Current Applications of Liquid Biopsy in Gastrointestinal Cancer Disease—From Early Cancer Detection to Individualized Cancer Treatment
Simple Summary
Abstract
1. Introduction
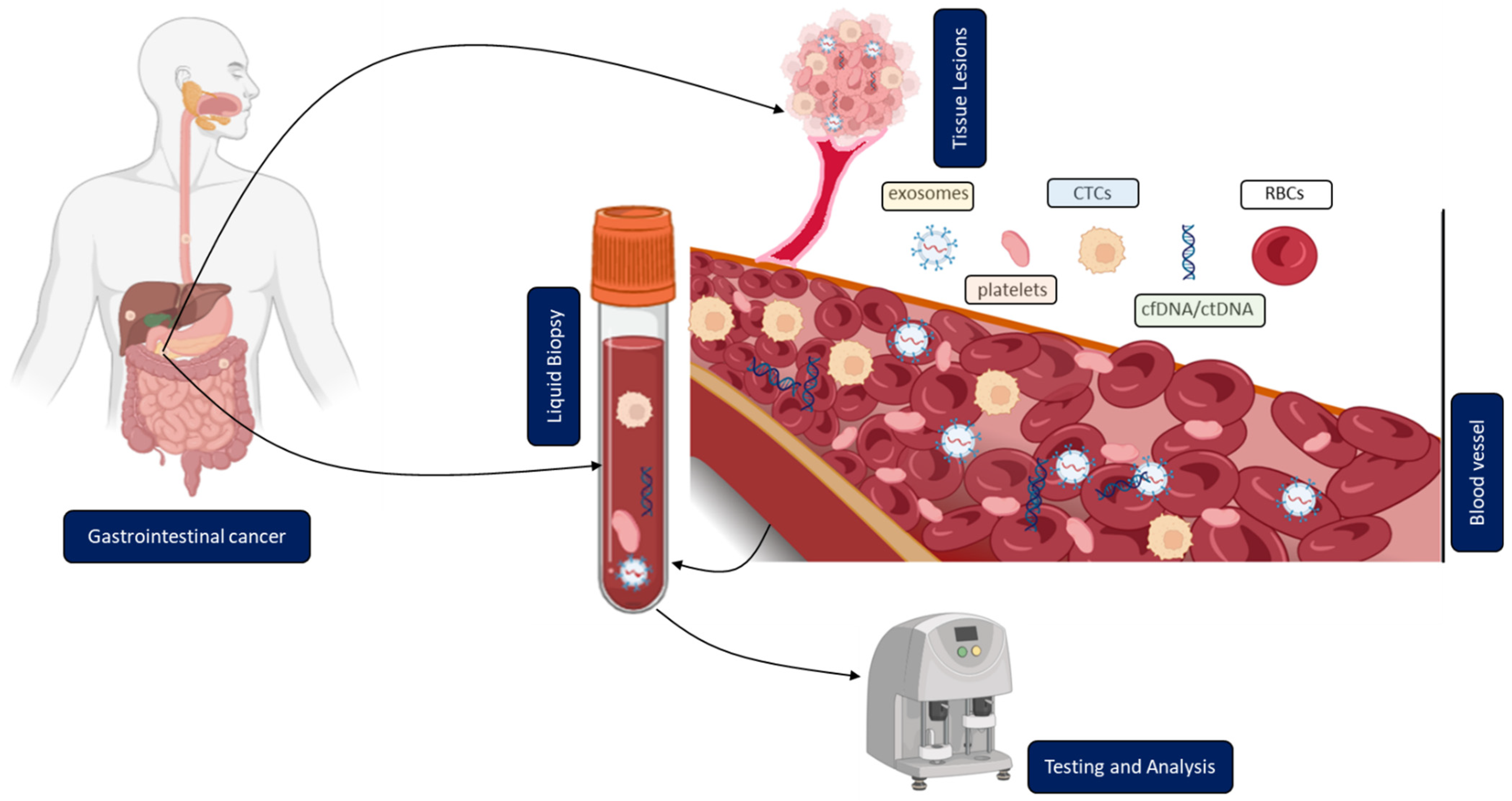
2. Overview of Different Methodologies and Their Current Clinical Application
2.1. Circulating Tumor Cells (CTCs)
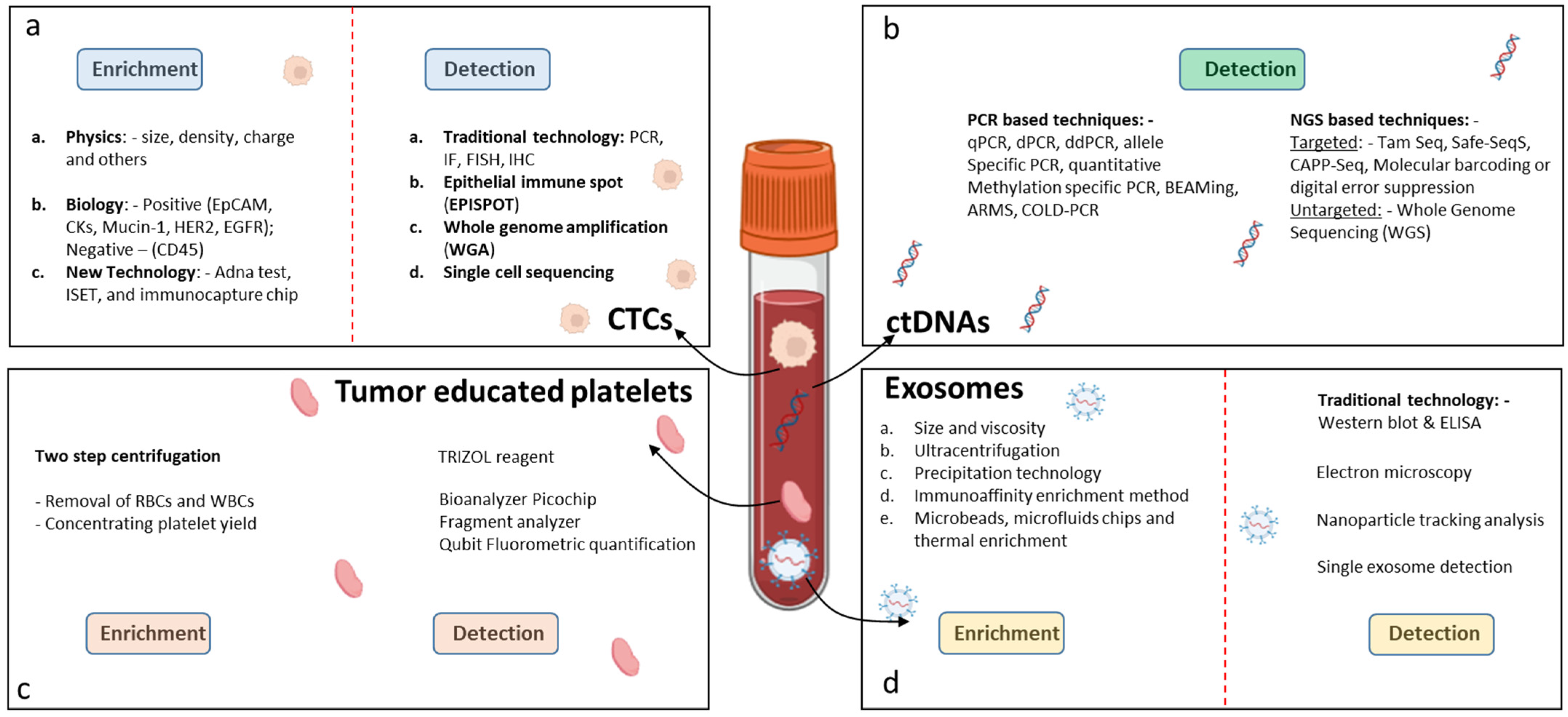
2.1.1. Isolation and Enrichment of CTCs
2.1.2. Clinical Application/Relevance of CTCs
2.2. Circulating Tumor DNA (ctDNA)
2.2.1. Detection and Analysis of ctDNA
2.2.2. Clinical Application/Relevance of ctDNA
2.3. Circulating Extracellular Vesicles (Tumor Exosomes)
2.3.1. Isolation of Tumor Exosomes
2.3.2. Clinical Application/Relevance of Tumor Exosomes
2.4. Tumor-Educated Blood Platelets (TEPs)
2.4.1. Isolation and Detection of Tumor-Educated Platelets
2.4.2. Clinical Application/Relevance of Tumor-Educated Blood Platelets
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AdSCs | adult stem cells |
| ALI | air-liquid-interface |
| CA19-9 | carbohydrate Antigen 19-9 |
| CEACAMs | carcinoembryonic antigen-related cell adhesion molecules |
| cfDNA | cell-free DNA |
| ctDNA | circulating tumor DNA |
| CRC | colorectal cancer |
| CRML | colorectal liver metastases |
| circRNA | circular RNA |
| CTCs | circulating tumor cells |
| ctDNA | circulating tumor DNA |
| DC | differential centrifugation |
| ddPCR | droplet digital PCR |
| DG | density gradient ultracentrifugation |
| ECM | extracellular matrix |
| EpCAM | epithelial cell adhesion molecule |
| EUS | endoscopic ultrasound |
| EVs | extracellular vesicles |
| FDA | the United States Food and Drug Administration |
| FISH | fluorescence in situ hybridization |
| GPC-1 | glypican 1 |
| GI | gastrointestinal |
| LB | liquid biopsy |
| LncRNA | long non-coding RNA |
| mRNA | messenger RNA |
| MVBs | multivesicular bodies |
| miRNA | microRNAs |
| NGS | next generation sequencing |
| PC | pancreatic cancer |
| PDAC | pancreatic ductal adenocarcinoma |
| PDO | patient-derived cancer organoid |
| PSCs | pluripotent stem cells |
| RT-qPCR | real-time quantitative polymerase chain reaction |
| SEC | size exclusion chromatography |
| TEPs | tumor-educated blood platelets |
| TME | tumor microenvironment |
| UC | ultracentrifugation |
References
- International Agency for Research on Cancer. Estimated Number of New Cases and Deaths of Cancer in 2020. 2020. Available online: https://gco.iarc.fr/today/home (accessed on 15 February 2022).
- Le, Q.; Wang, C.; Shi, Q. Meta-Analysis on the Improvement of Symptoms and Prognosis of Gastrointestinal Tumors Based on Medical Care and Exercise Intervention. J. Healthc. Eng. 2021, 2021, 5407664. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-D.; Zhang, P.-F.; Xi, H.-Q.; Wei, B.; Chen, L.; Tang, Y. Recent Advances in the Diagnosis, Staging, Treatment, and Prognosis of Advanced Gastric Cancer: A Literature Review. Front. Med. 2021, 8, 744839. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020, 159, 335–349.e15. [Google Scholar] [CrossRef]
- Saini, A.; Pershad, Y.; Albadawi, H.; Kuo, M.; Alzubaidi, S.; Naidu, S.; Knuttinen, M.-G.; Oklu, R. Liquid Biopsy in Gastrointestinal Cancers. Diagnostics 2018, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Niimi, K.; Ishibashi, R.; Mitsui, T.; Aikou, S.; Kodashima, S.; Yamashita, H.; Yamamichi, N.; Hirata, Y.; Fujishiro, M.; Seto, Y.; et al. Laparoscopic and endoscopic cooperative surgery for gastrointestinal tumor. Ann. Transl. Med. 2017, 5, 187. [Google Scholar] [CrossRef] [PubMed]
- Garraway, L.A.; Jänne, P.A. Circumventing Cancer Drug Resistance in the Era of Personalized Medicine. Cancer Discov. 2012, 2, 214–226. [Google Scholar] [CrossRef]
- Goyal, L.; Saha, S.K.; Liu, L.Y.; Siravegna, G.; Leshchiner, I.; Ahronian, L.G.; Lennerz, J.K.; Vu, P.; Deshpande, V.; Kambadakone, A.; et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion–Positive Cholangiocarcinoma. Cancer Discov. 2017, 7, 252–263. [Google Scholar] [CrossRef]
- Russo, M.; Siravegna, G.; Blaszkowsky, L.S.; Corti, G.; Crisafulli, G.; Ahronian, L.G.; Mussolin, B.; Kwak, E.L.; Buscarino, M.; Lazzari, L.; et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016, 6, 147–153. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Hazar-Rethinam, M.; Kleyman, M.; Han, G.C.; Liu, D.; Ahronian, L.G.; Shahzade, H.A.; Chen, L.; Parikh, A.R.; Allen, J.N.; Clark, J.W.; et al. Convergent Therapeutic Strategies to Overcome the Heterogeneity of Acquired Resistance in BRAF(V600E) Colorectal Cancer. Cancer Discov. 2018, 8, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.; Overman, M.; Dasari, A.; Kazmi, S.; Mazard, T.; Vilar, E.; Morris, V.; Lee, M.; Herron, D.; Eng, C.; et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann. Oncol. 2015, 26, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Emoto, S.; Ishigami, H.; Yamashita, H.; Yamaguchi, H.; Kaisaki, S.; Kitayama, J. Clinical significance of CA125 and CA72-4 in gastric cancer with peritoneal dissemination. Gastric Cancer 2011, 15, 154–161. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Lu, M.; Shen, L. Predictive value of serum CEA, CA19-9 and CA72.4 in early diagnosis of recurrence after radical resection of gastric cancer. Hepatogastroenterology 2011, 58, 2166–2170. [Google Scholar] [PubMed]
- Shimada, H.; Noie, T.; Ohashi, M.; Oba, K.; Takahashi, Y. Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer 2013, 17, 26–33. [Google Scholar] [CrossRef]
- Williams, S.C. Circulating tumor cells. Proc. Natl. Acad. Sci. USA 2013, 110, 4861. [Google Scholar] [CrossRef]
- Nel, I.; David, P.; Gerken, G.G.H.; Schlaak, J.F.; Hoffmann, A.-C. Role of circulating tumor cells and cancer stem cells in hepatocellular carcinoma. Hepatol. Int. 2014, 8, 321–329. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Klymkowsky, M.W.; Savagner, P. Epithelial-mesenchymal transition: A cancer researcher’s conceptual friend and foe. Am. J. Pathol. 2009, 174, 1588–1593. [Google Scholar] [CrossRef]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, M.; Christofori, G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009, 28, 15–33. [Google Scholar] [CrossRef]
- Barrallo-Gimeno, A.; Nieto, M.A. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development 2005, 132, 3151–3161. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Denève, E.; Riethdorf, S.; Ramos, J.; Nocca, D.; Coffy, A.; Daurès, J.-P.; Maudelonde, T.; Fabre, J.-M.; Pantel, K.; Alix-Panabières, C. Capture of Viable Circulating Tumor Cells in the Liver of Colorectal Cancer Patients. Clin. Chem. 2013, 59, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Tien, Y.W.; Kuo, H.C.; Ho, B.I.; Chang, M.C.; Chang, Y.T.; Cheng, M.F.; Chen, H.-L.; Liang, T.-Y.; Wang, C.-F.; Huang, C.-Y.; et al. A High Circulating Tumor Cell Count in Portal Vein Predicts Liver Metastasis from Periampullary or Pancreatic Cancer: A High Portal Venous CTC Count Predicts Liver Metastases. Medicine 2016, 95, e3407. [Google Scholar] [CrossRef] [PubMed]
- Bissolati, M.; Sandri, M.T.; Burtulo, G.; Zorzino, L.; Balzano, G.; Braga, M. Portal vein-circulating tumor cells predict liver metastases in patients with resectable pancreatic cancer. Tumor Biol. 2014, 36, 991–996. [Google Scholar] [CrossRef]
- Chapman, C.G.; Waxman, I. EUS-Guided Portal Venous Sampling of Circulating Tumor Cells. Curr. Gastroenterol. Rep. 2019, 21, 68. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Shahneh, F.Z. Sensitive antibody-based CTCs detection from peripheral blood. Hum. Antibodies 2013, 22, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Uenosono, Y.; Arigami, T.; Kozono, T.; Yanagita, S.; Hagihara, T.; Haraguchi, N.; Matsushita, D.; Hirata, M.; Arima, H.; Funasako, Y.; et al. Clinical significance of circulating tumor cells in peripheral blood from patients with gastric cancer. Cancer 2013, 119, 3984–3991. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, J.; Kim, S.T.; Park, S.H.; Park, J.O.; Park, Y.S.; Lim, H.Y.; Kang, W.K. Circulating Tumor Cells are Predictive of Poor Response to Chemotherapy in Metastatic gastric cancer. Int. J. Biol. Markers 2015, 30, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Bang-Christensen, S.R.; Katerov, V.; Jørgensen, A.M.; Gustavsson, T.; Choudhary, S.; Theander, T.G.; Salanti, A.; Allawi, H.T.; Agerbæk, M. Detection of VAR2CSA-Captured Colorectal Cancer Cells from Blood Samples by Real-Time Reverse Transcription PCR. Cancers 2021, 13, 5881. [Google Scholar] [CrossRef]
- Lim, M.; Park, S.; Jeong, H.-O.; Park, S.H.; Kumar, S.; Jang, A.; Lee, S.; Kim, D.U.; Cho, Y.-K. Circulating Tumor Cell Clusters Are Cloaked with Platelets and Correlate with Poor Prognosis in Unresectable Pancreatic Cancer. Cancers 2021, 13, 5272. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; McFaul, S.M.; Duffy, S.P.; Deng, X.; Tavassoli, P.; Black, P.C.; Ma, H. Technologies for label-free separation of circulating tumor cells: From historical foundations to recent developments. Lab A Chip 2013, 14, 32–44. [Google Scholar] [CrossRef]
- Nordgård, O.; Tjensvoll, K.; Gilje, B.; Søreide, K. Circulating tumour cells and DNA as liquid biopsies in gastrointestinal cancer. Br. J. Surg. 2018, 105, e110–e120. [Google Scholar] [CrossRef]
- Dizdar, L.; Fluegen, G.; van Dalum, G.; Honisch, E.; Neves, R.P.; Niederacher, D.; Neubauer, H.; Fehm, T.; Rehders, A.; Krieg, A.; et al. Detection of circulating tumor cells in colorectal cancer patients using the GILUPI CellCollector: Results from a prospective, single-center study. Mol. Oncol. 2019, 13, 1548–1558. [Google Scholar] [CrossRef]
- Bork, U.; Rahbari, N.N.; Schölch, S.; Reissfelder, C.; Kahlert, C.; Büchler, M.W.; Weitz, J.; Koch, M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: A prospective study. Br. J. Cancer 2015, 112, 1306–1313. [Google Scholar] [CrossRef]
- Van Dalum, G.; Stam, G.J.; Scholten, L.F.; Mastboom, W.J.; Vermes, I.; Tibbe, A.G.; De Groot, M.R.; Terstappen, L.W.M.M. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int. J. Oncol. 2015, 46, 1361–1368. [Google Scholar] [CrossRef]
- Sotelo, M.J.; Sastre, J.; Maestro, M.L.; Veganzones, S.; Viéitez, J.M.; Alonso, V.; Grávalos, C.; Escudero, P.; Vera, R.; Aranda, E.; et al. Role of circulating tumor cells as prognostic marker in resected stage III colorectal cancer. Ann. Oncol. 2014, 26, 535–541. [Google Scholar] [CrossRef]
- Lu, C.-Y.; Tsai, H.-L.; Uen, Y.-H.; Hu, H.-M.; Chen, C.-W.; Cheng, T.-L.; Lin, S.-R.; Wang, J.-Y. Circulating tumor cells as a surrogate marker for determining clinical outcome to mFOLFOX chemotherapy in patients with stage III colon cancer. Br. J. Cancer 2013, 108, 791–797. [Google Scholar] [CrossRef]
- Lu, Y.-J.; Wang, P.; Peng, J.; Wang, X.; Zhu, Y.-W.; Shen, N. Meta-analysis Reveals the Prognostic Value of Circulating Tumour Cells Detected in the Peripheral Blood in Patients with Non-Metastatic Colorectal Cancer. Sci. Rep. 2017, 7, 905. [Google Scholar] [CrossRef]
- Huang, X.; Gao, P.; Song, Y.; Sun, J.; Chen, X.; Zhao, J.; Xu, H.; Wang, Z. Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch System in colorectal cancer. BMC Cancer 2015, 15, 202. [Google Scholar] [CrossRef] [PubMed]
- Aranda, E.; Viéitez, J.M.; Gómez-España, A.; Calle, S.G.; Salud-Salvia, A.; Graña, B.; Garcia-Alfonso, P.; Rivera, F.; Quintero-Aldana, G.A.; Reina-Zoilo, J.J.; et al. FOLFOXIRI plus bevacizumab versus FOLFOX plus bevacizumab for patients with metastatic colorectal cancer and >/=3 circulating tumour cells: The randomised phase III VISNU-1 trial. ESMO Open 2020, 5, e000944. [Google Scholar] [CrossRef] [PubMed]
- Hardingham, J.E.; Grover, P.; Winter, M.; Hewett, P.J.; Price, T.J.; Thierry, B. Detection and Clinical Significance of Circulating Tumor Cells in Colorectal Cancer—20 Years of Progress. Mol. Med. 2015, 21 (Suppl. S1), S25–S31. [Google Scholar] [CrossRef]
- Krebs, M.G.; Renehan, A.G.; Backen, A.; Gollins, S.; Chau, I.; Hasan, J.; Valle, J.W.; Morris, K.; Beech, J.; Ashcroft, L.; et al. Circulating Tumor Cell Enumeration in a Phase II Trial of a Four-Drug Regimen in Advanced Colorectal Cancer. Clin. Color. Cancer 2014, 14, 115–122.e2. [Google Scholar] [CrossRef]
- Steinert, G.; Schölch, S.; Koch, M.; Weitz, J. Biology and significance of circulating and disseminated tumour cells in colorectal cancer. Langenbeck’s Arch. Surg. 2012, 397, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Pernot, S.; Badoual, C.; Terme, M.; Castan, F.; Cazes, A.; Bouche, O.; Bennouna, J.; Francois, E.; Ghiringhelli, F.; De La Fouchardiere, C.; et al. Dynamic evaluation of circulating tumour cells in patients with advanced gastric and oesogastric junction adenocarcinoma: Prognostic value and early assessment of therapeutic effects. Eur. J. Cancer 2017, 79, 15–22. [Google Scholar] [CrossRef]
- Kang, H.M.; Kim, G.H.; Jeon, H.K.; Kim, D.H.; Jeon, T.Y.; Park, D.Y.; Jeong, H.; Chun, W.J.; Kim, M.-H.; Park, J.; et al. Circulating tumor cells detected by lab-on-a-disc: Role in early diagnosis of gastric cancer. PLoS ONE 2017, 12, e0180251. [Google Scholar] [CrossRef]
- Cheng, B.; Tong, G.; Wu, X.; Cai, W.; Li, Z.; Tong, Z.; He, L.; Yu, S.; Wang, S. Enumeration Aand Characterization Of Circulating Tumor Cells And Its Application In Advanced Gastric Cancer. OncoTargets Ther. 2019, 12, 7887–7896. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, R.; Sun, X.; Liu, Y.; Wang, Z.; Yan, J.; Kong, X.; Liang, S.; Liu, Q.; Zhao, T.; et al. Prognostic significance of PD-L1 expression on cell-surface vimentin-positive circulating tumor cells in gastric cancer patients. Mol. Oncol. 2020, 14, 865–881. [Google Scholar] [CrossRef]
- Jhi, J.H.; Kim, G.H.; Park, S.J.; Kim, D.U.; Lee, M.W.; Lee, B.E.; Kwon, C.H.; Cho, Y.-K. Circulating Tumor Cells and TWIST Expression in Patients with Metastatic Gastric Cancer: A Preliminary Study. J. Clin. Med. 2021, 10, 4481. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhu, S.; Luo, Y.; Li, G.; Pu, Y.; Cai, B.; Zhang, C. Application of Circulating Tumor Cells and Circulating Free DNA from Peripheral Blood in the Prognosis of Advanced Gastric Cancer. J. Oncol. 2022, 2022, 9635218. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, S.; Zhang, W.; Wang, J.; Wang, M.; Hu, X.; Liu, F.; Zhang, Y.; Jiang, B.; Yuan, H. Circulating tumor cells as an independent prognostic factor in advanced colorectal cancer: A retrospective study in 121 patients. Int. J. Color. Dis. 2019, 34, 589–597. [Google Scholar] [CrossRef]
- Bahnassy, A.A.; Salem, S.E.; Mohanad, M.; Abulezz, N.Z.; Abdellateif, M.S.; Hussein, M.; Zekri, C.A.; Zekri, A.-R.N.; Allahloubi, N.M. Prognostic significance of circulating tumor cells (CTCs) in Egyptian non-metastatic colorectal cancer patients: A comparative study for four different techniques of detection (Flowcytometry, CellSearch, Quantitative Real-time PCR and Cytomorphology). Exp. Mol. Pathol. 2018, 106, 90–101. [Google Scholar] [CrossRef]
- Messaritakis, I.; Sfakianaki, M.; Papadaki, C.; Koulouridi, A.; Vardakis, N.; Koinis, F.; Hatzidaki, D.; Georgoulia, N.; Kladi, A.; Kotsakis, A.; et al. Prognostic significance of CEACAM5mRNA-positive circulating tumor cells in patients with metastatic colorectal cancer. Cancer Chemother. Pharmacol. 2018, 82, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Iinuma, H.; Watanabe, T.; Mimori, K.; Adachi, M.; Hayashi, N.; Tamura, J.; Matsuda, K.; Fukushima, R.; Okinaga, K.; Sasako, M.; et al. Clinical Significance of Circulating Tumor Cells, Including Cancer Stem-Like Cells, in Peripheral Blood for Recurrence and Prognosis in Patients with Dukes’ Stage B and C Colorectal Cancer. J. Clin. Oncol. 2011, 29, 1547–1555. [Google Scholar] [CrossRef]
- Tol, J.; Koopman, M.; Miller, M.C.; Tibbe, A.; Cats, A.; Creemers, G.J.M.; Vos, A.H.; Nagtegaal, I.; Terstappen, L.; Punt, C.J.A. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann. Oncol. 2009, 21, 1006–1012. [Google Scholar] [CrossRef]
- Cohen, S.J.A.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Prasoppokakorn, T.; Buntho, A.; Ingrungruanglert, P.; Tiyarattanachai, T.; Jaihan, T.; Kulkraisri, K.; Ariyaskul, D.; Phathong, C.; Israsena, N.; Rerknimitr, R.; et al. Circulating tumor cells as a prognostic biomarker in patients with hepatocellular carcinoma. Sci. Rep. 2022, 12, 1–9. [Google Scholar] [CrossRef]
- Hamaoka, M.; Kobayashi, T.; Tanaka, Y.; Mashima, H.; Ohdan, H. Clinical significance of glypican-3-positive circulating tumor cells of hepatocellular carcinoma patients: A prospective study. PLoS ONE 2019, 14, e0217586. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Yang, X.R.; Sun, Y.F.; Shen, M.N.; Ma, X.L.; Wu, J.; Zhang, C.-Y.; Zhou, Y.; Xu, Y.; Hu, B.; et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin. Cancer Res. 2014, 20, 4794–4805. [Google Scholar] [CrossRef]
- Xu, W.; Cao, L.; Chen, L.; Li, J.; Zhang, X.-F.; Qian, H.-H.; Kang, X.-Y.; Zhang, Y.; Liao, J.; Shi, L.-H.; et al. Isolation of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma Using a Novel Cell Separation Strategy. Clin. Cancer Res. 2011, 17, 3783–3793. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Li, F.; Li, T.T.; Zhang, J.T.; Shi, X.J.; Huang, X.Y.; Zhou, J.; Tang, Z.-Y.; Huang, Z.-L. A clinically feasible circulating tumor cell sorting system for monitoring the progression of advanced hepatocellular carcinoma. J. Nanobiotechnol. 2023, 21, 25. [Google Scholar] [CrossRef]
- Winograd, P.; Hou, S.; Court, C.M.; Lee, Y.-T.; Chen, P.-J.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Teng, P.-C.; Wang, J.J.; et al. Hepatocellular Carcinoma–Circulating Tumor Cells Expressing PD-L1 Are Prognostic and Potentially Associated with Response to Checkpoint Inhibitors. Hepatol. Commun. 2020, 4, 1527–1540. [Google Scholar] [CrossRef]
- Li, J.; Shi, L.; Zhang, X.; Sun, B.; Yang, Y.; Ge, N.; Liu, H.; Yang, X.; Chen, L.; Qian, H.; et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget 2015, 7, 2646–2659. [Google Scholar] [CrossRef] [PubMed]
- Gemenetzis, G.; Groot, V.; Yu, J.; Ding, D.; Teinor, J.; Javed, A.; Wood, L.; Burkhart, R.; Cameron, J.; Makary, M.; et al. Circulating tumor cells dynamics in pancreatic adenocarcinoma correlate with disease status: Results of the prospective cluster study. HPB 2019, 21, S1. [Google Scholar] [CrossRef]
- Soeth, E.; Grigoleit, U.; Moellmann, B.; Röder, C.; Schniewind, B.; Kremer, B.; Kalthoff, H.; Vogel, I. Detection of tumor cell dissemination in pancreatic ductal carcinoma patients by CK 20 RT-PCR indicates poor survival. J. Cancer Res. Clin. Oncol. 2005, 131, 669–676. [Google Scholar] [CrossRef]
- Gao, Y.; Zhu, Y.; Zhang, Z.; Zhang, C.; Huang, X.; Yuan, Z. Clinical significance of pancreatic circulating tumor cells using combined negative enrichment and immunostaining-fluorescence in situ hybridization. J. Exp. Clin. Cancer Res. 2016, 35, 66. [Google Scholar] [CrossRef]
- Court, C.; Ankeny, J.S.; Sho, S.; Winograd, P.; Hou, S.; Song, M.; Wainberg, Z.A.; Girgis, M.D.; Graeber, T.G.; Agopian, V.G.; et al. Circulating Tumor Cells Predict Occult Metastatic Disease and Prognosis in Pancreatic Cancer. Ann. Surg. Oncol. 2018, 25, 1000–1008. [Google Scholar] [CrossRef]
- Franses, J.W.; Philipp, J.; Missios, P.; Bhan, I.; Liu, A.; Yashaswini, C.; Tai, E.; Zhu, H.; Ligorio, M.; Nicholson, B.; et al. Pancreatic circulating tumor cell profiling identifies LIN28B as a metastasis driver and drug target. Nat. Commun. 2020, 11, 3303. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Metais, P. Nuclear Acids in Human Blood Plasma. Comptes Rendus Seances Soc. Biol. Ses Fil. 1948, 142, 241–243. [Google Scholar]
- Lo, Y.M.; Lau, T.K.; Zhang, J.; Leung, T.N.; Chang, A.M.; Hjelm, N.M.; Elmes, R.S.; Bianchi, D.W. Increased fetal DNA concentrations in the plasma of pregnant women carrying fetuses with trisomy 21. Clin. Chem. 1999, 45, 1747–1751. [Google Scholar] [CrossRef] [PubMed]
- Grumaz, S.; Stevens, P.; Grumaz, C.; Decker, S.O.; Weigand, M.A.; Hofer, S.; Brenner, T.; von Haeseler, A.; Sohn, K. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, Y.; Mouliere, F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA. Cancer Cell 2019, 36, 350–368. [Google Scholar] [CrossRef]
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. 2018, 93, 1649–1683. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of Double-stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and p53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef]
- Nagata, S. Apoptotic DNA fragmentation. Exp. Cell Res. 2000, 256, 12–18. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2007, 1775, 181–232. [Google Scholar] [CrossRef]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Lyberopoulou, A.; Aravantinos, G.; Efstathopoulos, E.P.; Nikiteas, N.; Bouziotis, P.; Isaakidou, A.; Papalois, A.; Marinos, E.; Gazouli, M. Mutational Analysis of Circulating Tumor Cells from Colorectal Cancer Patients and Correlation with Primary Tumor Tissue. PLoS ONE 2015, 10, e0123902. [Google Scholar] [CrossRef] [PubMed]
- Kinugasa, H.; Nouso, K.; Miyahara, K.; Morimoto, Y.; Dohi, C.; Tsutsumi, K.; Kato, H.; Okada, H.; Yamamoto, K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015, 121, 2271–2280. [Google Scholar] [CrossRef]
- Lebofsky, R.; Decraene, C.; Bernard, V.; Kamal, M.; Blin, A.; Leroy, Q.; Frio, T.R.; Pierron, G.; Callens, C.; Bieche, I.; et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol. Oncol. 2014, 9, 783–790. [Google Scholar] [CrossRef]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Riva, F.; Dronov, O.I.; Khomenko, D.I.; Huguet, F.; Louvet, C.; Mariani, P.; Stern, M.-H.; Lantz, O.; Proudhon, C.; Pierga, J.-Y.; et al. Clinical applications of circulating tumor DNA and circulating tumor cells in pancreatic cancer. Mol. Oncol. 2016, 10, 481–493. [Google Scholar] [CrossRef]
- Iqbal, M.; Kasi, P.M.; Mody, K. Feasibility and clinical value of circulating tumor DNA testing in patients with gastroesophageal adenocarcinoma. J. Clin. Oncol. 2018, 36, e16045. [Google Scholar] [CrossRef]
- Moran, S.; Martínez-Cardús, A.; Sayols, S.; Musulén, E.; Balañá, C.; Estival-Gonzalez, A.; Moutinho, C.; Heyn, H.; Diaz-Lagares, A.; de Moura, M.C.; et al. Epigenetic profiling to classify cancer of unknown primary: A multicentre, retrospective analysis. Lancet Oncol. 2016, 17, 1386–1395. [Google Scholar] [CrossRef]
- Tham, C.; Chew, M.; Soong, R.; Lim, J.; Ang, M.; Tang, C.; Zhao, Y.; Ong, S.Y.K.; Liu, Y. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer 2014, 120, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Siveke, J.T.; Lueong, S.; Hinke, A.; Herbst, A.; Tannapfel, A.; Reinacher-Schick, A.C.; Kolligs, F.T.; Hegewisch-Becker, S. Serial analysis of mutant KRAS in circulation cell-free DNA (cfDNA) of patients with KRAS mutant metastatic colorectal cancer: A translational study of the KRK0207 trial. J. Clin. Oncol. 2018, 36, e15599. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224. [Google Scholar] [CrossRef]
- Liu, M.; Klein, E.; Hubbell, E.; Maddala, T.; Aravanis, A.; Beausang, J.; Filippova, D.; Gross, S.; Jamshidi, A.; Kurtzman, K.; et al. Plasma cell-free DNA (cfDNA) assays for early multi-cancer detection: The circulating cell-free genome atlas (CCGA) study. Ann. Oncol. 2018, 29, viii14. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Payne, S.R.; Church, T.R.; Wandell, M.; Rösch, T.; Osborn, N.; Snover, D.; Day, R.W.; Ransohoff, D.F.; Rex, D.K. Endoscopic Detection of Proximal Serrated Lesions and Pathologic Identification of Sessile Serrated Adenomas/Polyps Vary on the Basis of Center. Clin. Gastroenterol. Hepatol. 2013, 12, 1119–1126. [Google Scholar] [CrossRef]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castaños-Vélez, E.; Blumenstein, B.A.; Rösch, T.; Osborn, N.; et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2013, 63, 317–325. [Google Scholar] [CrossRef]
- Taryma-Leśniak, O.; Sokolowska, K.E.; Wojdacz, T.K. Current status of development of methylation biomarkers for in vitro diagnostic IVD applications. Clin. Epigenet. 2020, 12, 100. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef]
- Taniguchi, H.; Nakamura, Y.; Kotani, D.; Yukami, H.; Mishima, S.; Sawada, K.; Shirasu, H.; Ebi, H.; Yamanaka, T.; Aleshin, A.; et al. CIRCULATE-Japan: Circulating tumor DNA–guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021, 112, 2915–2920. [Google Scholar] [CrossRef]
- Folprecht, G.; Reinacher-Schick, A.; Tannapfel, A.; Weitz, J.; Kossler, T.; Weiss, L.; Aust, D.E.; Von Bubnoff, N.; Kramer, M.; Thiede, C. Circulating tumor DNA-based decision for adjuvant treatment in colon cancer stage II evaluation: (CIRCULATE-trial) AIO-KRK-0217. J. Clin. Oncol. 2020, 38, TPS273. [Google Scholar] [CrossRef]
- Taïeb, J.; Benhaim, L.; Puig, P.L.; Le Malicot, K.; Emile, J.F.; Geillon, F.; Tougeron, D.; Manfredi, S.; Chauvenet, M.; Taly, V.; et al. Decision for adjuvant treatment in stage II colon cancer based on circulating tumor DNA: The CIRCULATE-PRODIGE 70 trial? Dig. Liver Dis. 2020, 52, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Nors, J.; Henriksen, T.V.; Gotschalck, K.A.; Juul, T.; Søgaard, J.; Iversen, L.H.; Andersen, C.L. IMPROVE-IT2: Implementing noninvasive circulating tumor DNA analysis to optimize the operative and postoperative treatment for patients with colorectal cancer—Intervention trial 2. Study protocol. Acta Oncol. 2020, 59, 336–341. [Google Scholar] [CrossRef]
- Tjensvoll, K.; Lapin, M.; Buhl, T.; Oltedal, S.; Berry, K.S.-O.; Gilje, B.; Søreide, J.A.; Javle, M.; Nordgård, O.; Smaaland, R. Abstract 5241: Clinical relevance of circulating tumor DNA in plasma from pancreatic cancer patients. Cancer Res. 2015, 75, 5241. [Google Scholar] [CrossRef]
- Liu, X.; Guo, S.; Ma, C.; Li, Y.; Liu, X.; Zhang, Z.; Yu, L.; Dai, M.; Shen, S.; Wu, H.M.; et al. Circulating tumor DNA methylation as markers for early detection of pancreatic ductal adenocarcinoma (PDAC). J. Clin. Oncol. 2021, 39, 395. [Google Scholar] [CrossRef]
- Yang, J.; Gong, Y.; Lam, V.K.; Shi, Y.; Guan, Y.; Zhang, Y.; Ji, L.; Chen, Y.; Zhao, Y.; Qian, F.; et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020, 11, 346. [Google Scholar] [CrossRef]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.-M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef]
- Willis, J.; Lefterova, M.I.; Artyomenko, A.; Kasi, P.M.; Nakamura, Y.; Mody, K.; Catenacci, D.V.; Fakih, M.; Barbacioru, C.; Zhao, J.; et al. Validation of Microsatellite Instability Detection Using a Comprehensive Plasma-Based Genotyping Panel. Clin. Cancer Res. 2019, 25, 7035–7045. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.-T.; Chen, M.-H.; Fang, W.-L.; Hsieh, C.-C.; Lin, C.-H.; Jhang, F.-Y.; Yang, S.-H.; Lin, J.-K.; Chen, W.-S.; Jiang, J.-K.; et al. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget 2016, 8, 3009–3017. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Ju, S.; Qi, J.; Zhao, J.; Shen, X.; Jing, R.; Yu, J.; Li, L.; Shi, Y.; Zhang, L.; et al. Alu-based cell-free DNA: A novel biomarker for screening of gastric cancer. Oncotarget 2016, 8, 54037–54045. [Google Scholar] [CrossRef]
- Kim, K.; Shin, D.G.; Park, M.K.; Baik, S.H.; Kim, T.H.; Kim, S.; Lee, S. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: Diagnostic validity and significant reduction of cfDNA after surgical resection. Ann. Surg. Treat. Res. 2014, 86, 136–142. [Google Scholar] [CrossRef]
- Eissa, M.A.L.; Lerner, L.; Abdelfatah, E.; Shankar, N.; Canner, J.K.; Hasan, N.M.; Yaghoobi, V.; Huang, B.; Kerner, Z.; Takaesu, F.; et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin. Epigenet. 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Bernard, V.; Kim, D.U.; Lucas, F.A.S.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated with Outcomes of Patients with Pancreatic Cancer. Gastroenterology 2019, 156, 108–118.e4. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D.; et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2018, 68, 663–671. [Google Scholar] [CrossRef]
- Uesato, Y.; Sasahira, N.; Ozaka, M.; Sasaki, T.; Takatsuki, M.; Zembutsu, H. Evaluation of circulating tumor DNA as a biomarker in pancreatic cancer with liver metastasis. PLoS ONE 2020, 15, e0235623. [Google Scholar] [CrossRef]
- Pietrasz, D.; Pécuchet, N.; Garlan, F.; Didelot, A.; Dubreuil, O.; Doat, S.; Imbert-Bismut, F.; Karoui, M.; Vaillant, J.-C.; Taly, V.; et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin. Cancer Res. 2017, 23, 116–123. [Google Scholar] [CrossRef]
- Patel, H.; Okamura, R.; Fanta, P.; Patel, C.; Lanman, R.B.; Raymond, V.M.; Kato, S.; Kurzrock, R. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J. Hematol. Oncol. 2019, 12, 130. [Google Scholar] [CrossRef]
- Takai, E.; Totoki, Y.; Nakamura, H.; Morizane, C.; Nara, S.; Hama, N.; Suzuki, M.; Furukawa, E.; Kato, M.; Hayashi, H.; et al. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci. Rep. 2015, 5, 18425. [Google Scholar] [CrossRef]
- Chen, I.; Raymond, V.M.; Geis, J.A.; Collisson, E.A.; Jensen, B.V.; Hermann, K.L.; Erlander, M.G.; Tempero, M.; Johansen, J.S. Ultrasensitive plasma ctDNA KRAS assay for detection, prognosis, and assessment of therapeutic response in patients with unresectable pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 97769–97786. [Google Scholar] [CrossRef]
- Botrus, G.; Kosirorek, H.; Sonbol, M.B.; Kusne, Y.; Uson, P.L.S.; Borad, M.J.; Ahn, D.H.; Kasi, P.M.; Drusbosky, L.M.; Dada, H.; et al. Circulating Tumor DNA-Based Testing and Actionable Findings in Patients with Advanced and Metastatic Pancreatic Adenocarcinoma. Oncol. 2021, 26, 569–578. [Google Scholar] [CrossRef]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Kay Li, Q.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lipton, L.; Cohen, J.; Tie, J.; Javed, A.; Li, L.; Goldstein, D.; Burge, M.; Cooray, P.; Nagrial, A.; et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann. Oncol. 2019, 30, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Théry, C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 15. [Google Scholar] [CrossRef]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- Rak, J. Microparticles in Cancer. Semin. Thromb. Hemost. 2010, 36, 888–906. [Google Scholar] [CrossRef]
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes Released by Melanoma Cells Prepare Sentinel Lymph Nodes for Tumor Metastasis. Cancer Res. 2011, 71, 3792–3801. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, T.; Zheng, M.; Liu, Y.; Chen, Z. Exosomal proteins as potential markers of tumor diagnosis. J. Hematol. Oncol. 2017, 10, 175. [Google Scholar] [CrossRef]
- Liangsupree, T.; Multia, E.; Riekkola, M.-L. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A 2020, 1636, 461773. [Google Scholar] [CrossRef]
- Cai, X.; Janku, F.; Zhan, Q.; Fan, J.-B. Accessing Genetic Information with Liquid Biopsies. Trends Genet. 2015, 31, 564–575. [Google Scholar] [CrossRef]
- Xu, R.; Greening, D.W.; Zhu, H.-J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Che, X.; Qu, J.; Hou, K.; Wen, T.; Li, Z.; Li, C.; Wang, S.; Xu, L.; Liu, Y.; et al. Exosomal PD-L1 Retains Immunosuppressive Activity and is Associated with Gastric Cancer Prognosis. Ann. Surg. Oncol. 2019, 26, 3745–3755. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Hernandez, J.M.; Doussot, A.; Bojmar, L.; Zambirinis, C.P.; Costa-Silva, B.; Van Beek, E.J.A.H.; Mark, M.T.; Molina, H.; Askan, G.; et al. Extracellular matrix proteins and carcinoembryonic antigen-related cell adhesion molecules characterize pancreatic duct fluid exosomes in patients with pancreatic cancer. HPB 2018, 20, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Liu, P.; Wu, Y.; Meng, X.; Wu, M.; Han, J.; Tan, X. Exosomal zinc transporter ZIP4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer. Cancer Sci. 2018, 109, 2946–2956. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Lin, K.; Baenke, F.; Lai, X.; Schneider, M.; Helm, D.; Polster, H.; Rao, V.S.; Ganig, N.; Wong, F.C.; Seifert, L.; et al. Comprehensive proteomic profiling of serum extracellular vesicles in patients with colorectal liver metastases identifies a signature for non-invasive risk stratification and early-response evaluation. Mol. Cancer 2022, 21, 91. [Google Scholar] [CrossRef]
- Tao, L.; Zhou, J.; Yuan, C.; Zhang, L.; Li, D.; Si, D.; Xiu, D.; Zhong, L. Metabolomics identifies serum and exosomes metabolite markers of pancreatic cancer. Metabolomics 2019, 15, 86. [Google Scholar] [CrossRef]
- Kitagawa, T.; Taniuchi, K.; Tsuboi, M.; Sakaguchi, M.; Kohsaki, T.; Okabayashi, T.; Saibara, T. Circulating pancreatic cancer exosomal RNA s for detection of pancreatic cancer. Mol. Oncol. 2018, 13, 212–227. [Google Scholar] [CrossRef]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef]
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S.; et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1627–1636. [Google Scholar] [CrossRef]
- Batista, I.A.; Melo, S.A. Exosomes and the Future of Immunotherapy in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 567. [Google Scholar] [CrossRef]
- Tang, G.; Wang, J.; Dong, W.; Dai, K.; Du, J. Exosomal miRNA Expression Profiling and the Roles of Exosomal miR-4741, miR-32, miR-3149, and miR-6727 on Gastric Cancer Progression. BioMed Res. Int. 2022, 2022, 1263812. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Shen, W.; Zou, H.; Lv, Q.; Shao, P. Circulating exosomal long non-coding RNA H19 as a potential novel diagnostic and prognostic biomarker for gastric cancer. J. Int. Med. Res. 2020, 48, 300060520934297. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Gao, H.; Xie, X.; Lu, P. Plasma Exosomal hsa_circ_0015286 as a Potential Diagnostic and Prognostic Biomarker for Gastric Cancer. Pathol. Oncol. Res. 2022, 28, 1610446. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Yang, H.; Zhang, X.; Wang, B.; Mao, J.; Li, X.; Wang, M.; Zhang, B.; Sun, Z.; Qian, H.; et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J. Exp. Clin. Cancer Res. 2018, 37, 162. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Geng, B.; Xu, Y.; Miao, X.; Chen, L.; Mu, X.; Pan, J.; Zhang, C.; Zhao, T.; Wang, C.; et al. Helicobacter pylori -induced exosomal MET educates tumour-associated macrophages to promote gastric cancer progression. J. Cell. Mol. Med. 2018, 22, 5708–5719. [Google Scholar] [CrossRef]
- Ostenfeld, M.S.; Jensen, S.G.; Jeppesen, D.K.; Christensen, L.L.; Thorsen, S.B.; Stenvang, J.; Nielsen, H.J.; Thomsen, A.; Mouritzen, P.; Rasmussen, M.H.; et al. miRNA profiling of circulating EpCAM+ extracellular vesicles: Promising biomarkers of colorectal cancer. J. Extracell Vesicles 2016, 5, 31488. [Google Scholar] [CrossRef]
- Julich-Haertel, H.; Urban, S.K.; Krawczyk, M.; Willms, A.; Jankowski, K.; Patkowski, W.; Kruk, B.; Krasnodębski, M.; Ligocka, J.; Schwab, R.; et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 2017, 67, 282–292. [Google Scholar] [CrossRef]
- Yan, L.; Dong, X.; Gao, J.; Liu, F.; Zhou, L.; Sun, Y.; Zhao, X. A novel rapid quantitative method reveals stathmin-1 as a promising marker for esophageal squamous cell carcinoma. Cancer Med. 2018, 7, 1802–1813. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, L.; Gong, M.; Su, G.; Zhu, S.; Zhang, W.; Wang, S.; Li, Z.; Chen, C.; Li, L.; et al. Protein Profiling and Sizing of Extracellular Vesicles from Colorectal Cancer Patients via Flow Cytometry. ACS Nano 2018, 12, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Bavisotto, C.C.; Cappello, F.; Macario, A.J.; de Macario, E.C.; Logozzi, M.; Fais, S.; Campanella, C. Exosomal HSP60: A potentially useful biomarker for diagnosis, assessing prognosis, and monitoring response to treatment. Expert Rev. Mol. Diagn. 2017, 17, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, Y.; Guo, X.; Zhou, L.; Jia, Z.; Peng, Z.; Tang, Y.; Liu, W.; Zhu, B.; Wang, L.; et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J. Cell. Mol. Med. 2017, 21, 838–847. [Google Scholar] [CrossRef]
- Sun, B.; Li, Y.; Zhou, Y.; Ng, T.K.; Zhao, C.; Gan, Q.; Gu, X.; Xiang, J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J. Cell. Physiol. 2018, 234, 1416–1425. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.T.; Corken, A.; Ware, J. Platelets at the interface of thrombosis, inflammation, and cancer. Blood 2015, 126, 582–588. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Menter, D.G.; Kopetz, S.; Hawk, E.; Sood, A.K.; Loree, J.M.; Gresele, P.; Honn, K.V. Platelet "first responders" in wound response, cancer, and metastasis. Cancer Metastasis Rev. 2017, 36, 199–213. [Google Scholar] [CrossRef]
- Naderi-Meshkin, H.; Ahmadiankia, N. Cancer metastasis versus stem cell homing: Role of platelets. J. Cell. Physiol. 2018, 233, 9167–9178. [Google Scholar] [CrossRef]
- Haemmerle, M.; Stone, R.L.; Menter, D.G.; Afshar-Kharghan, V.; Sood, A.K. The Platelet Lifeline to Cancer: Challenges and Opportunities. Cancer Cell 2018, 33, 965–983. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.; Cedervall, J. The pro-inflammatory role of platelets in cancer. Platelets 2018, 29, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.R.; Yousef, G.M.; Ni, H. Cancer and platelet crosstalk: Opportunities and challenges for aspirin and other antiplatelet agents. Blood 2018, 131, 1777–1789. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, H.; Zhu, Q.; Zhan, P.; Zhu, S.; Zhang, J.; Lv, T.; Yong, S. Patterns and functional implications of platelets upon tumor "education". Int. J. Biochem. Cell Biol. 2017, 90, 68–80. [Google Scholar] [CrossRef]
- Mancuso, M.E.; Santagostino, E. Platelets: Much more than bricks in a breached wall. Br. J. Haematol. 2017, 178, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Hisada, Y.; Mackman, N. Cancer-associated pathways and biomarkers of venous thrombosis. Blood 2017, 130, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Kanikarla-Marie, P.; Lam, M.; Menter, D.G.; Kopetz, S. Platelets, circulating tumor cells, and the circulome. Cancer Metastasis Rev. 2017, 36, 235–248. [Google Scholar] [CrossRef]
- Leblanc, R.; Peyruchaud, O. Metastasis: New functional implications of platelets and megakaryocytes. Blood 2016, 128, 24–31. [Google Scholar] [CrossRef]
- Kuznetsov, H.S.; Marsh, T.; Markens, B.A.; Castaño, Z.; Greene-Colozzi, A.; Hay, S.A.; Brown, V.E.; Richardson, A.L.; Signoretti, S.; Battinelli, E.M.; et al. Identification of Luminal Breast Cancers That Establish a Tumor-Supportive Macroenvironment Defined by Proangiogenic Platelets and Bone Marrow–Derived Cells. Cancer Discov. 2012, 2, 1150–1165. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; Kooi, I.E.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef]
- Tjon-Kon-Fat, L.-A.; Lundholm, M.; Schröder, M.; Wurdinger, T.; Thellenberg-Karlsson, C.; Widmark, A.; Wikström, P.; Nilsson, R.J.A. Platelets harbor prostate cancer biomarkers and the ability to predict therapeutic response to abiraterone in castration resistant patients. Prostate 2017, 78, 48–53. [Google Scholar] [CrossRef]
- Xiao, R.; Liu, C.; Zhang, B.; Ma, L. Tumor-Educated Platelets as a Promising Biomarker for Blood-Based Detection of Renal Cell Carcinoma. Front. Oncol. 2022, 12, 844520. [Google Scholar] [CrossRef]
- Best, M.G.; Veld, S.G.J.G.I.; Sol, N.; Wurdinger, T. RNA sequencing and swarm intelligence–enhanced classification algorithm development for blood-based disease diagnostics using spliced blood platelet RNA. Nat. Protoc. 2019, 14, 1206–1234. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jiang, Q.; Li, D.-Z.; Zhou, X.; Yu, D.-S.; Zhong, J. TIMP1 mRNA in tumor-educated platelets is diagnostic biomarker for colorectal cancer. Aging 2019, 11, 8998–9012. [Google Scholar] [CrossRef]
- Asghar, S.; Waqar, W.; Umar, M.; Manzoor, S. Tumor educated platelets, a promising source for early detection of hepatocellular carcinoma: Liquid biopsy an alternative approach to tissue biopsy. Clin. Res. Hepatol. Gastroenterol. 2020, 44, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Tsujimoto, H.; Sugasawa, H.; Kouzu, K.; Itazaki, Y.; Sugihara, T.; Harada, M.; Ito, N.; Kishi, Y.; Ueno, H. Prognostic value of platelet-related measures for overall survival in esophageal squamous cell carcinoma: A systematic review and meta-analysis. Crit. Rev. Oncol. 2021, 164, 103427. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zheng, X.; Wu, M.; Zhang, F.; Xu, S.; Wang, X.; Song, M.; You, C.; Zhang, T.; Jiang, M.; et al. Development and validation of postoperative and preoperative platelets ratio (PPR) to predict the prognosis of patients undergoing surgery for colorectal cancer: A dual-center retrospective cohort study. Cancer Med. 2022, 12, 111–121. [Google Scholar] [CrossRef]
- Zhang, L.-X.; Wei, Z.-J.; Xu, A.-M.; Zang, J.H. Can the neutrophil-lymphocyte ratio and platelet-lymphocyte ratio be beneficial in predicting lymph node metastasis and promising prognostic markers of gastric cancer patients? Tumor maker retrospective study. Int. J. Surg. 2018, 56, 320–327. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Sabrkhany, S.; Kuijpers, M.J.; van Kuijk, S.M.; Sanders, L.; Pineda, S.; Damink, S.O.; Dingemans, A.-M.C.; Griffioen, A.W.; Egbrink, M.G.O. A combination of platelet features allows detection of early-stage cancer. Eur. J. Cancer 2017, 80, 5–13. [Google Scholar] [CrossRef]
- Sabrkhany, S.; Kuijpers, M.J.E.; Knol, J.C.; Olde Damink, S.W.M.; Dingemans, A.C.; Verheul, H.M.; Piersma, S.R.; Pham, T.V.; Griffioen, A.W.; Oude Egbrink, M.G.A.; et al. Exploration of the platelet proteome in patients with early-stage cancer. J. Proteom. 2018, 177, 65–74. [Google Scholar] [CrossRef]
- Peterson, J.E.; Zurakowski, D.; Italiano, J.E., Jr.; Michel, L.V.; Connors, S.; Oenick, M.; D’Amato, R.J.; Klement, G.L.; Folkman, J. VEGF, PF4 and PDGF are elevated in platelets of colorectal cancer patients. Angiogenesis 2012, 15, 265–273. [Google Scholar] [CrossRef]
| Cancer Type | Threshold | Sample Size (Number) | Sensitivity | Specificity | AUC | Clinical Significance | References |
|---|---|---|---|---|---|---|---|
| Gastric cancer | 2 CTCs | 116 | 85.3 | 90.3 | 0.928 | Distinguish between GC patients and healthy controls and provide clinical output | [50] |
| Gastric cancer | CTC-PD-L1 | 32 with progressive GCs | Monitor prognosis and guide future individualized immunotherapy | [51] | |||
| Gastric cancer | CSV+PD-L1+CTCs | 70 | 71 | Predicts treatment response and prognosis in GC patients | [52] | ||
| Gastric cancer | CTCs and TWIST | 32 with metastatic cancer | 80.6 | As a prognostic marker | [53] | ||
| Gastric cancer | CTCs/cfDNA | 45 patients with progressive GC | 95.6 | Predicting the efficacy and prognosis of neoadjuvant chemotherapy for progressive GC | [54] | ||
| Colorectal cancer | ≥3 (chemotherapy and serum CEA) | 121 | Presence of CTCs might be valuable for predicting survival outcome | [55] | |||
| Non-metastatic colorectal cancer (NMCRC) | ≥4 (CS. CK19, MUC1, CD44, CD133 and ALDH1) | 63 | 68.3 | 95 | CTCs could be novel therapeutic targets for NMCRC | [56] | |
| Metastatic colorectal cancer (mCRC) | ≥1.92 (CEACAM) | 436 | Detection of peripheral blood CEACAM5 mRNA-positive CTCs as an adverse prognostic factor correlated with poor clinical outcome in patients with mCRC | [57] | |||
| Duke’s stage B and C colorectal cancer | (carcinoembryonic antigen CEA), cytokeratin (CK) 19, CK20, and/or CD133 (CEA/CK/CD133) | 735 | CTC as a detection marker in patients with Duke’s stage B and C | [58] | |||
| Advanced CRC | ≥3 CTCs (EpCAM, CK and CD45) | 467 | CTC count before and during chemotherapy treatment as an independent predictor of PFS and OS in advanced CRC patients | [59] | |||
| Colorectal cancer | ≥3 CTCs (EpCAM, CK and CD45) | 430 | CTC count before and during chemotherapy treatment as an independent predictor of PFS and OS during metastatic CRC patients | [60] | |||
| Hepatocellular carconoma | ≥5 CTCs EpCAM and mucin1 | 73 | CTCs could possibly be a novel prognostic biomarker in HCC | [61] | |||
| Hepatocellular carconoma | ≥2 CTCs, EpCAM, CD8/18/19 | 964 | The CellSearch system could determine the clinical utility of CTCs in HCC | [25] | |||
| Hepatocellular carconoma | ≥5 CTCs, Glypican-3 | 85 | GPC-3 as a useful biomarker for HCC patient outcomes | [62] | |||
| Hepatocellular carconoma | EpCAM | 299 | CTC detection by qPCR could be utilized in clinics for auxiliary diagnosis, treatment response assessment, and decision making | [63] | |||
| Hepatocellular carconoma | ASGPR, Hep Par 1 | 85 | A highly sensitive and specific CTC detection tool | [64] | |||
| Hepatocellular carconoma | (EpCAM)/vimentin/Glypican-3 (GPC3) | 44 | 96.94 | 98.12 | A convenient and feasible CTC capture system to predict clinical outcomes in HCC patients | [65] | |
| Hepatocellular carconoma | PD-L1 | 87 | 71.1 | 91.8 | Favorable response to anti-PD-1 therapy is associated with the presence of PD-L1+ CTCs | [66] | |
| Hepatocellular carconoma | pERK+/pAkt− CTCs | 109 | pERK+/pAkt− CTCs are sensible to sorafenib | [67] | |||
| Pancreatic cancer | ISET | 165 | Higher CTC counts correlate with earlier recurrence. Increase in CTC numbers after neoadjuvant treatment. CTC+ correlates with early recurrence and OS in the pretreated group. | [68] | |||
| Pancreatic cancer | CK20 | 172 | CTC predicts poor OS | [69] | |||
| Pancreatic cancer | CK20 | 25 | 88 | 90 | CTCs predict the prognosis of pancreatic cancer | [70] | |
| Pancreatic cancer | NanoVelcro CTC assay (CK) | 100 | CTC as a promising prognostic biomarker for PDAC patients | [71] | |||
| Pancreatic cancer | LIN28B | 35 | Molecular characterization of CTCs provides a unique opportunity to correlate gene set metastatic profiles, identify drivers of dissemination, and develop therapies targeting the “seeds” of metastasis | [72] |
| Cancer Type | Sample Size | Sensitivity (%) | Specificity (%) | AUC | Clinical Significance | References |
|---|---|---|---|---|---|---|
| Gastric cancer | 46 patients with stage I–III GC | 39 | 100 | MRD with ctDNA testing identifies patients at high risk of recurrence | [106] | |
| Gastric cancer | 61 cases of partially metastatic GC | Associated with improved prognosis | [107] | |||
| Gastric cancer | 1145 | 87 | 0.984 | Potential to expand access to targetted therapies and immunotherapy to all patients with advanced cancer | [108] | |
| Gastric cancer | 428 | 68.9 | 95.8 | 0.98 | Predicts response to chemotherapy and surgery in patients with CRC; tumor recurrence should be considered in GC with persistently elevated cfDNAs levels after surgery | [109] |
| Gastric cancer | 124 | 78.96 | 91.81 | 0.94 | For early screening of GC | [110] |
| Gastric cancer | 30 | 96.67 | 94.11 | 0.991 | For early detection of cancer and assessment of tumor load | [111] |
| Pancreatic cancer | 39 | 97.3 | 91.6 | Minimal invasive blood-based biomarker panel which could potentially be used as a diagnostic and screening tool in a select subset of high-risk populations | [112] | |
| Pancreatic cancer | 194 | ctDNA in combination with exosomal DNA provides both predictive and prognostic information relevant to therapeutic stratification | [113] | |||
| Colorectal cancer | 455 | Reduced the usage of adjuvant chemotherapy | [99] | |||
| Colorectal cancer | 250 | Detection of residual disease | [114] | |||
| Locally advanced rectal cancer (LARC) | 462 | ctDNA analysis as a useful guide for adjuvant chemotherapy selection in LARC patients | [115] | |||
| Pancreatic cancer with liver metastasis | 104 | Use of circulating tumor DNA as an independent prognostic marker for advanced pancreatic cancer | [116] | |||
| Pancreatic cancer | 135 | ctDNA as an independent prognostic marker in advanced PDAC as well as an indicator of shorter disease-free survival in resected patients when detected after surgery | [117] | |||
| Pancreatic cancer | 112 | Increased ctDNA levels were a poor prognostic factor for survival. | [118] | |||
| Pancreatic cancer | 259 | Plasma cfDNA might provide a prognostic and diagnostic tool to assist surgical decision-making in PDAC patients | [119] | |||
| Pancreatic cancer | 189 | Longitudinal ctDNA KRAS assists in therapeutical decision-making and provides a kinetically robust and quantitative measurement of patient response. | [120] | |||
| Pancreatic cancer | 171 | 86 | 88 | ctDNA methylation approach to discriminate PDAC plasma from non-malignant diseases | [105,121] | |
| Pancreatic cancer | 101 | ctDNA as genetic predictors of result in pancreatic cancer and might open new avenues of therapeutic intervention. | [122] | |||
| Pancreatic cancer | 112 | ctDNA-guided approach intensified the treatment strategies for pancreatic cancer patients. | [123] |
| Cancer Type | Biomarkers | Sample Type | Expression | Clinical Significance | References |
|---|---|---|---|---|---|
| Gastric cancer | miRNA-4741, miR-32, miR-3149 and miR-6727 | tissue and plasma | miR-4741—upregulated miR-32, miR-3149 and miR-6727—downregulated | Acts as a diagnostic marker for GC and an influential factor in inhibiting GC progression | [144] |
| Gastric cancer | LncRNAH19GC | serum | downregulated | Possible biomarkers with diagnostic and prognostic value | [145] |
| Gastric cancer | hsa_circ_00115286 | tissue, plasma, and cells | upregulated | Possibly a non-invasive biomarker for GC diagnosis and prognostic assessment | [146] |
| Gastric cancer | TRIM3 | serum | downregulated | Inhibition of GC progression in vitro and in vivo | [147] |
| Gastric cancer | MET | cells | upregulated | Amplifies tumor growth and development in vitro and in vivo | [148] |
| Colorectal cancer | Exo-EpCAM | plasma | upregulated | May have potential as non-invasive biomarkers for detection of CRC | [149] |
| HCC/Colongiocarcinoma | EpCAM | serum | upregulated | A novel non-invasive biomarker to assess the presence and possible extent of cancers in patients with advanced liver disease | [150] |
| Esophageal cancer | Stathmin | serum | upregulated | A very promising diagnostic and predictive marker for SCC in the clinic, especially for ESCC | [151] |
| Colorectal cancer | CD147 | blood | upregulated | EV-mediated intercellular communication and the development of advanced diagnostic and therapeutic strategies | [152] |
| Colorectal liver metastasis (CRLM) | CXCL17 | serum | downregulated | EV-bound CXCL7 was found as a biomarker of early response in CRLM patients receiving systemic chemotherapy | [138] |
| HCC/Colongiocarcinoma | CD147 | serum | upregulated | A novel non-invasive biomarker to assess the presence and possible extent of cancers in patients with advanced liver disease | [150] |
| Colorectal cancer | Hsp60 | cells | upregulated | Biomarker for diagnostics, assessing prognosis, and monitoring disease progression and response to treatment, particularly in cancer | [153] |
| Colorectal cancer | Glypican-1 (GPC1) | plasma | upregulated | Specific markers for the diagnosis of CRC and targets for the therapy of CRC. | [154] |
| Colorectal cancer | CopineIII (CPNE3) | plasma | upregulated | Exosomal CPNE3 show potential implications in CRC diagnosis and prognosis. | [155] |
| Pancreatic cancer | CEACAMs | pancreatic fluid | upregulated | Exosome isolation is feasible from pancreatic duct fluid, and that exosomal proteins may be utilized to diagnose patients with PDAC. | [135] |
| Pancreatic cancer | Tenascin C | pancreatic fluid | upregulated | Exosome isolation is feasible from pancreatic duct fluid, and that exosomal proteins may be utilized to diagnose patients with PDAC. | [135] |
| Pancreatic cancer | Glypcan-1 (GCP-1) | serum | upregulated | GPC1+ crExos may serve as a potential non-invasive diagnostic and screening tool to detect early stages of pancreatic cancer to facilitate possible curative surgical therapy. | [137] |
| Pancreatic cancer | ZIP-4 | cell line | upregulated | Exosomal ZIP4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer | [136] |
| Pancreatic cancer | DNA MAFs | plasma | upregulated | Exosomal DNA in combination with ctDNA provides both predictive and prognostic information relevant to therapeutic stratification | [113] |
| HCC/Colongiocarcinoma | Annexin V | serum | upregulated | A novel non-invasive biomarker to assess the presence and possible extent of cancers in patients with advanced liver disease | [150] |
| Cancer Type | Sample Size (Number) | Sensitivity | Specificity | AUC | Clinical Significance | References |
|---|---|---|---|---|---|---|
| Gastric cancer | 904 | NLR is better to predict overall survival than PLR in gastric cancer patients | [177] | |||
| Stage I to III liver, stomach, pancreas, and esophagus | 1005 | 69–98% | 99% | CancerSEEK localized cancer to a small number of anatomic sites in a median of 83% of the patients | [178] | |
| Pancreatic cancer | 42 | 82.70% | Discriminate between patients with early-stage cancer and healthy individuals | [179] | ||
| Pancreatic cancer | 4 | Platelet proteome can be mined for potential biomarkers of cancer. | [180] | |||
| Pan cancer (colorectal cancer, pancreatic cancer, hepatobiliary cancer) | 90 | 81%, 71%, 58% | 0.996, 0.999, 1.00 | Provides a valuable platform that could potentially enable clinical advances in blood-based liquid biopsies | [169] | |
| Liver cancer | 127 | 96 | Provides a valuable platform that could potentially enable clinical advances in blood-based liquid biopsies | [169] | ||
| Colorectal cancer | 35 | 0.893 | Differences between cancer and control samples in this study, although statistically significant, were not clinically significant | [181] |
| Liquid Biopsy | Status | Cancer | Study Title | Study Type | Clinical Trial Identifier | Estimated Enrollment | Conditions | Interventions | Locations | |
|---|---|---|---|---|---|---|---|---|---|---|
| CTC-1 | CTC | Recruiting | Gastric cancer | Detection of CTC in the Diagnosis of Metastasis in Gastric Cancer | Observational | NCT05208372 | 200 | Stomach Neoplasms, Metastasis | Diagnostic test: CTC test | Liaoning, China |
| CTC-2 | CTC | Recruiting | Gastric cancer | Tumor Cell and DNA Detection in the Blood, Urine and Bone Marrow of Patients With Solid Cancers | Observational | NCT02838836 | 120 | Esophageal Cancer, Gastric Cancer, Pancreatic Cancer, Hepatocellular Cancer, Colorectal Cancer | Procedure: study sample collection | Missouri, United States |
| CTC-3 | CTC | Recruiting | Gastric cancer | Tumor Cell and DNA Detection in the Blood, Urine, and Bone Marrow | Observational | NCT03551951 | 320 | Esophageal Cancer, Gastric Cancer, Pancreatic Cancer, Hepatocellular Cancer, Colorectal Cancer | Diagnostic test: test for circulating tumor cells, DNA alterations | Missouri, United States |
| CTC-4 | CTC | Recruiting | Pancreatic cancer | Heat Shock Protein (HSP) 70 to Quantify and Characterize Circulating Tumor Cells (HSP70CTC) | Observational (Patient Registry) | NCT04628806 | 120 | Pancreatic Cancer Stage IV | Diagnostic Test: CTC isolation by HSP70 | Berlin, Germany |
| CTC-5 | CTC | Recruiting | Liver cancer | Prognostic Value of Liver Cancer CTCs Isolated by a Novel Microfluidic Platform | Observational (Patient Registry) | NCT05242237 | 300 | Hepatocellular Carcinoma, Circulating Tumor Cell, Whole Genome Sequencing | Chongqing, China | |
| CTC-6 | CTC | Recruiting | Liver cancer | Clinical Study for Combined Analysis of CTC and Exosomes on Predicting the Efficacy of Immunotherapy in Patients with Hepatocellular Carcinoma | Observational | NCT05575622 | 200 | HCC | Device: CTC PD-L1, exosomal PD-L1, and exosomal LAG-3 detection | Hubei, China |
| CTC-7 | CTC | Recruiting | Liver cancer | The Role of Circulating Tumor Cells As Markers of Advanced Disease and Prognosis In HCC | Observational | NCT04800497 | 200 | Hepatocellular Carcinoma, Recurrent Hepatocellular Carcinoma, Circulating Tumor Cell | Procedure: hepatic resection | 4 locations in Italy |
| CTC-8 | CTC | Recruiting | Colorectal cancer | Sample Collection Study for the CellMax Life Circulating Tumor Cell and Circulating Tumor DNA Platforms for the Early Detection of Colorectal Cancer and Adenomas | Observational | NCT05127096 | 100 | Colorectal Cancer Screening | Diagnostic test: FirstSight blood test | Alabama, California, United States |
| ctDNA-1 | ctDNA | Recruiting | Gastric cancer | Detection of ctDNA in the Diagnosis of Metastasis in Gastric Cancer | Observational | NCT05208372 | 200 | Stomach Neoplasms, Metastasis | Diagnostic test: ctDNA test | Liaoning, China |
| ctDNA-2 | ctDNA | Recruiting | Gastric cancer | ctDNA Screening in Advanced HER2 Positive Gastric Cancer | Observational | NCT04520295 | 100 | HER2-Positive Gastric Cancer | Genetic: ctDNA screening | Shanghai, China |
| ctDNA-3 | ctDNA | Recruiting | Gastric cancer | Monitoring Minimal Residual Disease in Gastric Cancer by Liquid Biopsy Study Description | Observational | NCT05029869 | 100 | Gastric Cancer, ctDNA | Diagnostic test: ctDNA | Ho Chi Minh City, Vietnam |
| ctDNA-4 | ctDNA | Recruiting | Gastric cancer | Potential Clinical Utilities of Circulating Tumor DNA in Advanced HER2 Negative Gastric Cancer | Observational | NCT05513144 | 30 | Gastric Cancer, ctDNA | Jiangsu, China | |
| ctDNA-5 | ctDNA | Recruiting | Gastric cancer | Detection of Plasma Circulating Tumor DNA in Gastric Cancer | Observational | NCT05027347 | 200 | ctDNA, Gastric Cancer | Diagnostic test: plasma circulating tumor DNA | Ho Chi Minh City, Vietnam |
| ctDNA-6 | ctDNA | Recruiting | Gastric cancer | Clinical Utility of Circulating Tumor DNA in Gastro-Esophageal Cancer (CURE) | Observational | NCT04576858 | 1950 | Esophageal Cancer, Gastric Cancer | Diagnostic test: circulating tumor DNA | Copenhagen, Denmark |
| ctDNA-7 | ctDNA | Recruiting | Pancreatic cancer | Observational Study of ctDNA in Resectable and Borderline Resectable Pancreatic Cancer | Observational | NCT05379907 | 30 | Pancreatic Cancer | Other: SIGNATERA™ ctDNA testing | Virginia, United States |
| ctDNA-8 | ctDNA | Recruiting | Pancreatic cancer | ctDNA Assay in Patients with Resectable Pancreatic Cancer | Observational | NCT05052671 | 50 | Pancreas Cancer | Oklahoma, United States | |
| ctDNA-9 | ctDNA | Recruiting | Pancreatic cancer | Liquid Biopsy for ctDNA in Peritoneal Lavage and Blood in Pancreatic Cancer | Observational (Patient Registry) | NCT05400681 | 200 | Pancreatic Cancer, Pancreatic Adenocarcinoma | Odense, Denmark | |
| ctDNA-10 | ctDNA | Recruiting | Pancreatic cancer | Prognostic Role of Circulating Tumor DNA in Resectable Pancreatic Cancer (PROJECTION) | Observational | NCT04246203 | 200 | Pancreatic Cancer | Other: liquid Biopsy | Bavaria, Berlin, Cologne, Germany |
| ctDNA-11 | ctDNA | Recruiting | Pancreatic cancer | DNA Mutation Detection in Circulating Tumor DNA and Tissue by mmADPS for Pancreatic Cancer | Observational (Patient Registry) | NCT05604573 | 150 | Pancreatic Cancer | Diagnostic Test: cell-free DNA in blood, genetic mutation in tissue | Seoul, South Korea |
| ctDNA-12 | ctDNA | Recruiting | Liver cancer | Tumor Cell and DNA Detection in the Blood, Urine and Bone Marrow of Patients with Solid Cancers | Observational | NCT02838836 | 120 | Esophageal Cancer, Gastric Cancer, Pancreatic Cancer, Hepatocellular Cancer, Colorectal Cancer | Procedure: study sample collection | Missouri, United States |
| ctDNA-13 | ctDNA | Recruiting | Liver cancer | Cohort Study of Patients with Hepatocellular Carcinoma and Circulating Tumor DNA Monitoring of Chemoembolization (Mona-Lisa) | Observational | NCT05390112 | 167 | Circulating Tumor DNA Hepatocellular Carcinoma Non-resectable | Biological: DNA | Rouen, France |
| ctDNA-14 | ctDNA | Recruiting | Colorectal cancer | Comparison of Diagnostic Sensitivity Between ctDNA Methylation and CEA in Colorectal Cancer | Observational (Patient Registry) | NCT05558436 | 712 | Colorectal Cancer | Diagnostic test: detection of ctDNA methylation | Guangdong, China |
| ctDNA-15 | ctDNA | Recruiting | Colorectal cancer | Role of Circulating Tumour DNA Testing in Assessing for Alterations of Primary Anti-EGFR Resistance in RAS/RAF Wild-type Metastatic Colorectal Cancer Patients | Observational | NCT05051592 | 40 | Colorectal Cancer | Singapore, Singapore | |
| ctDNA-16 | ctDNA | Recruiting | Colorectal cancer | Circulating Tumor DNA Analysis to Optimize Treatment for Patients with Colorectal Cancer | Observational | NCT03637686 | 1800 | Colorectal Cancer | 10 locations in Denmark | |
| ctDNA-17 | ctDNA | Recruiting | Colorectal cancer | Tracking Mutations in Cell Free Tumour DNA to Predict Relapse in Early Colorectal Cancer (TRACC) | Observational | NCT04050345 | 1000 | Colorectal Cancer | 36 locations in United Kingdom | |
| ctDNA-18 | ctDNA | Recruiting | Colorectal cancer | Circulating Tumour DNA (ctDNA) as a Prognostic and Predictive Marker in Colorectal Cancer—a Pilot Study | Observational | NCT04726800 | 300 | Colorectal Cancer | 8 locations in Sweden and Norway | |
| ctDNA-19 | ctDNA | Recruiting | Colorectal cancer | Sample Collection Study for the CellMax Life Circulating Tumor Cell and Circulating Tumor DNA Platforms for the Early Detection of Colorectal Cancer and Adenomas | Observational | NCT05127096 | 1000 | Colorectal Cancer Screening | Diagnostic test: FirstSight blood test | 15 locations in United States |
| ctDNA-20 | ctDNA | Recruiting | Colorectal cancer | Dynamic Monitoring of ctDNA Methylation to Predict Relapse in Colorectal Cancer after Radical Resection | Observational (Patient Registry) | NCT03737539 | 300 | Colorectal Cancer, ctDNA, Surveillance, Methylation | Diagnostic test: multigene methylation detection | Shanghai, China |
| ctDNA-21 | ctDNA | Recruiting | Colorectal cancer | Epidemiological Study to Monitor Study Participants With Resected Stage II (High Risk) or Stage III Colorectal Cancer for Circulating Tumor DNA before, during and after Their Treatment with Adjuvant Chemotherapy | Observational | NCT04813627 | 1500 | Colorectal Cancer Stage II and III | Procedure: regular blood sample collection for ctDNA assessment | 67 locations in United States |
| ctDNA-22 | ctDNA | Recruiting | Colorectal cancer | BESPOKE Study of ctDNA Guided Immunotherapy | Observational | NCT04761783 | 1539 | Colorectal Cancer | California, United States | |
| exo-1 | Exosomes | Recruiting | Gastric cancer | Use of Circulating Exosomal LncRNA-GC1 to Monitor Gastric Cancer | Observational | NCT05397548 | 700 | Gastric Cancer | Diagnostic test: measurement of levels of circulating exosomal lncRNA-GC1 | Beijing, China |
| exo-2 | Exosomes | Recruiting | Pancreatic cancer | Interrogation of Exosome-mediated Intercellular Signaling in Patients with Pancreatic Cancer | Observational | NCT02393703 | 111 | Pancreatic Cancer, Benign Pancreatic Disease | New York, United States | |
| exo-3 | Exosomes | Recruiting | Pancreatic cancer | New Biomarkers in Pancreatic Cancer Using EXPEL Concept (PANEXPEL) | Observational | NCT03791073 | 200 | Oncology | Montpellier, France | |
| exo-4 | Exosomes | Recruiting | Pancreatic cancer | A Pancreatic Cancer Screening Study in Hereditary High Risk Individuals | Observational | NCT03250078 | 100 | Pancreatic Neoplasms | Diagnostic test: MRI/MRCP | Connecticut, United States |
| exo-5 | Exosomes | Recruiting | Pancreatic cancer | A Study of Blood Based Biomarkers for Pancreas Adenocarcinoma | Observational | NCT03334708 | 700 | Pancreatic Cancer, Pancreatic Diseases, Pancreatitis, Pancreatic Cyst | Diagnostic test: blood draw, diagnostic test: tumor tissue collection, diagnostic test: cyst fluid | 13 locations in United States |
| exo-6 | Exosomes | Recruiting | Liver cancer | A Study of Imaging, Blood, and Tissue Samples to Guide Treatment of Colon Cancer and Related Liver Tumors | Observational | NCT03432806 | 80 | Colon Cancer, Liver Tumor | Other: blood draws, procedure: colectomy or hepatectomy, diagnostic test: Fibroscan test | New Jersey, New York, United States |
| exo-7 | Exosomes | Recruiting | Liver cancer | Clinical Study for Combined Analysis of CTC and Exosomes on Predicting the Efficacy of Immunotherapy in Patients with Hepatocellular Carcinoma | Observational | NCT05575622 | 200 | HCC | Device: CTC PD-L1, exosomal PD-L1, and exosomal LAG-3 detection | Hubei, China |
| TEP-1 | Tumor educated Platelets (TEP) | Recruiting | Gastric cancer | Project CADENCE (CAncer Detected Early caN be CurEd) | Observational | NCT05633342 | 15,000 | Liver Cancer, Gastric Cancer, Colorectal Cancer, Esophageal Cancer, Pancreatic Cancer | Singapore, Singapore | |
| TEP-2 | Tumor educated Platelets (TEP) | Not recruiting yet | Pancreatic cancer | ITGA2b and SELP Expression in Cancer Pancreas and Biliary Tract Cancer | Observational (Patient Registry) | NCT05493878 | 128 | Pancreatic Cancer | Diagnostic test: mRNA expression | Assiut, Egypt |
| TEP-3 | Tumor educated Platelets (TEP) | Recruiting | Pancreatic cancer | Pre- and Post-operative TEG Indices in Patients with or without Adenocarcinoma Undergoing Surgical Resection | Observational | NCT05517811 | 400 | Liver Cancer, Esophageal Cancer, Colorectal Cancer, Pancreas Cancer, Biliary Cancer | Diagnostic test: TEG indices | Colorado, United States |
| Liquid Biopsy | Status | Cancer | Study Title | Study Type | Clinical Trial Identifier | Estimated Enrollment | Conditions | Interventions | Locations | |
|---|---|---|---|---|---|---|---|---|---|---|
| CTC-1 | CTC | Recruiting | Gastric cancer | Liquid Biopsy in Monitiring the Neoadjuvant Chemotherapy and Operation in Gastric Cancer | Interventional (Clinical Trial) | NCT03957564 | 40 | Gastric Cancer, Gastro-Esophageal Junction Cancer | Detection of imaging data and level of CTCs | Qinghai, China |
| CTC-2 | CTC | Recruiting | Gastric cancer | Phase III Randomised Trial to Evaluate Folfox with or without Docetaxel (tfox) as 1st Line Chemotherapy for Locally Advanced or Metastatic Oesophago-Gastric carcinoma | Interventional (Clinical Trial) | NCT03006432 | 506 | Esophago-Gastric Carcinoma | Drug testing and CTC level | 98 locations in France |
| CTC-3 | CTC | Recruiting | Gastric cancer | RegoNivo vs. Standard of Care Chemotherapy in AGOC | Interventional (Clinical Trial) | NCT04879368 | 450 | Gastro-Esophageal cancer | Drug: regorafenib, biological: nivolumab, drug: docetaxel, drug: paclitaxel, drug: irinotecan, drug: trifluridine/tipracil | 75 locations in United States |
| CTC-4 | CTC | Recruiting | Gastric cancer | Avelumab + Paclitaxel/Ramucirumab (RAP) as Second Line Treatment in Gastro-esophageal Adenocarcinoma (AIO-STO-0218) | Interventional (Clinical Trial) | NCT03966118 | 59 | Gastroesophageal Junction Adenocarcinoma, Adenocarcinoma of the Stomach | Drug: avelumab, drug: ramucirumab, drug: paclitaxel | Berlin, Germany |
| CTC-5 | CTC | Recruiting | Gastric cancer | Ascending Doses of Ceralasertib in Combination with Chemotherapy and/or Novel Anti Cancer Agents | Interventional (Clinical Trial) | NCT02264678 | 330 | Gastric Cancer | Drug: administration of ceralasertib in combination with carboplatin, drug: administration of ceralasertib, drug: administration of ceralasertib in combination with olaparib, drug: administation of ceralasertib in combination with durvalumab | 27 locations in United States |
| CTC-6 | CTC | Recruiting | Pancreaticcancer | Liquid Biopsy and Pancreas Cancer: Detection of AXL(+) CTCs (CTC-AXL-PANC) | Interventional (Clinical Trial) | NCT05346536 | 63 | Pancreatic Ductal Adenocarcinoma Metastatic Pancreatic Cancer Circulating Tumor Cell | Other: detection of circulating tumor cells expressing Axl: CTC-AXL(+) | Montpellier, France |
| CTC-7 | CTC | Recruiting | Pancreatic cancer | EUS-guided PORtal Vein Sampling for Circulating Tumor Cells in Pancreatic Cancer Patients | Interventional (Clinical Trial) | NCT05247164 | 70 | Pancreatic Cancer, Pancreatic Adenocarcinoma | Procedure: EUS-guided portal vein sampling | Milan, Italy |
| CTC-8 | CTC | Recruiting | Pancreatic cancer | Echo-endoscopy Biopsy Impact on the Circulating Tumor Cell Level | Interventional (Clinical Trial) | NCT04677244 | 42 | Cancer of Pancreas | Procedure: blood sample in portal vein | Marseille, France |
| CTC-9 | CTC | Recruiting | Liver cancer | A Trial of Adjuvant Therapy after Hepatocarcinoma Resection Based on Folate Receptor-positive Circulating Tumor Cells | Interventional (Clinical Trial) | NCT04521491 | 184 | HCC | Drug: FOLFOX4 (infusional fluorouracil [FU], leucovorin [LV], and oxaliplatin [OXA]). | Shanghai, China |
| CTC-10 | CTC | Recruiting | Colorectal cancer | Influence of Opioid Analgesia on Circulating Tumor Cells in Open Colorectal Cancer Surgery | Interventional (Clinical Trial) | NCT03700411 | 120 | Colorectal Cancer, Circulating Tumor Cell | Drug: morphine, piritramid, epidural | 3 locations in Czech Republic |
| CTC-11 | CTC | Recruiting | Colorectal cancer | Tumoral Circulating Cells and Colorectal Cancer Progression | Interventional (Clinical Trial) | NCT03256084 | 120 | Colorectal Cancer | Procedure: blood and tumor samples | Marseille, France |
| ctDNA-1 | ctDNA | Recruiting | Gastric cancer | Liquid Biopsy in Monitoring the Neoadjuvant Chemotherapy and Operation in Gastric Cancer | Interventional (Clinical Trial) | NCT03957564 | 40 | Gastric Cancer, Gastro-Esophageal Junction Cancer | Drug: neoadjuvant chemotherapy with PSOX regimen. Other: detect the imaging data and levels of CTC, ctDNA, cfDNA, CEA, CA19-9, CA72-4 in plasma. Other: detect the tumor-related DNA in pathological tissues after operation. Other: follow-up of DFS and OS in patients with gastric cancer after operation. | Qinghai, China |
| ctDNA-2 | ctDNA | Recruiting | Gastric cancer | MR-guided Pre-operative RT in Gastric Cancer | Interventional (Clinical Trial) | NCT04162665 | 36 | Gastric Adenocarcinoma | Radiation: MR-guided radiation therapy, procedure: blood for ctDNA | Missouri, United States, Seoul, South Korea |
| ctDNA-3 | ctDNA | Recruiting | Gastric cancer | Peritoneal Carcinomatosis Leveraging ctDNA Guided Treatment in GI Cancer Study (PERICLES Study) | Interventional (Clinical Trial) | NCT04929015 | 30 | Gastric Cancer, ctDNA | Colorectal carcinoma by AJCC V8 stage, digestive system neoplasm, esophageal carcinoma by AJCC V8 stage, gastric carcinoma by AJCC V8 stage, liver and intrahepatic bile duct carcinoma, peritoneal carcinomatosis | New Jersey, United States |
| ctDNA-4 | ctDNA | Recruiting | Pancreatic cancer | Pilot Comparing ctDNA IDV vs. SPV Sample in Pts Undergoing Biopsies for Hepatobiliary and Pancreatic Cancers | Interventional (Clinical Trial) | NCT05497531 | 15 | Hepatobiliary Cancer, Pancreatic Cancer, Hepatocellular Carcinoma, Cholangiocarcinoma, Ampullary Cancer, Pancreatic Carcinoma | Diagnostic test: ctDNA blood collection | California, United States |
| ctDNA-5 | ctDNA | Recruiting | Pancreatic cancer | PLATON—Platform for Analyzing Targetable Tumor Mutations (Pilot-study) | Interventional (Clinical Trial) | NCT04484636 | 200 | Hepatocellular Cancer, Cholangiocarcinoma, Gallbladder Cancer, Pancreatic Cancer, Esophageal Cancer, Stomach Cancer | Diagnostic test: FoundationOne®CDx and FoundationOne®Liquid | 30 locations in Germany |
| ctDNA-6 | ctDNA | Recruiting | Liver cancer | ctDNA-Directed Post-Hepatectomy Chemotherapy for Patients With Resectable Colorectal Liver Metastases | Interventional (Clinical Trial) | NCT05062317 | 120 | Liver Metastases | Drug: leucovorin drug: 5-FLUOROURACIL, drug: oxaliplatin, drug: irinotecan, drug: capecitabine, drug: bevacizumab | Texas, United States |
| ctDNA-7 | ctDNA | Recruiting | Hepatocellular, Gastric | Risk factors of Immune—ChEckpoint inhibitors MEdiated Liver, gastrointestinal, endocrine and skin Toxicity (ICEMELT) | Interventionl (Clinical Trial) | NCT04631731 | 200 | Gastric Cancer, Hepatocellular Carcinoma | Diagnostic test: blood screening, diagnostic test: tissue screening | New South Wales, Australia |
| ctDNA-8 | ctDNA | Recruiting | Colorectal cancer | A Phase II Randomized Therapeutic Optimization Trial for Subjects with Refractory Metastatic Colorectal Cancer Using ctDNA: Rapid 1 Trial | Interventional (Clinical Trial) | NCT04786600 | 78 | Metastatic Colorectal cancer | Device: Signatera ctDNA assay, drug: pre-specified sequence of FDA-approved drugs and drug combinations | Florida, United States |
| ctDNA-9 | ctDNA | Recruiting | Colorectal cancer | Initial Attack on Latent Metastasis Using TAS-102 for ct DNA Identified Colorectal Cancer Patients after Curative Resection (ALTAIR) | Interventional (Clinical Trial) | NCT04457297 | 240 | Colorectal Neoplasms, Trifluridine, and Tipiracil, Circulating Tumor DNA | Drug: trifluridine and tipiracil, drug: placebo | 39 locations in Japan |
| ctDNA-10 | ctDNA | Recruiting | Colorectal cancer | IMPROVE Intervention Trial Implementing Non-invasive Circulating Tumor DNA Analysis to Optimize the Operative and Postoperative Treatment for Patients with Colorectal Cancer | Interventional (Clinical Trial) | NCT03748680 | 64 | Colorectal Cancer, Circulating Tumor DANN, Adjuvant Chemotherapy, Progression Free Survival | Drug: Capox (or FOLFOX) including fluoropyrimidine and oxaliplatin combination chemotherapy | 4 locations in Denmark |
| ctDNA-11 | ctDNA | Recruiting | Colorectal cancer | Circulating Tumor DNA Analysis to Optimize the Operative and Postoperative Treatment for Patients with Colorectal Cancer—Intervention Trial 2 (IMPROVE-IT2) | Interventional (Clinical Trial) | NCT04084249 | 254 | Colorectal Cancer, Colo-rectal Cancer, ctDNA, Gastro-Intestinal Disorder, Colorectal Neoplasms, Gastrointestinal Cancer, Gastrointestinal Neoplasms, Digestive System Disease, Digestive System Neoplasm, Colonic Diseases, Colonic Neoplasms, Colonic Cancer, | Diagnostic test: ctDNA-analysis, other: intensified follow-up schedule | Randers, Denmark |
| ctDNA-12 | ctDNA | Recruiting | Colorectal cancer | Circulating Cell-Free Tumor DNA Testing in Guiding Treatment for Patients with Advanced or Metastatic Colorectal Cancer | Interventional (Clinical Trial) | NCT03844620 | 100 | Refractory Colorectal Carcinoma, Stage III Colorectal Cancer AJCC v8, Stage IIIA Colorectal Cancer AJCC v8, Stage IIIB Colorectal Cancer AJCC v8, Stage IIIC Colorectal Cancer AJCC v8, Stage IV Colorectal Cancer AJCC v8, Stage IVA Colorectal Cancer AJCC v8, Stage IVB Colorectal Cancer AJCC v8 Stage, IVC Colorectal Cancer AJCC v8 | Other: best practice, other: laboratory procedure, other: quality-of-life assessment, other: questionnaire administration, drug: regorafenib, drug: trifluridine and tipiracil hydrochloride | Texas, United States |
| ctDNA-13 | ctDNA | Recruiting | Colorectal cancer | A Phase II Clinical Trial Comparing the Efficacy of RO7198457 Versus Watchful Waiting in Patients With ctDNA-positive, Resected Stage II (High Risk) and Stage III Colorectal Cancer | Interventional (Clinical Trial) | NCT04486378 | 201 | Colorectal Cancer Stage II and III | Drug: RO7198457 intravenous (i.v.), other: observational group (no intervention) | 53 locations in United States |
| ctDNA-14 | ctDNA | Recruiting | Colorectal cancer | TAS-102 in ctDNA-defined Minimal Residual Disease in Colorectal Cancer after Completion of Adjuvant Chemotherapy | Interventional (Clinical Trial) | NCT05343013 | 15 | Colorectal Cancer | Drug: TAS-102 | Texas, United States |
| ctDNA-15 | ctDNA | Recruiting | Colon cancer | Colon Adjuvant Chemotherapy Based on Evaluation of Residual Disease (CIRCULATE-US) | Interventional (Clinical Trial) | NCT05174169 | 1912 | Stage III Colon Cancer | Device: Signatera test, Drug: mFOLFOX6 3–6 month, drug: CAPOX 3 month, drug: mFOLFIRINOX, drug: mFOLFOX6 6 month, drug: CAPOX 6 month | Pennsylvania, United States |
| ctDNA-16 | ctDNA | Recruiting | Colon cancer | DYNAMIC-III: Circulating Tumour DNA Analysis Informing Adjuvant Chemotherapy in Stage III Colon Cancer: A Multi-centre Phase II/III Randomised Controlled Study (Protocol No: ctDNA-08) | Interventional (Clinical Trial) | ACTRN12617001566325 | 356/1000 | Stage III Colon Cancer | Pre-operative combined chemotherapy and radiotherapy, post-operative combined chemotherapy and radiotherapy, 3 months of fluoropyrimidine adjuvant chemotherapy, ECOG performance status 0–2 | NSW, NT, QLD, SA, TAS, WA, VIC, Australia |
| exo-1 | Exosomes | Recruiting | Gastric cancer | A Study of exoASO-STAT6 (CDK-004) in Patients With Advanced Hepatocellular Carcinoma (HCC) and Patients With Liver Metastases From Primary Gastric Cancer and Colorectal Cancer (CRC) | Interventional (Clinical Trial) | NCT05375604 | 30 | Advanced Hepatocellular Carcinoma (HCC), Gastric Cancer Metastatic to Liver, Colorectal Cancer Metastatic to Liver | Drug: CDK-004 | California, New York, Tennessee, United States |
| exo-2 | Exosomes | Recruiting | Pancreatic cancer | iExosomes in Treating Participants with Metastatic Pancreas Cancer with KrasG12D Mutation | Interventional (Clinical Trial) | NCT03608631 | 28 | KRAS NP_004976.2:p.G12DMetastatic Pancreatic AdenocarcinomaPancreatic Ductal AdenocarcinomaStage IV Pancreatic Cancer AJCC v8 | Drug: mesenchymal stromal cells-derived exosomes with KRAS G12D siRNA | Texas, United States |
| exo-3 | Exosomes | Recruiting | Pancreatic cancer | Ultra-High Resolution Optical Coherence Tomography in Detecting Micrometer Sized Early Stage Pancreatic Cancer in Participants with Pancreatic Cancer | Interventional (Clinical Trial) | NCT03711890 | 75 | Pancreatic Carcinoma, Pancreatic Intraductal Papillary Mucinous Neoplasm, Pancreatobiliary-Type | Procedure: Optical Coherence TomographyProcedure: Therapeutic Conventional SurgeryDiagnostic Test: Laboratory Evaluation | Ohio, United States |
| exo-4 | Exosomes | Recruiting | Liver cancer | A Study of exoASO-STAT6 (CDK-004) in Patients with Advanced Hepatocellular Carcinoma (HCC) and Patients with Liver Metastases from Primary Gastric Cancer and Colorectal Cancer (CRC) | Interventional (Clinical Trial) | NCT05375604 | 30 | Advanced Hepatocellular Carcinoma (HCC), Gastric Cancer Metastatic to Liver, Colorectal Cancer Metastatic to Liver | Drug: CDK-004 | 3 locations in United States |
| TEP-1 | Tumor educated Platelets (TEP) | Recruiting | Pancreatic cancer | Serial Measurements of Molecular and Architectural Responses to Therapy (SMMART) PRIME Trial | Interventional (Clinical Trial) | NCT03878524 | 40 | Stage II Pancreatic Cancer AJCC v8, Stage III Pancreatic Cancer AJCC v8, Stage IV Pancreatic Cancer AJCC v8, Stage IV AJCC v8, Unresectable Pancreatic Adenocarcinoma | Drug testing | Oregon, United States |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
David, P.; Mittelstädt, A.; Kouhestani, D.; Anthuber, A.; Kahlert, C.; Sohn, K.; Weber, G.F. Current Applications of Liquid Biopsy in Gastrointestinal Cancer Disease—From Early Cancer Detection to Individualized Cancer Treatment. Cancers 2023, 15, 1924. https://doi.org/10.3390/cancers15071924
David P, Mittelstädt A, Kouhestani D, Anthuber A, Kahlert C, Sohn K, Weber GF. Current Applications of Liquid Biopsy in Gastrointestinal Cancer Disease—From Early Cancer Detection to Individualized Cancer Treatment. Cancers. 2023; 15(7):1924. https://doi.org/10.3390/cancers15071924
Chicago/Turabian StyleDavid, Paul, Anke Mittelstädt, Dina Kouhestani, Anna Anthuber, Christoph Kahlert, Kai Sohn, and Georg F. Weber. 2023. "Current Applications of Liquid Biopsy in Gastrointestinal Cancer Disease—From Early Cancer Detection to Individualized Cancer Treatment" Cancers 15, no. 7: 1924. https://doi.org/10.3390/cancers15071924
APA StyleDavid, P., Mittelstädt, A., Kouhestani, D., Anthuber, A., Kahlert, C., Sohn, K., & Weber, G. F. (2023). Current Applications of Liquid Biopsy in Gastrointestinal Cancer Disease—From Early Cancer Detection to Individualized Cancer Treatment. Cancers, 15(7), 1924. https://doi.org/10.3390/cancers15071924






