Antibody Surface Profiling Identifies Glycoforms in Multiple Myeloma as Targets for Immunotherapy: From Antibody Derivatives to Mimetic Peptides for Killing Tumor Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Peptides
2.3. Cancer Cell Lines
2.4. Isolation of Blood Cells
2.5. Affinity Selection of scFv Antibody Fragments
2.6. Phage Amplification
2.7. Cloning and Expression of the Selected scFv–Fc Fusion Proteins
2.8. Flow Cytometry Analysis
2.9. ELISA
2.10. Competition Experiments
2.11. Confocal Microscopy Analysis
2.12. Analysis of NK Cell Degranulation and Activation
2.13. Cytotoxicity Assay
2.14. Syndecan-1 Knockdown and Overexpression in HEK293T Cells
2.15. Statistical Analysis
3. Results
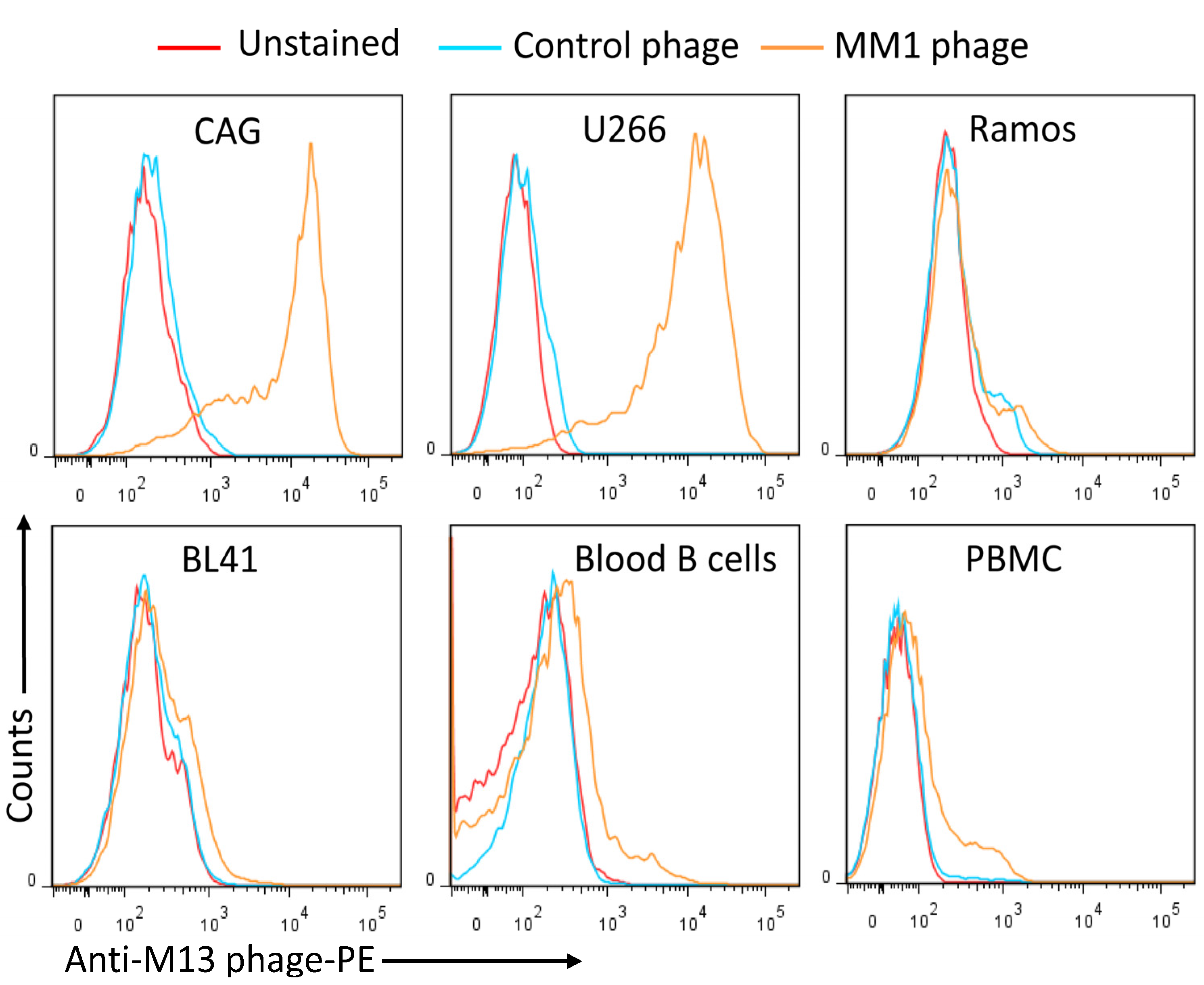
3.1. Isolation of MM-Specific scFv Antibody Fragments
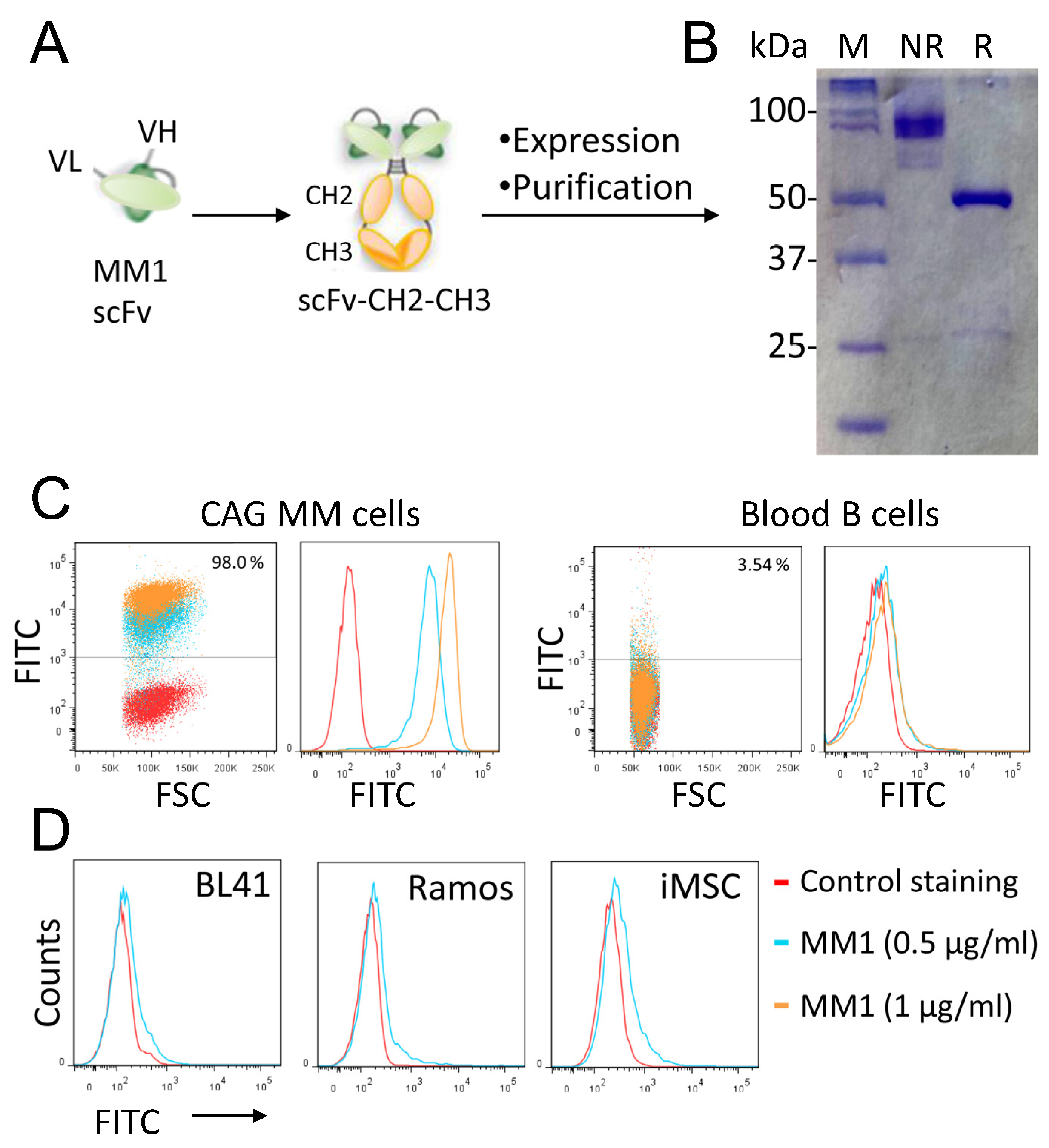
3.2. Conversion of the MM1 scFv into a scFv-Fc Antibody
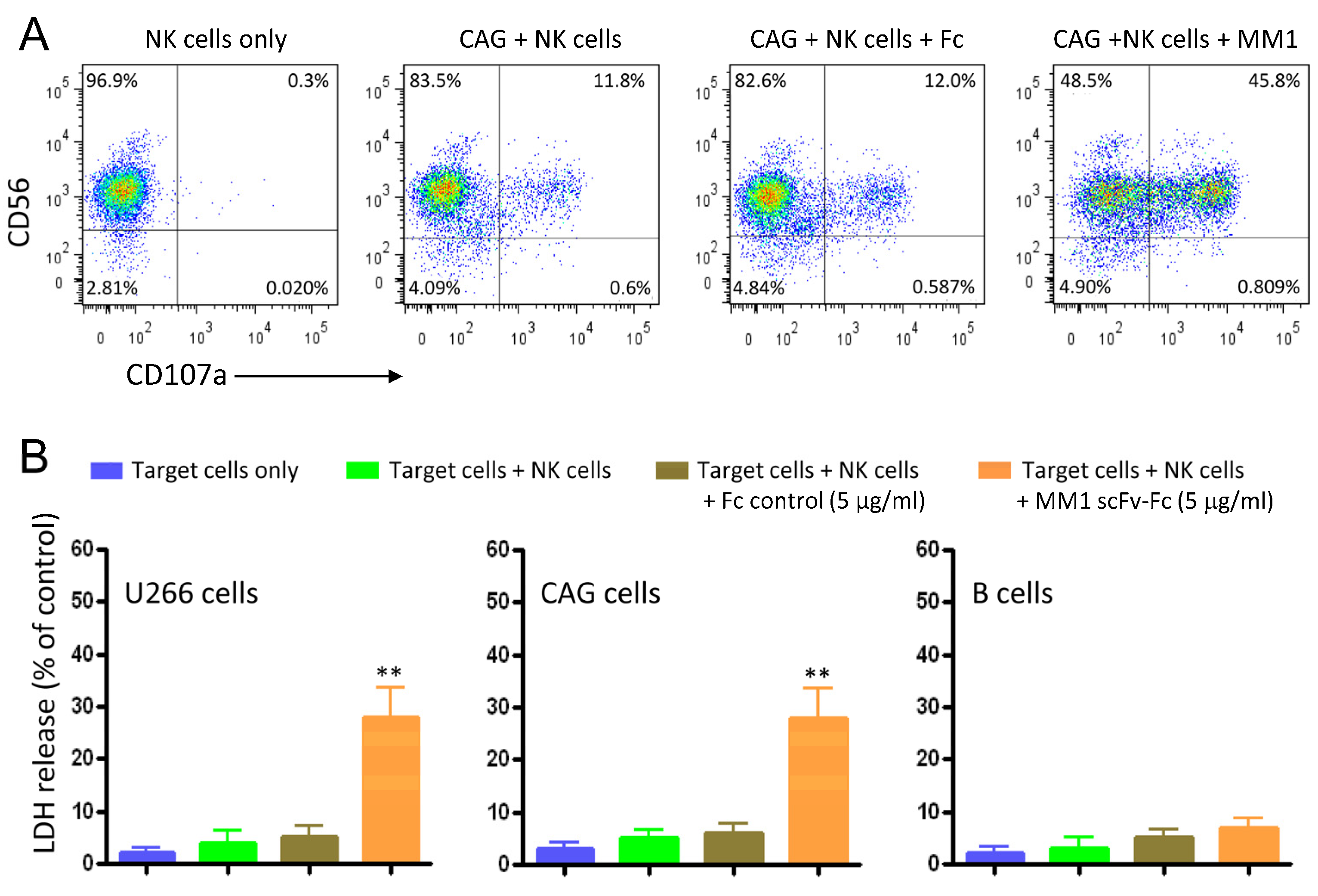
3.3. Activation of NK Cells and Induction of ADCC against Myeloma Cell Lines
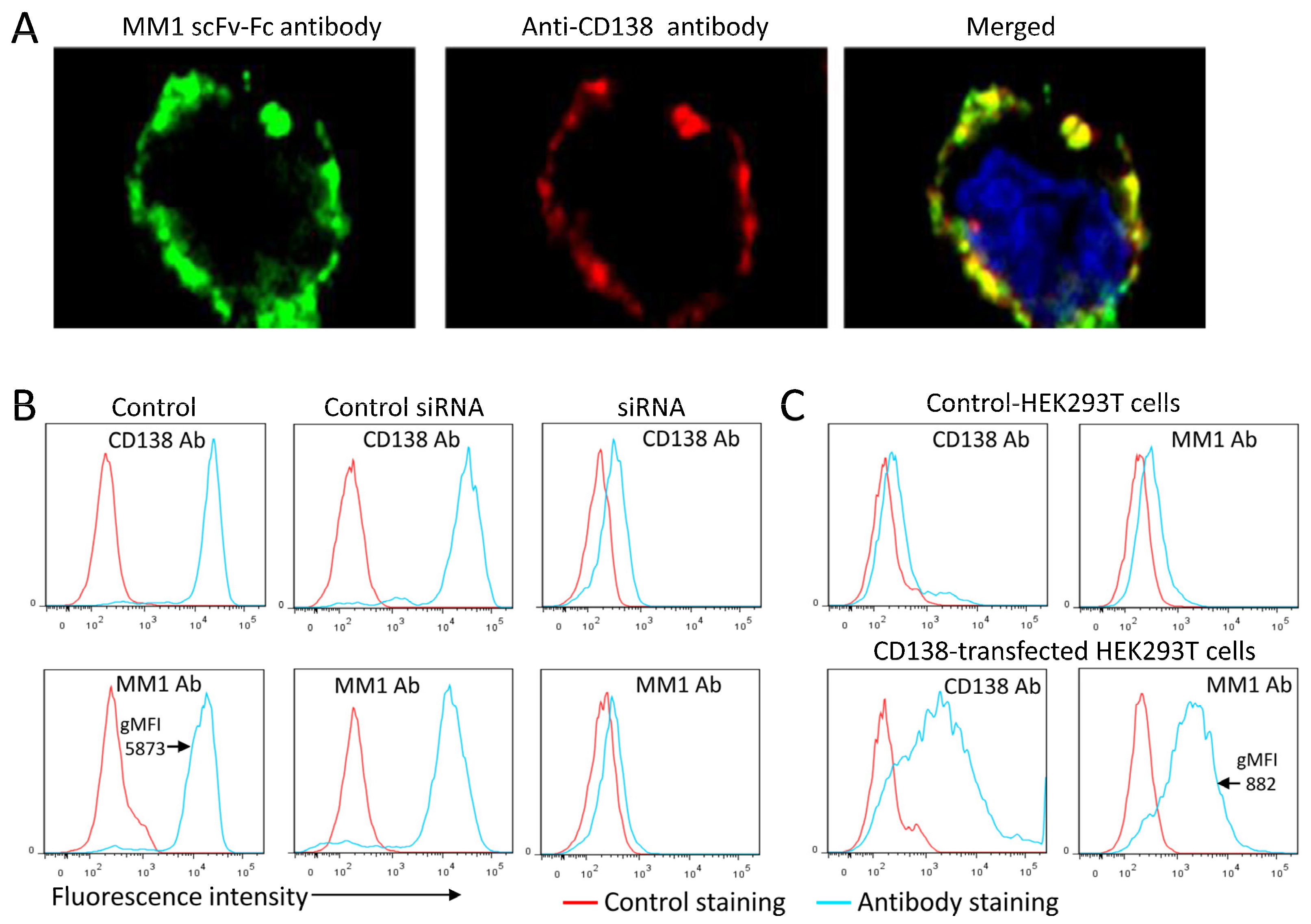
3.4. Evidence for the Interaction of the scFv-Fc with Cell-Surface Heparan Sulfate
3.5. Involvement of Syndecan-1 Associated Heparan Sulfate
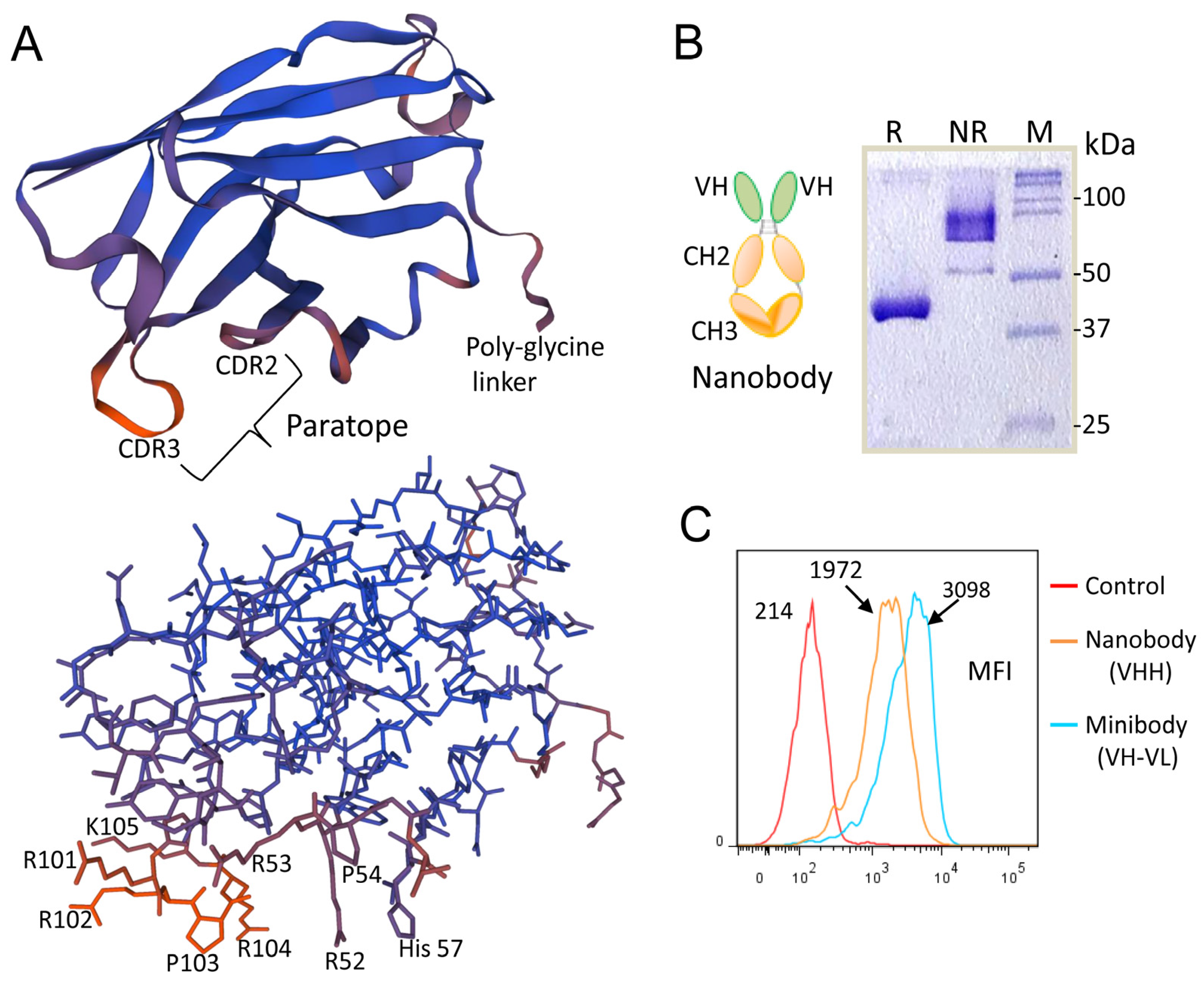
3.6. The Role of the CDRs of the scFv-Fc Heavy Chain in Recognition of MM Cells
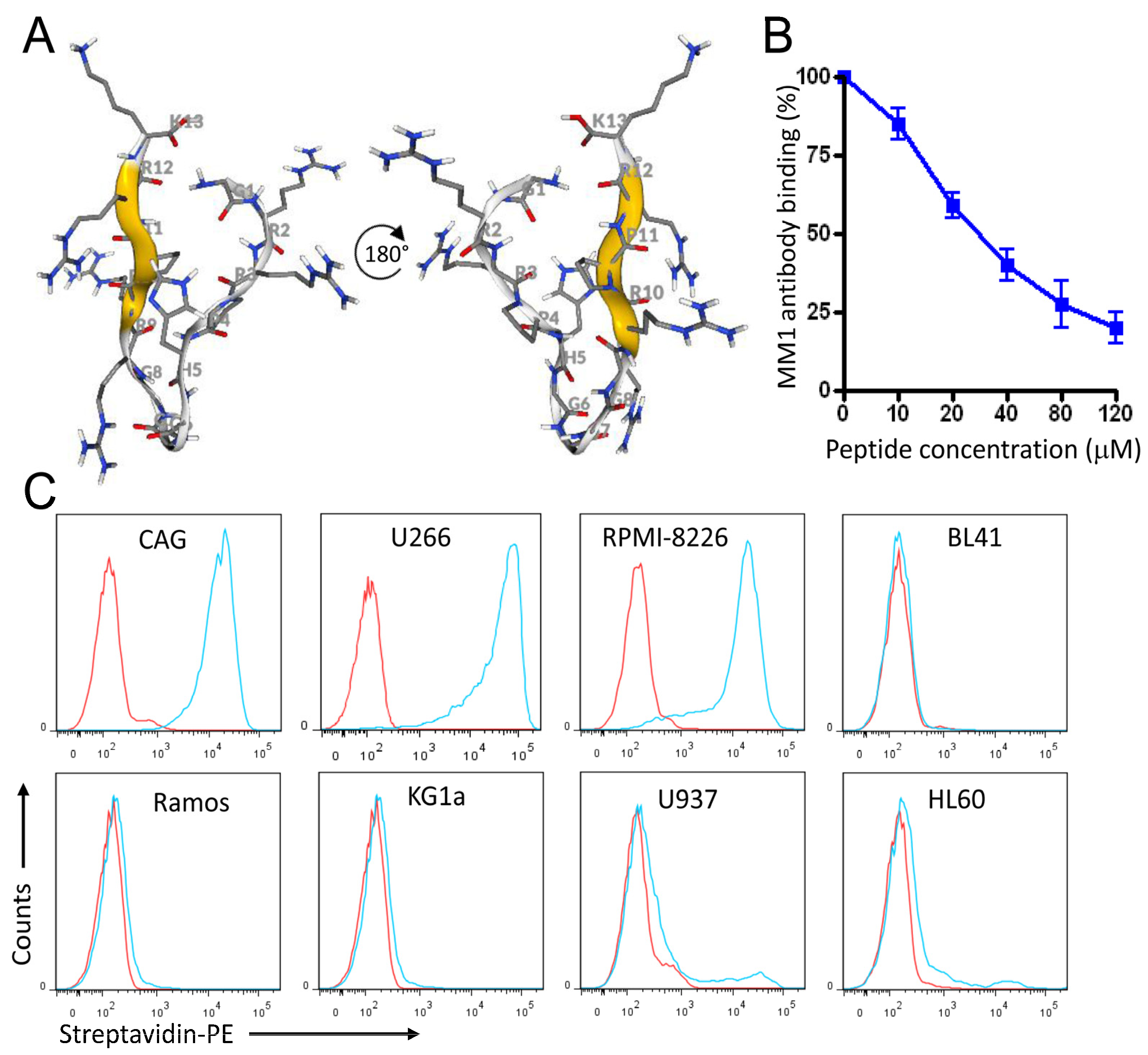
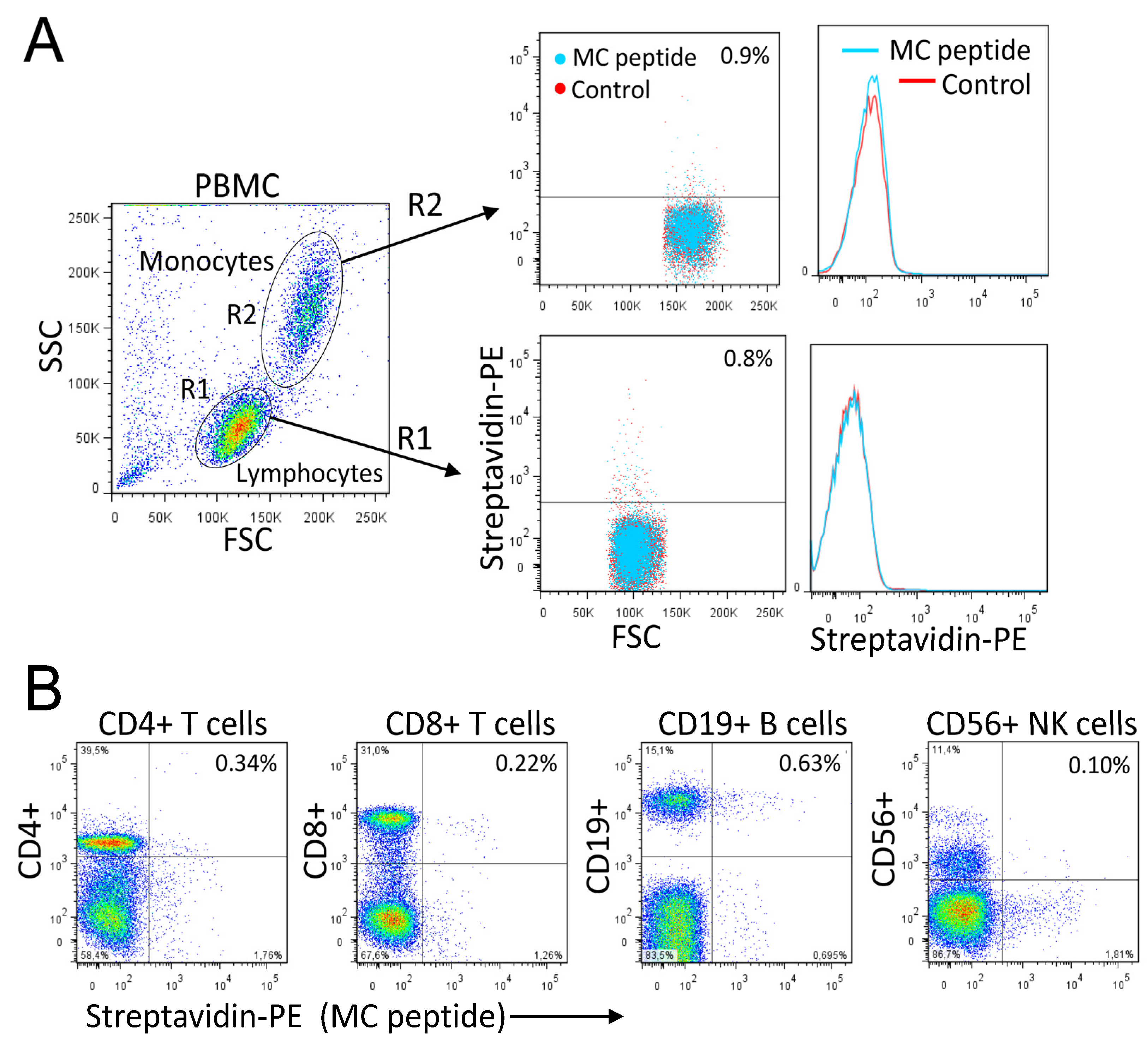
3.7. Rational Design of a Peptide That Mimics the Selected Heparan Sulfate scFvs
3.8. A Pro-Apoptotic Peptide Conjugated to MC Peptide Selectively Kills MM Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADCC | Antibody-dependent cellular cytotoxicity |
| ADP | Antibody-dependent phagocytosis |
| CDC | Complement-dependent cytotoxicity |
| CDR | Complementarity-determining regions |
| HS | Heparan sulfate |
| HSPGs | Heparan sulfate peptidoglycans |
| MM | Multiple myeloma |
| PBMCs | Peripheral blood mononuclear cells |
| PE | Phycoerythrin |
| PEG | Polyethylene glycol |
| PI | Propidium iodide |
| scFv | Single-chain variable fragment |
| siRNA | Small interfering RNA |
| VH | Heavy variable chain |
| VL | Light variable chain |
References
- Mayes, P.A.; Hance, K.W.; Hoos, A. The promise and challenges of immune agonist antibody development in cancer. Nat. Rev. Drug Discov. 2018, 17, 509–527. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, T.; Shen, S.; Wang, J.; Abdou, P.; Gu, Z.; Mo, R. Advances in Engineering Cells for Cancer Immunotherapy. Theranostics 2019, 9, 7889–7905. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Jenkins, R.W.; Sullivan, R.J. Mechanisms of Resistance to Immune Checkpoint Blockade. Am. J. Clin. Dermatol. 2019, 20, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Kalbasi, A.; Ribas, A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020, 20, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M. Releasing the Immune System Brakes Using siRNAs Enhances Cancer Immunotherapy. Cancers 2019, 11, 176. [Google Scholar] [CrossRef] [PubMed]
- Rosenblatt, J.; Avigan, D. Targeting the PD-1/PD-L1 axis in multiple myeloma: A dream or a reality? Blood 2017, 129, 275–279. [Google Scholar] [CrossRef]
- Suen, H.; Brown, R.F.; Yang, S.; Ho, P.J.; Gibson, J.; Joshua, D. The failure of immune checkpoint blockade in multiple myeloma with PD-1 inhibitors in a phase 1 study. Leukemia 2015, 29, 1621–1622. [Google Scholar] [CrossRef]
- Kapoor, S.; Champion, G.; Basu, A.; Mariampillai, A.; Olnes, M.J. Immune Therapies for Myelodysplastic Syndromes and Acute Myeloid Leukemia. Cancers 2021, 13, 5026. [Google Scholar] [CrossRef]
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef]
- Zanwar, S.; Abeykoon, J.P.; Kapoor, P. Challenges and Strategies in the Management of Multiple Myeloma in the Elderly Population. Curr. Hematol. Malig. Rep. 2019, 14, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Musto, P.; La Rocca, F. Monoclonal antibodies in relapsed/refractory myeloma: Updated evidence from clinical trials, real-life studies, and meta-analyses. Expert Rev. Hematol. 2020, 13, 331–349. [Google Scholar] [CrossRef] [PubMed]
- van de Donk, N.; Usmani, S.Z. CD38 Antibodies in Multiple Myeloma: Mechanisms of Action and Modes of Resistance. Front. Immunol. 2018, 9, 2134. [Google Scholar] [CrossRef]
- Bonello, F.; Mina, R.; Boccadoro, M.; Gay, F. Therapeutic Monoclonal Antibodies and Antibody Products: Current Practices and Development in Multiple Myeloma. Cancers 2019, 12, 15. [Google Scholar] [CrossRef]
- Storti, P.; Costa, F.; Marchica, V.; Burroughs-Garcia, J.; Dalla Palma, B.; Toscani, D.; Eufemiese, R.A.; Giuliani, N. Novel Ap-proaches to Improve Myeloma Cell Killing by Monoclonal Antibodies. J. Clin. Med. 2020, 9, 2864. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, M.A.; Jakubowiak, A.J.; McCarthy, P.L.; Orlowski, R.Z.; Attal, M.; Bladé, J.; Goldschmidt, H.; Weisel, K.C.; Ramasamy, K.; Zweegman, S.; et al. Developments in continuous therapy and maintenance treatment approaches for patients with newly diagnosed multiple myeloma. Blood Cancer J. 2020, 10, 17. [Google Scholar] [CrossRef]
- Winter, G.; Griffiths, A.D.; Hawkins, R.E.; Hoogenboom, H.R. Making Antibodies by Phage Display Technology. Annu. Rev. Immunol. 1994, 12, 433–455. [Google Scholar] [CrossRef]
- Dübel, S.; Stoevesandt, O.; Taussig, M.J.; Hust, M. Generating recombinant antibodies to the complete human proteome. Trends Biotechnol. 2010, 28, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M. Phage Display Libraries: From Binders to Targeted Drug Delivery and Human Therapeutics. Mol. Biotechnol. 2019, 61, 286–303. [Google Scholar] [CrossRef] [PubMed]
- Shadidi, M.; Sioud, M. An Anti-leukemic Single Chain Fv Antibody Selected from a Synthetic Human Phage Antibody Library. Biochem. Biophys. Res. Commun. 2001, 280, 548–552. [Google Scholar] [CrossRef]
- Sioud, M.; Westby, P.; Vasovic, V.; Fløisand, Y.; Peng, Q. Development of a new high-affinity human antibody with antitumor activity against solid and blood malignancies. FASEB J. 2018, 32, 5063–5077. [Google Scholar] [CrossRef] [PubMed]
- Bator, J.; Reading, C. Measurement of antibody affinity for cell surface antigens using an enzyme-linked immunosorbent assay. J. Immunol. Methods 1989, 125, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Flatekval, G.F.; Sioud, M. Modulation of dendritic cell maturation and function with mono- and bifunctional small interfering RNAs targeting indoleamine 2,3-dioxygenase. Immunology 2009, 128 (Suppl. 1), e837–e848. [Google Scholar] [CrossRef]
- Sanseviero, E. NK Cell-Fc Receptors Advance Tumor Immunotherapy. J. Clin. Med. 2019, 8, 1667. [Google Scholar] [CrossRef] [PubMed]
- Afratis, N.; Gialeli, C.; Nikitovic, D.; Tsegenidis, T.; Karousou, E.; Theocharis, A.D.; Pavão, M.S.; Tzanakakis, G.N.; Karamanos, N.K. Glycosaminoglycans: Key players in cancer cell biology and treatment. FEBS J. 2012, 279, 1177–1197. [Google Scholar] [CrossRef] [PubMed]
- Annaval, T.; Wild, R.; Crétinon, Y.; Sadir, R.; Vivès, R.R.; Lortat-Jacob, H. Heparan Sulfate Proteoglycans Biosynthesis and Post Synthesis Mechanisms Combine Few Enzymes and Few Core Proteins to Generate Extensive Structural and Functional Di-versity. Molecules 2020, 25, 4215. [Google Scholar] [CrossRef]
- Sioud, M.; Olberg, A. Oslo University Hospital-Radiumhospitalet, Oslo, Norway. 2021; unpublished data. [Google Scholar]
- Baeuerle, P.A.; Huttner, W.B. Chlorate—A potent inhibitor of protein sulfation in intact cells. Biochem. Biophys. Res. Commun. 1986, 141, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, E.; Rhodes, J.M.; Simons, M. Syndecans: Newkids onthe signaling block. Circ. Res. 2005, 96, 488–500. [Google Scholar] [CrossRef]
- O’Connell, F.P.; Pinkus, J.L.; Pinkus, G.S. CD138 (syndecan-1), a plasma cell marker immunohistochemical profile in hema-topoietic and nonhematopoietic neoplasms. Am. J. Clin. Pathol. 2004, 121, 254–263. [Google Scholar] [CrossRef]
- Sela-Culang, I.; Kunik, V.; Ofran, Y. The structural basis of antibody-antigen recognition. Front. Immunol. 2013, 4, 302. [Google Scholar] [CrossRef]
- Davidkova, G.; Pettersson, S.; Holmberg, D.; Lundkvist, I. Selective usage of VH genes in adult human B lymphocyte reper-toires. Scand. J. Immunol. 1997, 45, 62–73. [Google Scholar] [CrossRef]
- Griffin, L.M.; Snowden, J.R.; Lawson, A.D.; Wernery, U.; Kinne, J.; Baker, T.S. Analysis of heavy and light chain sequences of conventional camelid antibodies from Camelus dromedarius and Camelus bactrianus species. J. Immunol. Methods 2014, 405, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Papo, N.; Shai, Y. Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. 2005, 62, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M.; Mobergslien, A. Selective killing of cancer cells by peptide-targeted delivery of an anti-microbial peptide. Biochem. Pharmacol. 2012, 84, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Barash, U.; Cohen-Kaplan, V.; Dowek, I.; Sanderson, R.D.; Ilan, N.; Vlodavsky, I. Proteoglycans in health and disease: New concepts for heparanase function in tumor progression and metastasis. FEBS J. 2010, 277, 3890–3903. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, A.; Malvi, P.; Wajapeyee, N. Heparan Sulfate and Heparan Sulfate Proteoglycans in Cancer Initiation and Pro-gression. Front. Endocrinol. 2018, 9, 483. [Google Scholar] [CrossRef] [PubMed]
- Faria-Ramos, I.; Poças, J.; Marques, C.; Santos-Antunes, J.; Macedo, G.; Reis, C.A.; Magalhães, A. Heparan Sulfate Glycosa-minoglycans: (Un)Expected Allies in Cancer Clinical Management. Biomolecules 2021, 11, 136. [Google Scholar] [CrossRef]
- Akl, M.R.; Nagpal, P.; Ayoub, N.M.; Prabhu, S.A.; Gliksman, M.; Tai, B.; Hatipoglu, A.; Goy, A.; Suh, K.S. Molecular and clinical profiles of syndecan-1 in solid and hematological cancer for prognosis and precision medicine. Oncotarget 2015, 6, 28693–28715. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.C.; Multhaupt, H.A.; Couchman, J.R. Cell surface heparan sulfate proteoglycans control adhesion and invasion of breast carcinoma cells. Mol. Cancer 2015, 14, 15. [Google Scholar] [CrossRef]
- Olberg, A.; Sioud, M. Oslo University Hospital-Radiumhospitalet, Oslo, Norway. 2021; unpublished data. [Google Scholar]
- Muyldermans, S.; Smider, V.V. Distinct antibody species: Structural differences creating therapeutic opportunities. Curr. Opin. Immunol. 2016, 40, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Barthelemy, P.A.; Raab, H.; Appleton, B.A.; Bond, C.J.; Wu, P.; Wiesmann, C.; Sidhu, S.S. Comprehensive Analysis of the Factors Contributing to the Stability and Solubility of Autonomous Human VH Domains. J. Biol. Chem. 2008, 283, 3639–3654. [Google Scholar] [CrossRef] [PubMed]
- Williams, W.V.; Kieber-Emmons, T.; Vonfeldt, J.; Greene, M.; Weiner, D.B. Design of bioactive peptides based on antibody hypervariable region structures. Development of conformationally constrained and dimeric peptides with enhanced affinity. J. Biol. Chem. 1991, 266, 5182–5190. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Li, Y.; Zhang, W.; Shen, B. Rational design of potent mimic peptide derived from monoclonal antibody: Antibody mimic design. Immunol. Lett. 2005, 98, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Mahtouk, K.; Cremer, F.W.; Rème, T.; Jourdan, M.; Baudard, M.; Moreaux, J.; Requirand, G.; Fiol, G.; De Vos, J.; Moos, M.; et al. Heparan sulphate proteoglycans are essential for the myeloma cell growth activity of EGF-family ligands in multiple myeloma. Oncogene 2006, 25, 7180–7191. [Google Scholar] [CrossRef] [PubMed]
- Filla, M.S.; Dam, P.; Rapraeger, A.C. The cell surface proteoglycan syndecan-1 mediates fibroblast growth factor-2 binding and activity. J. Cell. Physiol. 1998, 174, 310–321. [Google Scholar] [CrossRef]
- Bartolini, B.; Caravà, E.; Caon, I.; Parnigoni, A.; Moretto, P.; Passi, A.; Vigetti, D.; Viola, M.; Karousou, E. Heparan Sulfate in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Hideshima, T.; Fulciniti, M.; Lutz, R.J.; Yasui, H.; Okawa, Y.; Kiziltepe, T.; Vallet, S.; Pozzi, S.; Santo, L.; et al. The monoclonal antibody nBT062 conjugated to cytotoxic Maytansinoids has selective cytotoxicity against CD138-positive multiple myeloma cells in vitro and in vivo. Clin. Cancer Res. 2009, 15, 4028–4037. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.R.; Ailawadhi, S.; Siegel, D.S.; Heffner, L.T.; Somlo, G.; Jagannath, S.; Zimmerman, T.M.; Munshi, N.C.; Madan, S.; Chanan-Khan, A.; et al. Indatuximab ravtansine plus dexamethasone with lenalidomide or pomalidomide in relapsed or refractory multiple myeloma: A multicentre, phase 1/2a study. Lancet Haematol. 2021, 8, e794–e807. [Google Scholar] [CrossRef]
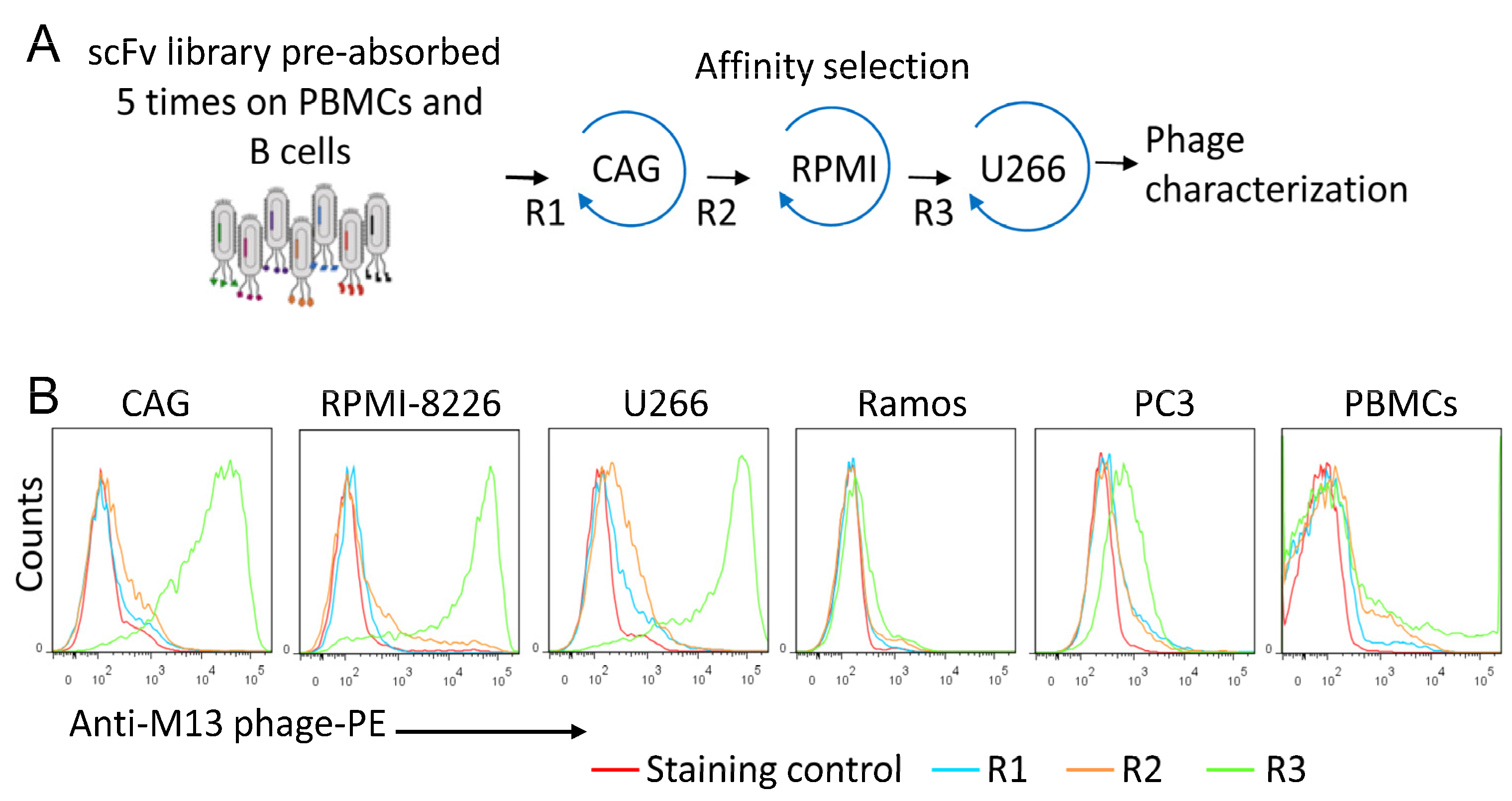

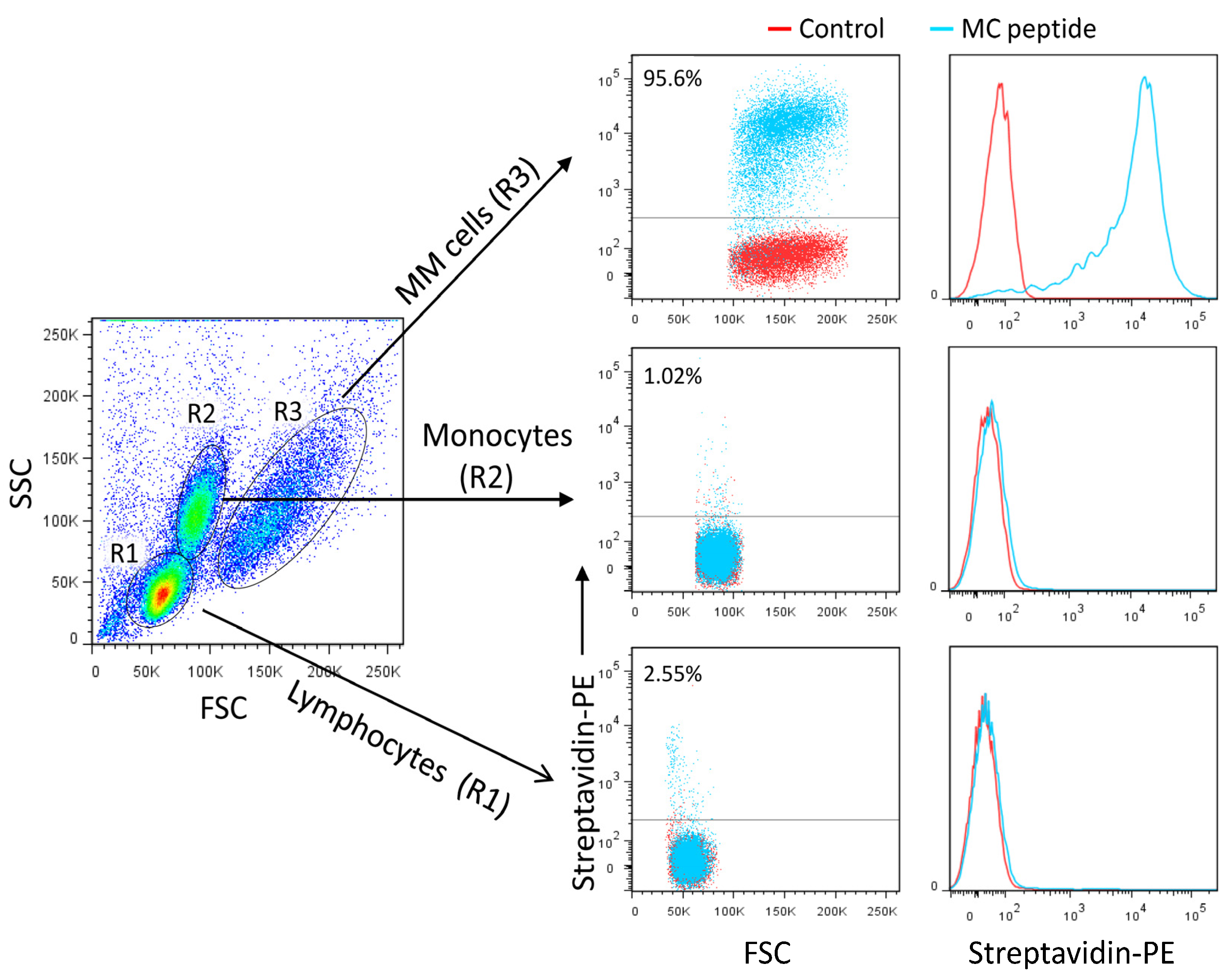
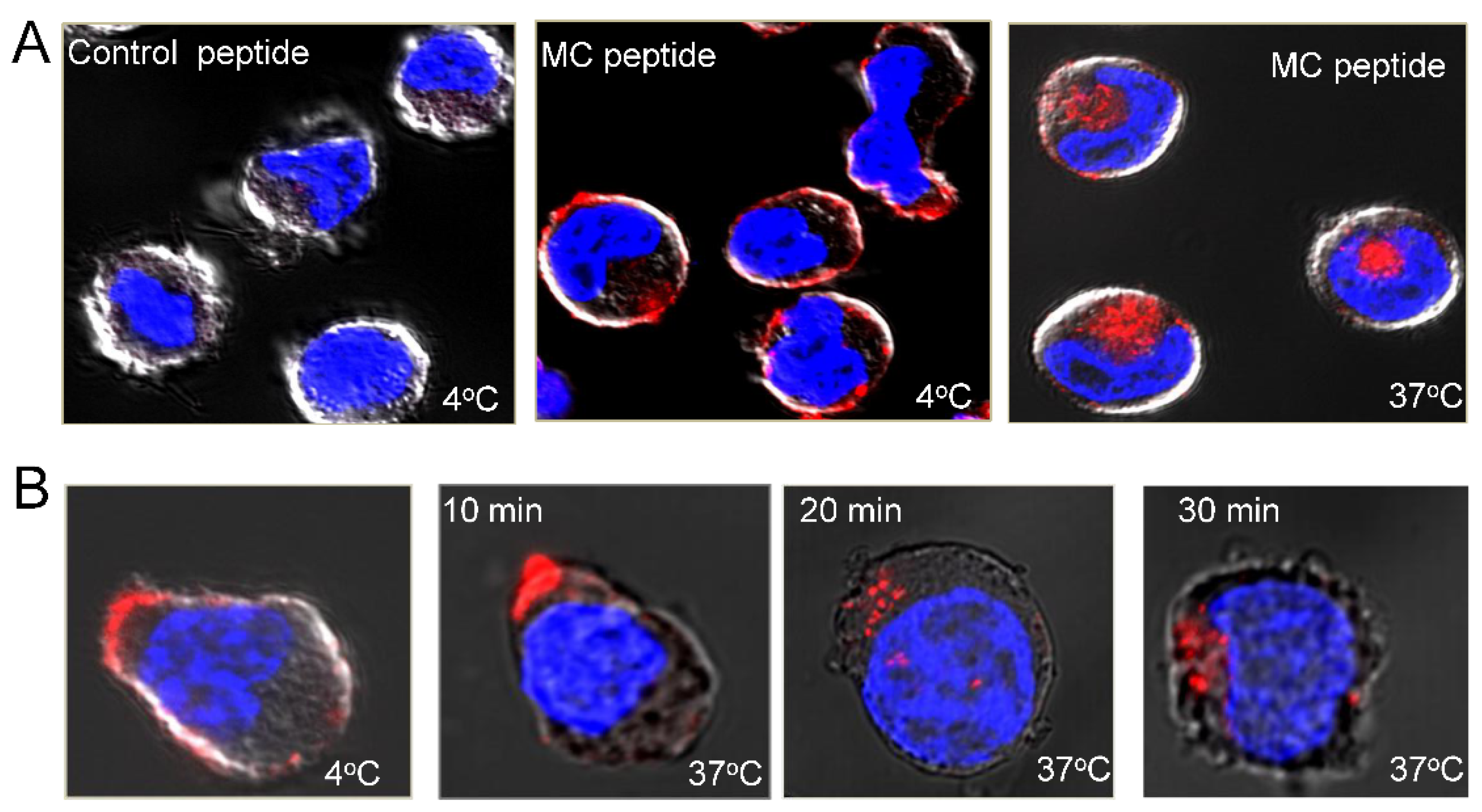
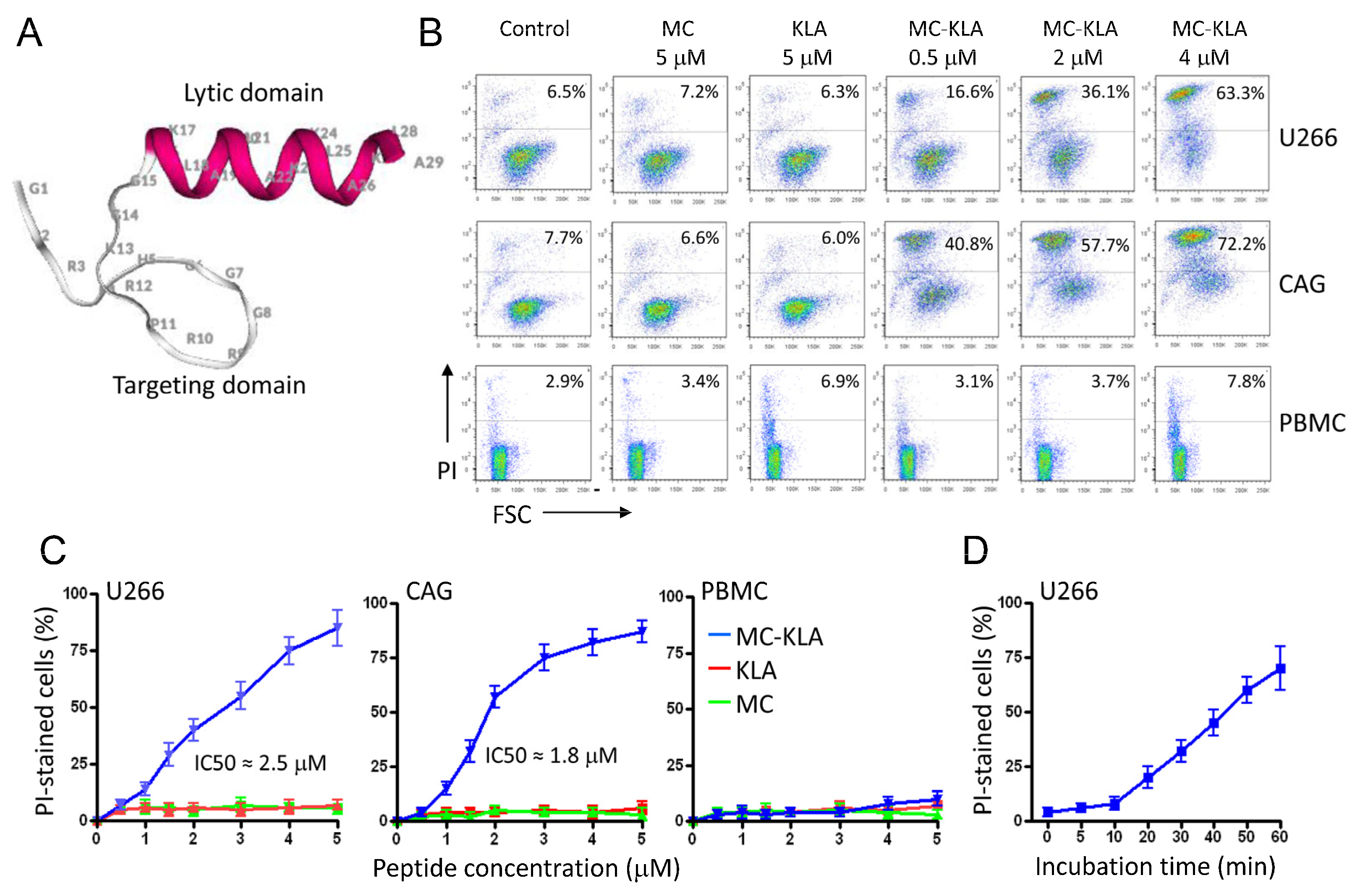
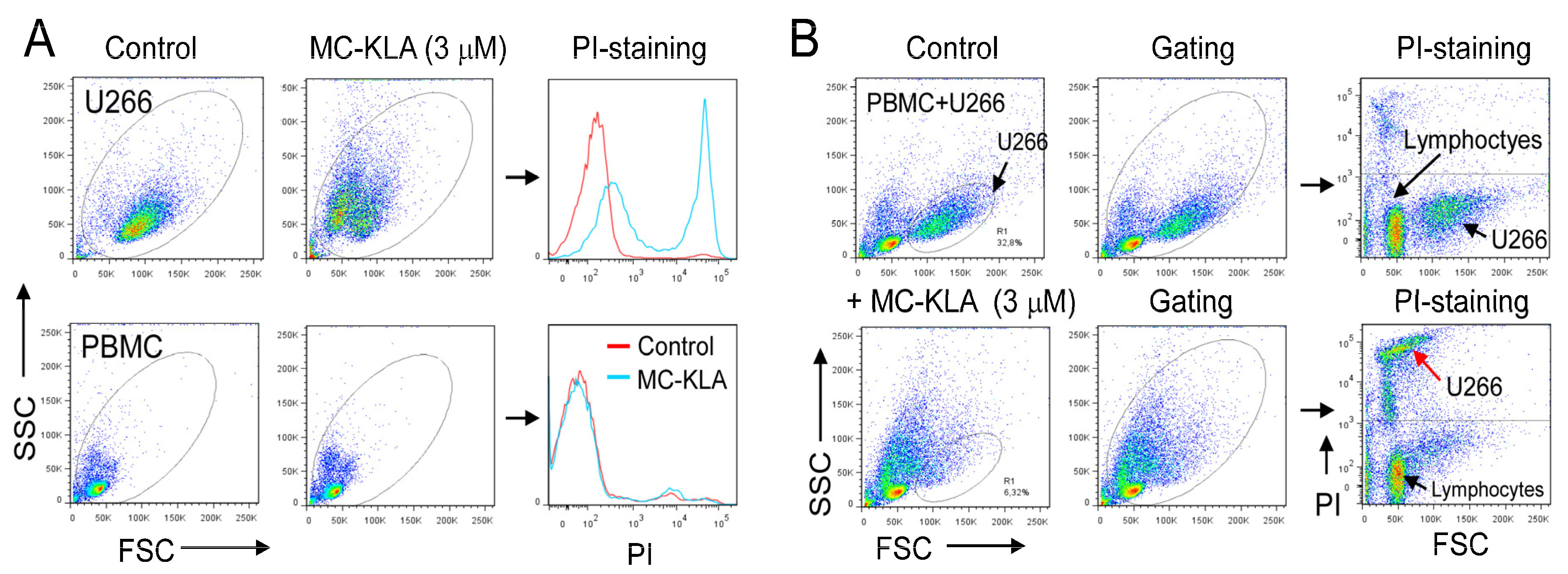
| Sequential Affinity Selection on Cancer Cell Lines | Cell Number | Input Number of Phages (TU) | Recovered Number of Phages (TU) | Enrichment over Previous Round * |
|---|---|---|---|---|
| CAG | 1 × 107 | 1 × 1010 | 2.0 × 103 | 0 |
| RPMI 8226 | 1 × 107 | 1 × 1010 | 1.5 × 104 | 5 |
| U266 | 1 × 107 | 1 × 1010 | 1.2 × 107 | 800 |
| Clone | VH CDR2 | VH CDR3 | VL CDR2 | VL CDR3 | Frequency * |
|---|---|---|---|---|---|
| MM1 | AIRHPGLHTEY | AKGGRRFDY | RASRLQS | QQANSPPPT | 21/30 |
| MM10 | TIRRQGGNTEY | AKSARVFDY | TASRLRS | QQWTAKPGT | 2/30 |
| MM12 | AIRRPHLNTEY | AKGRRPRKFDY | RASHLQS | QQPNAPAPT | 7/30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sioud, M.; Olberg, A. Antibody Surface Profiling Identifies Glycoforms in Multiple Myeloma as Targets for Immunotherapy: From Antibody Derivatives to Mimetic Peptides for Killing Tumor Cells. Cancers 2023, 15, 1934. https://doi.org/10.3390/cancers15071934
Sioud M, Olberg A. Antibody Surface Profiling Identifies Glycoforms in Multiple Myeloma as Targets for Immunotherapy: From Antibody Derivatives to Mimetic Peptides for Killing Tumor Cells. Cancers. 2023; 15(7):1934. https://doi.org/10.3390/cancers15071934
Chicago/Turabian StyleSioud, Mouldy, and Anniken Olberg. 2023. "Antibody Surface Profiling Identifies Glycoforms in Multiple Myeloma as Targets for Immunotherapy: From Antibody Derivatives to Mimetic Peptides for Killing Tumor Cells" Cancers 15, no. 7: 1934. https://doi.org/10.3390/cancers15071934
APA StyleSioud, M., & Olberg, A. (2023). Antibody Surface Profiling Identifies Glycoforms in Multiple Myeloma as Targets for Immunotherapy: From Antibody Derivatives to Mimetic Peptides for Killing Tumor Cells. Cancers, 15(7), 1934. https://doi.org/10.3390/cancers15071934





