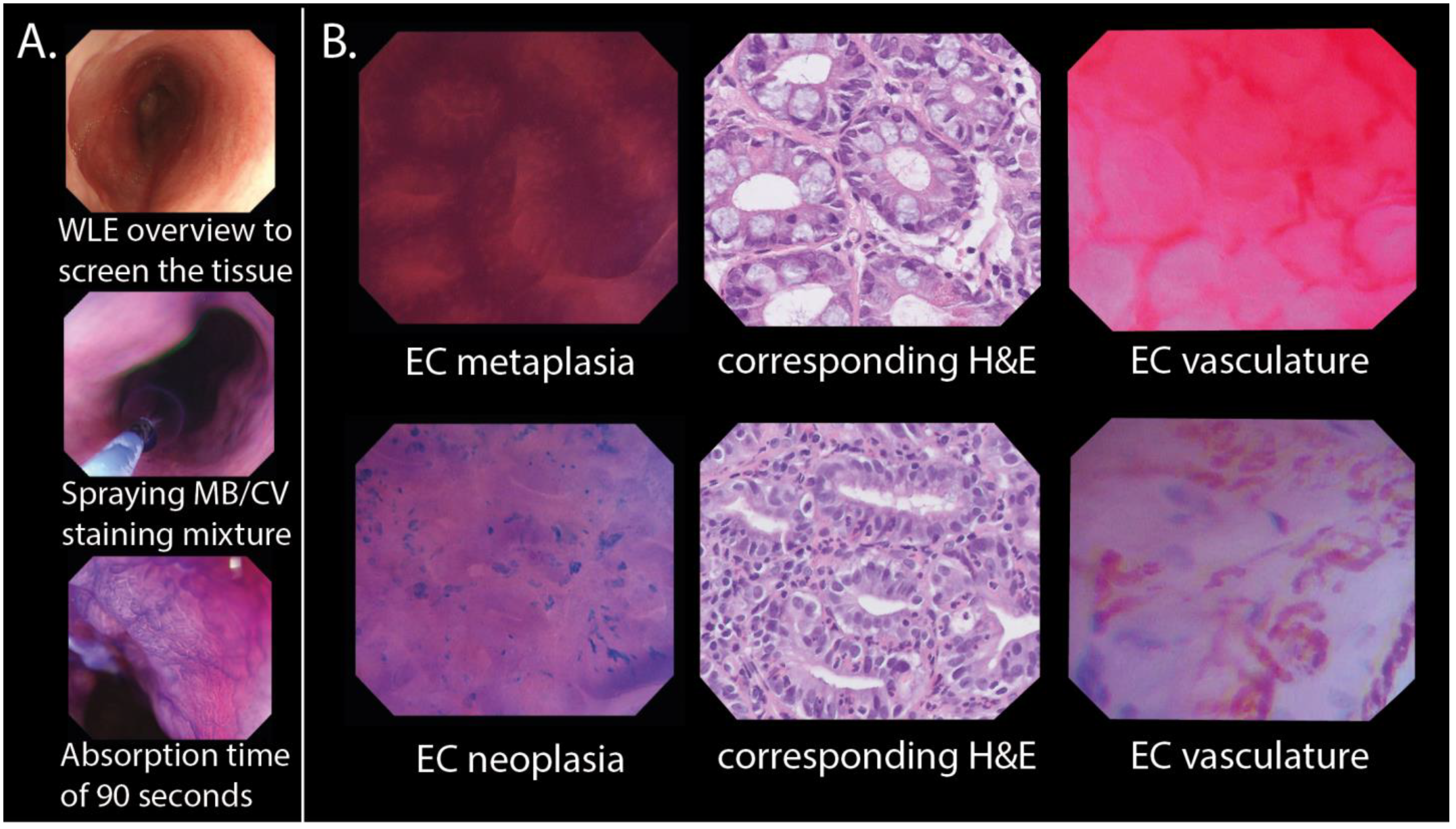
2.1. Study Setting and Population
2.2. Online Platform with Training and Testing Modules
2.2.1. Participants in the Online Modules
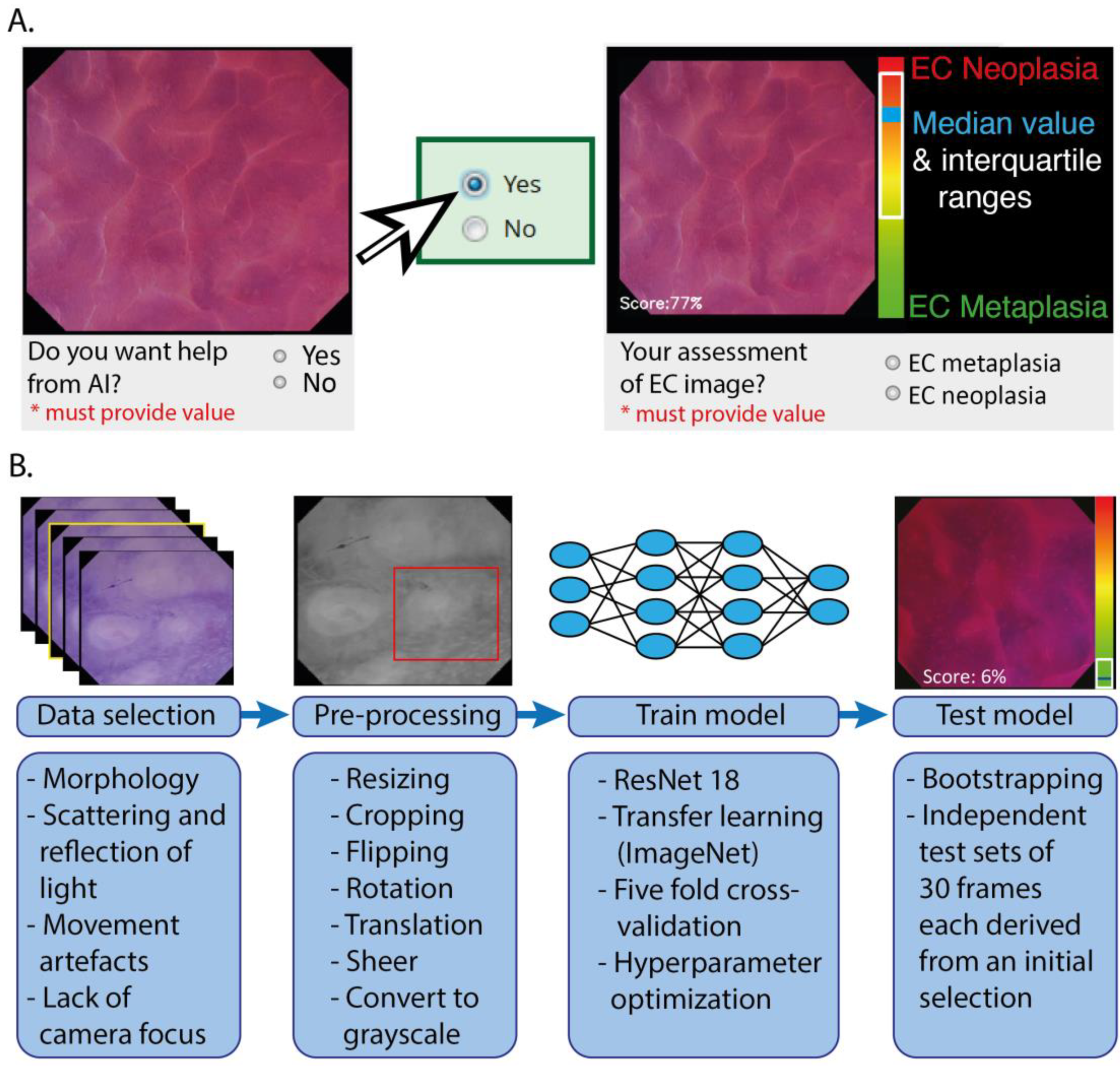
2.2.2. Training and Testing Modules
2.3. Training, Validating, and Testing of the CNN Architecture
2.4. Statistical Analysis
3. Results
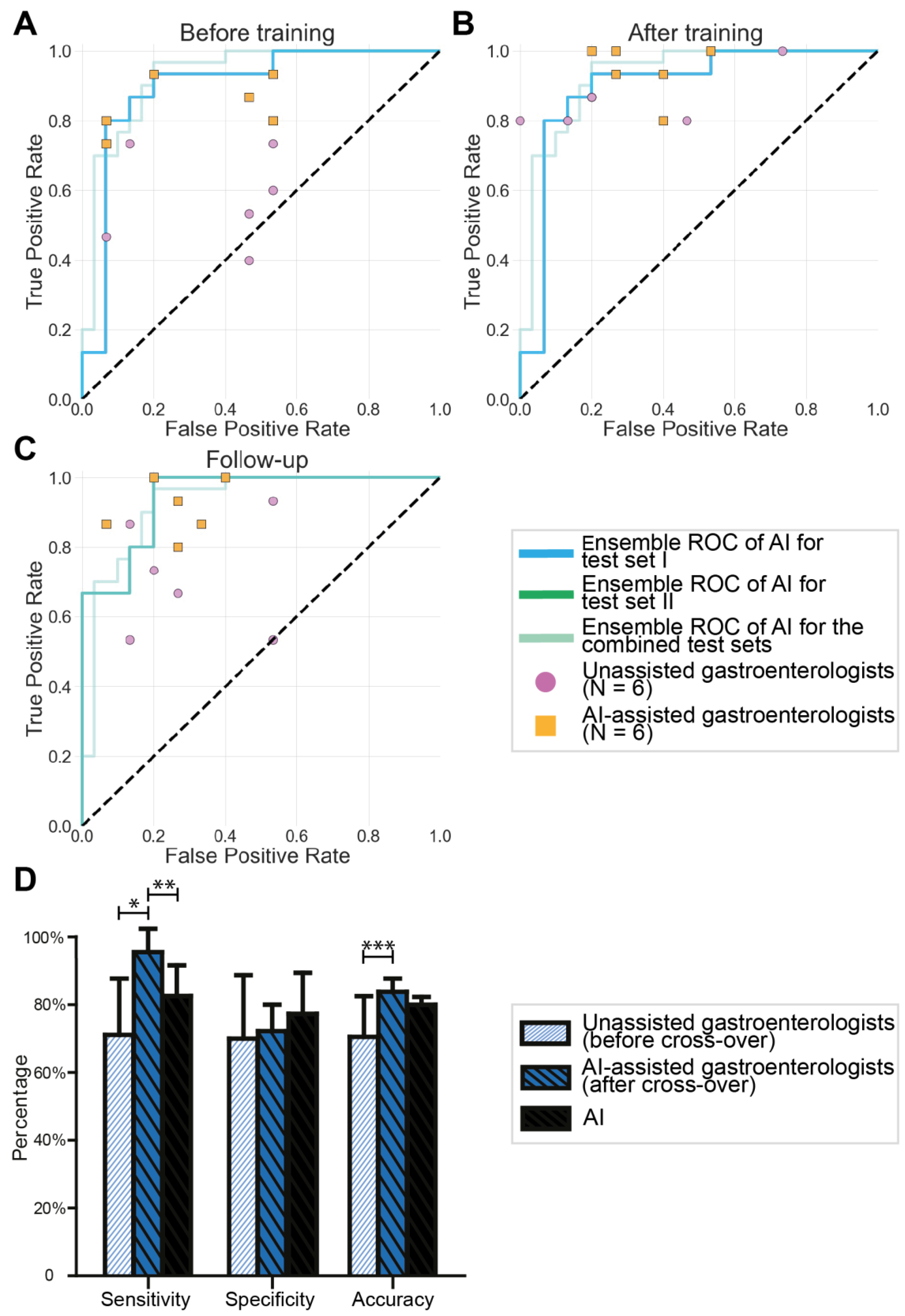
3.1. Validation of the Endocytoscopic BE Classification System by Endoscopists
3.2. Results of Testing the AI Algorithm
3.3. AI-Assisted Gastroenterologists versus Unassisted Gastroenterologists
3.4. Cross-Over of Unassisted Gastroenterologists to AI Assistance
3.5. Human–Machine Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coleman, H.G.; Xie, S.H.; Lagergren, J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology 2018, 154, 390–405. [Google Scholar] [CrossRef]
- El-Serag, H.B.; Naik, A.D.; Duan, Z.; Shakhatreh, M.; Helm, A.; Pathak, A.; Hinojosa-Lindsey, M.; Hou, J.; Nguyen, T.; Chen, J.; et al. Surveillance endoscopy is associated with improved outcomes of oesophageal adenocarcinoma detected in patients with Barrett’s oesophagus. Gut 2016, 65, 1252–1260. [Google Scholar] [CrossRef]
- Van Munster, S.; Nieuwenhuis, E.; Weusten, B.L.; Herrero, L.A.; Bogte, A.; Alkhalaf, A.; Schenk, B.E.; Schoon, E.J.; Curvers, W.; Koch, A.D.; et al. Long-term outcomes after endoscopic treatment for Barrett’s neoplasia with radiofrequency ablation ± endoscopic resection: Results from the national Dutch database in a 10-year period. Gut 2021, 71, 265–276. [Google Scholar] [CrossRef]
- Fitzgerald, R.C.; Di Pietro, M.; Ragunath, K.; Ang, Y.; Kang, J.Y.; Watson, P.; Trudgill, N.; Patel, P.; Kaye, P.V.; Sanders, S.; et al. British Society of Gastroenterology guidelines on the diagnosis and management of Barrett’s oesophagus. Gut 2014, 63, 7–42. [Google Scholar] [CrossRef]
- Shaheen, N.J.; Falk, G.W.; Iyer, P.G.; Gerson, L.B. ACG Clinical Guideline: Diagnosis and Management of Barrett’s Esophagus. Am. J. Gastroenterol. 2016, 111, 30–50. [Google Scholar] [CrossRef]
- Badreddine, R.J.; Wang, K.K. Barrett esophagus: An update. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 369–378. [Google Scholar] [CrossRef]
- Nagengast, W.B.; Hartmans, E.; Garcia-Allende, P.B.; Peters, F.T.M.; Linssen, M.D.; Koch, M.; Koller, M.; Tjalma, J.; Karrenbeld, A.; Jorritsma-Smit, A.; et al. Near-infrared fluorescence molecular endoscopy detects dysplastic oesophageal lesions using topical and systemic tracer of vascular endothelial growth factor A. Gut 2019, 68, 7–10. [Google Scholar] [CrossRef]
- Vithayathil, M.; Modolell, I.; Ortiz-Fernandez-Sordo, J.; Oukrif, D.; Pappas, A.; Januszewicz, W.; O’Donovan, M.; Hadjinicolaou, A.; Bianchi, M.; Blasko, A.; et al. Image-Enhanced Endoscopy and Molecular Biomarkers Vs Seattle Protocol to Diagnose Dysplasia in Barrett’s Esophagus. Clin. Gastroenterol. Hepatol. 2022, 20, 2514–2523.e3. [Google Scholar] [CrossRef]
- Visrodia, K.; Singh, S.; Krishnamoorthi, R.; Ahlquist, D.A.; Wang, K.K.; Iyer, P.G.; Katzka, D.A. Magnitude of Missed Esophageal Adenocarcinoma After Barrett’s Esophagus Diagnosis: A Systematic Review and Meta-analysis. Gastroenterology 2016, 150, 599–607.e7. [Google Scholar] [CrossRef]
- Kolb, J.M.; Davis, C.; Williams, J.L.; Holub, J.; Shaheen, N.; Wani, S. Wide Variability in Dysplasia Detection Rate and Adherence to Seattle Protocol and Surveillance Recommendations in Barrett’s Esophagus: A Population-Based Analysis Using the GIQuIC National Quality Benchmarking Registry. Am. J. Gastroenterol. 2023. [Google Scholar] [CrossRef]
- Neumann, H.; Fuchs, F.S.; Vieth, M.; Atreya, R.; Siebler, J.; Kiesslich, R.; Neurath, M. Review article: In vivo imaging by endocytoscopy. Aliment. Pharm. 2011, 33, 1183–1193. [Google Scholar] [CrossRef]
- Pohl, H.; Koch, M.; Khalifa, A.; Papanikolaou, I.; Scheiner, K.; Wiedenmann, B.; Rösch, T. Evaluation of endocytoscopy in the surveillance of patients with Barrett’s esophagus. Endoscopy 2007, 39, 492–496. [Google Scholar] [CrossRef]
- Tomizawa, Y.; Iyer, P.G.; Wongkeesong, L.M.; Buttar, N.S.; Lutzke, L.S.; Wu, T.T.; Wang, K.K. Assessment of the diagnostic performance and interobserver variability of endocytoscopy in Barrett’s esophagus: A pilot ex-vivo study. World J. Gastroenterol. 2013, 19, 8652–8658. [Google Scholar] [CrossRef]
- Xiong, Y.-Q.; Ma, S.-J.; Zhou, J.-H.; Zhong, X.-S.; Chen, Q. A meta-analysis of confocal laser endomicroscopy for the detection of neoplasia in patients with Barrett’s esophagus. J. Gastroenterol. Hepatol. 2016, 31, 1102–1110. [Google Scholar] [CrossRef]
- Wang, K.K.; Carr-Locke, D.L.; Singh, S.K.; Neumann, H.; Bertani, H.; Galmiche, J.P.; Arsenescu, R.I.; Caillol, F.; Chang, K.J.; Chaussade, S.; et al. Use of probe-based confocal laser endomicroscopy (pCLE) in gastrointestinal applications. A consensus report based on clinical evidence. United Eur. Gastroenterol. J. 2015, 3, 230–254. [Google Scholar] [CrossRef]
- Mori, Y.; Kudo, S.-E.; East, J.E.; Rastogi, A.; Bretthauer, M.; Misawa, M.; Sekiguchi, M.; Matsuda, T.; Saito, Y.; Ikematsu, H.; et al. Cost savings in colonoscopy with artificial intelligence-aided polyp diagnosis: An add-on analysis of a clinical trial (with video). Gastrointest. Endosc. 2020, 92, 905–911. [Google Scholar] [CrossRef]
- Sharma, P.; Savides, T.J.; Canto, M.I.; Corley, D.A.; Falk, G.W.; Goldblum, J.R.; Wang, K.K.; Wallace, M.B.; Wolfsen, H.C. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on imaging in Barrett’s Esophagus. Gastrointest. Endosc. 2012, 76, 252–254. [Google Scholar] [CrossRef]
- Dayyeh, B.K.; Thosani, N.; Konda, V.; Wallace, M.B.; Rex, D.K.; Chauhan, S.S.; Hwang, J.H.; Komanduri, S.; Manfredi, M.; Maple, J.T.; et al. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE Preservation and Incorporation of Valuable Endoscopic Innovations thresholds for adopting real-time imaging-assisted endoscopic targeted biopsy during endoscopic surveillance of Barrett’s esophagus. Gastrointest. Endosc. 2016, 83, 684–698.e7. [Google Scholar]
- Appannagari, A.; Soudagar, A.S.; Pietrzak, C.; Sharma, P.; Gupta, N. Are gastroenterologists willing to implement imaging-guided surveillance for Barrett’s esophagus? Results from a national survey. Endosc. Int. Open 2015, 3, E181–E185. [Google Scholar] [CrossRef]
- Machicado, J.D.; Han, S.; Yadlapati, R.H.; Simon, V.C.; Qumseya, B.J.; Sultan, S.; Kushnir, V.M.; Komanduri, S.; Rastogi, A.; Muthusamy, V.R.; et al. A Survey of Expert Practice and Attitudes Regarding Advanced Imaging Modalities in Surveillance of Barrett’s Esophagus. Dig. Dis. Sci. 2018, 63, 3262–3271. [Google Scholar] [CrossRef]
- Le Berre, C.; Sandborn, W.J.; Aridhi, S.; Devignes, M.-D.; Fournier, L.; Smaïl-Tabbone, M.; Danese, S.; Peyrin-Biroulet, L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology 2020, 158, 76–94.e2. [Google Scholar] [CrossRef]
- van der Sommen, F.; de Groof, J.; Struyvenberg, M.; van der Putten, J.; Boers, T.; Fockens, K.; Schoon, E.J.; Curvers, W.; Mori, Y.; Byrne, M.; et al. Machine learning in GI endoscopy: Practical guidance in how to interpret a novel field. Gut 2020, 69, 2035–2045. [Google Scholar] [CrossRef]
- Ruffle, J.K.; Farmer, A.D.; Aziz, Q. Artificial Intelligence-Assisted Gastroenterology-Promises and Pitfalls. Am. J. Gastroenterol. 2019, 114, 422–428. [Google Scholar] [CrossRef]
- van der Putten, J.; de Groof, J.; Struyvenberg, M.; Boers, T.; Fockens, K.; Curvers, W.; Schoon, E.; Bergman, J.; van der Sommen, F.; de With, P.H. Multi-stage domain-specific pretraining for improved detection and localization of Barrett’s neoplasia: A comprehensive clinically validated study. Artif. Intell. Med. 2020, 107, 101914. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; IEEE: New York, NY, USA, 2009. [Google Scholar]
- Smith, L.N. Cyclical Learning Rates for Training Neural Networks. In Proceedings of the Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; IEEE: New York, NY, USA, 2017. [Google Scholar]
- Zhang, Z. Improved Adam Optimizer for Deep Neural Networks. In Proceedings of the 26th International Symposium on Quality of Service (IWQoS), Banff, AB, Canada, 4–6 June 2018; IEEE: New York, NY, USA, 2019. [Google Scholar]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Mori, Y.; Kudo, S.-E.; Wakamura, K.; Misawa, M.; Ogawa, Y.; Kutsukawa, M.; Kudo, T.; Hayashi, T.; Miyachi, H.; Ishida, F.; et al. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest. Endosc. 2015, 81, 621–629. [Google Scholar] [CrossRef]
- Misawa, M.; Kudo, S.E.; Mori, Y.; Nakamura, H.; Kataoka, S.; Maeda, Y.; Kudo, T.; Hayashi, T.; Wakamura, K.; Miyachi, H.; et al. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology 2016, 150, 1531–1532.e3. [Google Scholar] [CrossRef]
- Takeda, K.; Kudo, S.-E.; Mori, Y.; Misawa, M.; Kudo, T.; Wakamura, K.; Katagiri, A.; Baba, T.; Hidaka, E.; Ishida, F.; et al. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy 2017, 49, 798–802. [Google Scholar] [CrossRef]
- Mori, Y.; Kudo, S.E.; Misawa, M.; Saito, Y.; Ikematsu, H.; Hotta, K.; Ohtsuka, K.; Urushibara, F.; Kataoka, S.; Ogawa, Y.; et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann. Intern. Med. 2018, 169, 357–366. [Google Scholar] [CrossRef]
- Kudo, S.-E.; Misawa, M.; Mori, Y.; Hotta, K.; Ohtsuka, K.; Ikematsu, H.; Saito, Y.; Takeda, K.; Nakamura, H.; Ichimasa, K.; et al. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin. Gastroenterol. Hepatol. 2020, 18, 1874–1881.e2. [Google Scholar] [CrossRef]
- Bajbouj, M.; Vieth, M.; Rösch, T.; Miehlke, S.; Becker, V.; Anders, M.; Pohl, H.; Madisch, A.; Schuster, T.; Schmid, R.; et al. Probe-based confocal laser endomicroscopy compared with standard four-quadrant biopsy for evaluation of neoplasia in Barrett’s esophagus. Endoscopy 2010, 42, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Meining, A.R.; Coron, E.; Lightdale, C.J.; Wolfsen, H.C.; Bansal, A.; Bajbouj, M.; Galmiche, J.P.; Abrams, J.A.; Rastogi, A.; et al. Real-time increased detection of neoplastic tissue in Barrett’s esophagus with probe-based confocal laser endomicroscopy: Final results of an international multicenter, prospective, randomized, controlled trial. Gastrointest. Endosc. 2011, 74, 465–472. [Google Scholar] [CrossRef]
- Sturm, M.B.; Piraka, C.; Elmunzer, B.J.; Kwon, R.S.; Joshi, B.P.; Appelman, H.D.; Turgeon, D.K.; Wang, T.D. In vivo molecular imaging of Barrett’s esophagus with confocal laser endomicroscopy. Gastroenterology 2013, 145, 56–58. [Google Scholar] [CrossRef]
- Di Pietro, M.; Bird-Lieberman, E.L.; Liu, X.; Nuckcheddy-Grant, T.; Bertani, H.; O’Donovan, M.; Fitzgerald, R.C. Autofluorescence-Directed Confocal Endomicroscopy in Combination with a Three-Biomarker Panel Can Inform Management Decisions in Barrett’s Esophagus. Am. J. Gastroenterol. 2015, 110, 1549–1558. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human-computer collaboration for skin cancer recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef]
- Seah, J.C.Y.; Tang, C.H.M.; Buchlak, Q.D.; Holt, X.G.; Wardman, J.B.; Aimoldin, A.; Esmaili, N.; Ahmad, H.; Pham, H.; Lambert, J.F.; et al. Effect of a comprehensive deep-learning model on the accuracy of chest X-ray interpretation by radiologists: A retrospective, multireader multicase study. Lancet Digit. Health 2021, 3, e496–e506. [Google Scholar] [CrossRef]
- de Groof, A.J.; Struyvenberg, M.R.; van der Putten, J.; van der Sommen, F.; Fockens, K.N.; Curvers, W.L.; Zinger, S.; Pouw, R.E.; Coron, E.; Baldaque-Silva, F.; et al. Deep-Learning System Detects Neoplasia in Patients with Barrett’s Esophagus with Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study with Benchmarking. Gastroenterology 2020, 158, 915–929.e4. [Google Scholar] [CrossRef]
- van der Putten, J.; de Groof, J.; van der Sommen, F.; Struyvenberg, M.; Zinger, S.; Curvers, W.; Schoon, E.; Bergman, J.; de With, P.H. Informative Frame Classification of Endoscopic Videos Using Convolutional Neural Networks and Hidden Markov Models. In Proceedings of the International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: New York, NY, USA, 2019. [Google Scholar]
- Boers, T.G.; van der Putten, J.; de Groof, J.; Struyvenberg, M.; Fockens, K.; Curvers, W.; Schoon, E.; van der Sommen, F.; Bergman, J. Detection of frame informativeness in endoscopic videos using image quality and recurrent neural networks. In Proceedings of the Medical Imaging 2020: Image Processing, SPIE, Houston, TX, USA, 15–20 February 2020. [Google Scholar]
- Holzinger, A. Interactive machine learning for health informatics: When do we need the human-in-the-loop? Brain Inform. 2016, 3, 119–131. [Google Scholar] [CrossRef]
- Keane, P.A.; Topol, E.J. AI-facilitated health care requires education of clinicians. Lancet 2021, 397, 1254. [Google Scholar] [CrossRef]
- Singh, R.; Yeap, S.P. Endoscopic imaging in Barrett’s esophagus. Expert Rev Gastroenterol Hepatol 2015, 9, 475–485. [Google Scholar] [CrossRef]
- Pohl, H.; Roesch, T.; Vieth, M.; Koch, M.; Becker, V.; Anders, M.; Khalifa, A.C.; Meining, A. Miniprobe confocal laser microscopy for the detection of invisible neoplasia in patients with Barrett’s oesophagus. Gut 2008, 57, 1648–1653. [Google Scholar] [CrossRef]
- Tofteland, N.; Singh, M.; Gaddam, S.; Wani, S.B.; Gupta, N.; Rastogi, A.; Bansal, A.; Kanakadandi, V.; McGregor, D.H.; Ulusarac, O.; et al. Evaluation of the updated confocal laser endomicroscopy criteria for Barrett’s esophagus among gastrointestinal pathologists. Dis. Esophagus. 2014, 27, 623–629. [Google Scholar] [CrossRef]
- Kara, M.A.; Ennahachi, M.; Fockens, P.; ten Kate, F.J.; Bergman, J.J. Detection and classification of the mucosal and vascular patterns (mucosal morphology) in Barrett’s esophagus by using narrow band imaging. Gastrointest. Endosc. 2006, 64, 155–166. [Google Scholar] [CrossRef]
- van der Wel, M.J.; Coleman, H.G.; Bergman, J.J.; Jansen, M.; Meijer, S.L. Histopathologist features predictive of diagnostic concordance at expert level among a large international sample of pathologists diagnosing Barrett’s dysplasia using digital pathology. Gut 2020, 69, 811–822. [Google Scholar] [CrossRef]
| Test Set 1 | Test Set 2 | ||
|---|---|---|---|
| Before Training (Test Set 1A) | After Training (Test Set 1B) | Follow-Up | |
| Sensitivity (95% CI) | 57.33% (48.86–65.80) | 90.0% (85.37–94.63) | 73.33% (63.03–83.84) |
| Specificity (95% CI) | 72.0% (57.47–86.53) | 65.33% (56.40–74.27) | 66.0% (56.09–75.92) |
| Accuracy (95% CI) | 64.67% (55.12–74.22) | 77.67% (73.03–82.31) | 69.67% (64.71–74.62) |
| Test Set 1 | Test Set 2 | ||||||
|---|---|---|---|---|---|---|---|
| Before Training (Test Set 1A) | p-Value | After Training (Test Set 1B) | p-Value | Follow-Up | p-Value | ||
| Sensitivity (95% CI) | Unassisted gastroenterologists | 57.78% (43.33–72.23) | 0.002 | 85.56% (77.38–93.73) | 0.076 | 71.11% (53.60–88.63) | 0.025 |
| AI-assisted gastroenterologists | 84.44% (76.97–92.91) | 94.44% (86.26–100) | 91.11% (82.64–99.58) | ||||
| Specificity (95% CI) | Unassisted gastroenterologists | 63.33% (41.32–85.35) | 0.668 | 71.11% (43.24–98.97) | 0.652 | 70.0% (50.34–89.66) | 0.631 |
| AI-assisted gastroenterologists | 68.89% (45.20–92.58) | 65.56% (52.72–78.39) | 74.44% (62.93–86.50) | ||||
| Accuracy (95% CI) | Unassisted gastroenterologists | 60.56% (47.58–73.54) | 0.033 | 78.33% (67.11–89.56) | 0.765 | 70.56% (57.96–83.15) | 0.050 |
| AI-assisted gastroenterologists | 76.67% (66.05–87.28) | 80.0% (71.72–88.28) | 82.78% (76.36–89.20) | ||||
| Test Set 1 | Test Set 2 | |||
|---|---|---|---|---|
| Before Training (Test Set 1A) | After Training (Test Set 1B) | Follow-Up | ||
| Kappa values (95% CI) | Unassisted gastroenterologists | 0.491 (0.437–0.545) | 0.562 (0.501–0.623) | 0.526 (0.469–0.583) |
| AI-assisted gastroenterologists | 0.597 (0.537–0.657) | 0.687 (0.622–0.752) | 0.696 (0.631–0.761) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der Laan, J.J.H.; van der Putten, J.A.; Zhao, X.; Karrenbeld, A.; Peters, F.T.M.; Westerhof, J.; de With, P.H.N.; van der Sommen, F.; Nagengast, W.B. Optical Biopsy of Dysplasia in Barrett’s Oesophagus Assisted by Artificial Intelligence. Cancers 2023, 15, 1950. https://doi.org/10.3390/cancers15071950
van der Laan JJH, van der Putten JA, Zhao X, Karrenbeld A, Peters FTM, Westerhof J, de With PHN, van der Sommen F, Nagengast WB. Optical Biopsy of Dysplasia in Barrett’s Oesophagus Assisted by Artificial Intelligence. Cancers. 2023; 15(7):1950. https://doi.org/10.3390/cancers15071950
Chicago/Turabian Stylevan der Laan, Jouke J. H., Joost A. van der Putten, Xiaojuan Zhao, Arend Karrenbeld, Frans T. M. Peters, Jessie Westerhof, Peter H. N. de With, Fons van der Sommen, and Wouter B. Nagengast. 2023. "Optical Biopsy of Dysplasia in Barrett’s Oesophagus Assisted by Artificial Intelligence" Cancers 15, no. 7: 1950. https://doi.org/10.3390/cancers15071950
APA Stylevan der Laan, J. J. H., van der Putten, J. A., Zhao, X., Karrenbeld, A., Peters, F. T. M., Westerhof, J., de With, P. H. N., van der Sommen, F., & Nagengast, W. B. (2023). Optical Biopsy of Dysplasia in Barrett’s Oesophagus Assisted by Artificial Intelligence. Cancers, 15(7), 1950. https://doi.org/10.3390/cancers15071950







