Predicting Breast Cancer Events in Ductal Carcinoma In Situ (DCIS) Using Generative Adversarial Network Augmented Deep Learning Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical and Histological Data
2.2. GAN Augmented DL Classification Model
2.3. Bayesian Network Analysis of BCE Risk Factors
3. Results
3.1. DL Network Performs Significantly Better when Augmented with L-GAN-Generated Aggressive Cancer Image Patches
3.2. Correlation of L-GAN and Clinicopathological Data
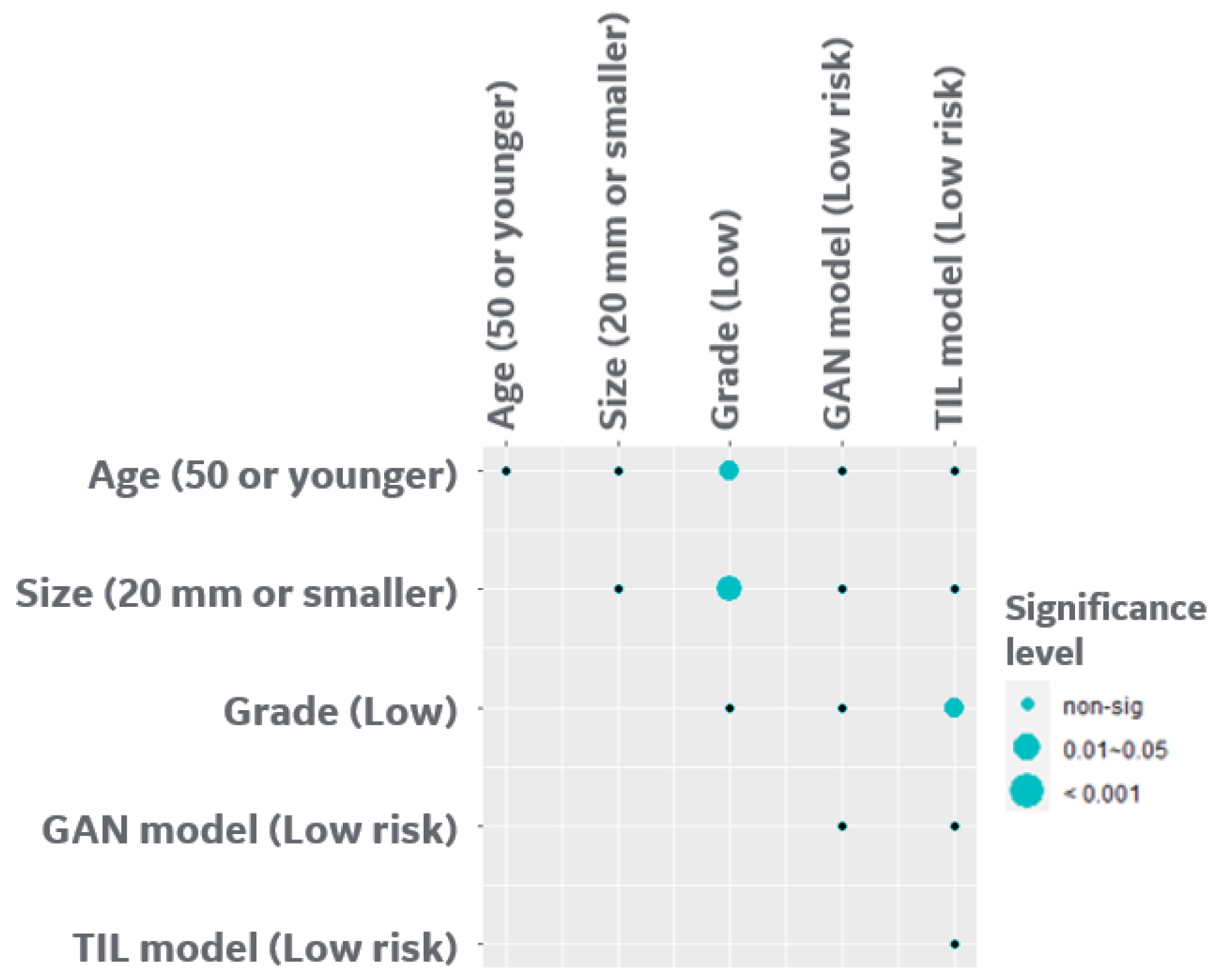
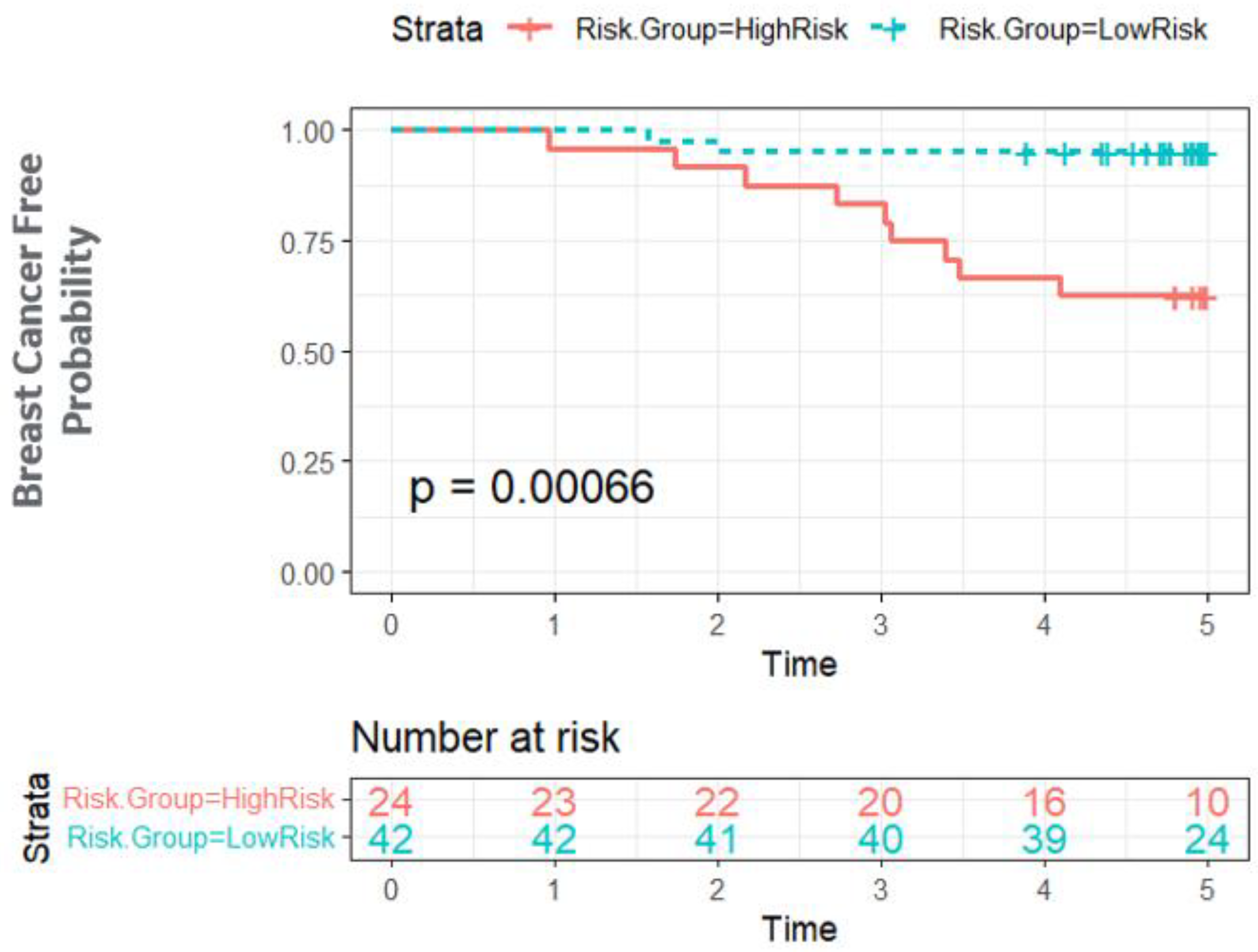
3.3. Bayesian Network Provides a Scalable Interpretable Framework for Combining Multi-Modal Risk Score
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Badve, S.S.; Gokmen-Polar, Y. Ductal carcinoma in situ of breast: Update 2019. Pathology 2019, 51, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Hophan, S.L.; Odnokoz, O.; Liu, H.; Luo, Y.; Khan, S.; Gradishar, W.; Zhou, Z.; Badve, S.; Torres, M.A.; Wan, Y. Ductal Carcinoma In Situ of Breast: From Molecular Etiology to Therapeutic Management. Endocrinology 2022, 163, bqac027. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group. Overview of the randomized trials of radiotherapy in ductal carcinoma in situ of the breast. J. Natl. Cancer Inst. Monogr. 2010, 2010, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, A.; Khoury, A.L.; Esserman, L.J. Precision surgery and avoiding over-treatment. Eur. J. Surg. Oncol. 2017, 43, 938–943. [Google Scholar] [CrossRef]
- Mukhtar, R.A.; Wong, J.M.; Esserman, L.J. Preventing Overdiagnosis and Overtreatment: Just the Next Step in the Evolution of Breast Cancer Care. J. Natl. Compr. Cancer Netw. 2015, 13, 737–743. [Google Scholar] [CrossRef]
- Esserman, L.J.; Thompson, I.M.; Reid, B.; Nelson, P.; Ransohoff, D.F.; Welch, H.G.; Hwang, S.; Berry, D.A.; Kinzler, K.W.; Black, W.C.; et al. Addressing overdiagnosis and overtreatment in cancer: A prescription for change. Lancet Oncol. 2014, 15, e234–e242. [Google Scholar] [CrossRef]
- Rudloff, U.; Jacks, L.M.; Goldberg, J.I.; Wynveen, C.A.; Brogi, E.; Patil, S.; Van Zee, K.J. Nomogram for predicting the risk of local recurrence after breast-conserving surgery for ductal carcinoma in situ. J. Clin. Oncol. 2010, 28, 3762–3769. [Google Scholar] [CrossRef]
- Lari, S.A.; Kuerer, H.M. Biological Markers in DCIS and Risk of Breast Recurrence: A Systematic Review. J. Cancer 2011, 2, 232–261. [Google Scholar] [CrossRef]
- Lehman, C.D.; Gatsonis, C.; Romanoff, J.; Khan, S.A.; Carlos, R.; Solin, L.J.; Badve, S.; McCaskill-Stevens, W.; Corsetti, R.L.; Rahbar, H.; et al. Association of Magnetic Resonance Imaging and a 12-Gene Expression Assay with Breast Ductal Carcinoma In Situ Treatment. JAMA Oncol. 2019, 5, 1036–1042. [Google Scholar] [CrossRef]
- Rakovitch, E.; Gray, R.; Baehner, F.L.; Sutradhar, R.; Crager, M.; Gu, S.; Nofech-Mozes, S.; Badve, S.S.; Hanna, W.; Hughes, L.L.; et al. Refined estimates of local recurrence risks by DCIS score adjusting for clinicopathological features: A combined analysis of ECOG-ACRIN E5194 and Ontario DCIS cohort studies. Breast Cancer Res. Treat. 2018, 169, 359–369. [Google Scholar] [CrossRef]
- Solin, L.J.; Gray, R.; Baehner, F.L.; Butler, S.M.; Hughes, L.L.; Yoshizawa, C.; Cherbavaz, D.B.; Shak, S.; Page, D.L.; Sledge, G.W., Jr.; et al. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J. Natl. Cancer Inst. 2013, 105, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Raldow, A.C.; Sher, D.; Chen, A.B.; Recht, A.; Punglia, R.S. Cost Effectiveness of the Oncotype DX DCIS Score for Guiding Treatment of Patients with Ductal Carcinoma In Situ. J. Clin. Oncol. 2016, 34, 3963–3968. [Google Scholar] [CrossRef] [PubMed]
- Klimov, S.; Miligy, I.M.; Gertych, A.; Jiang, Y.; Toss, M.S.; Rida, P.; Ellis, I.O.; Green, A.; Krishnamurti, U.; Rakha, E.A.; et al. A whole slide image-based machine learning approach to predict ductal carcinoma in situ (DCIS) recurrence risk. Breast Cancer Res. 2019, 21, 83. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Romo-Bucheli, D.; Wang, X.; Janowczyk, A.; Ganesan, S.; Gilmore, H.; Rimm, D.; Madabhushi, A. Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab. Investig. 2018, 98, 1438–1448. [Google Scholar] [CrossRef]
- Lee, G.; Veltri, R.W.; Zhu, G.; Ali, S.; Epstein, J.I.; Madabhushi, A. Nuclear Shape and Architecture in Benign Fields Predict Biochemical Recurrence in Prostate Cancer Patients Following Radical Prostatectomy: Preliminary Findings. Eur. Urol. Focus 2017, 3, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Hoque, A.; Lippman, S.M.; Boiko, I.V.; Atkinson, E.N.; Sneige, N.; Sahin, A.; Weber, D.M.; Risin, S.; Lagios, M.D.; Schwarting, R.; et al. Quantitative nuclear morphometry by image analysis for prediction of recurrence of ductal carcinoma in situ of the breast. Cancer Epidemiol. Biomark. Prev. 2001, 10, 249–259. [Google Scholar]
- Li, H.; Whitney, J.; Bera, K.; Gilmore, H.; Thorat, M.A.; Badve, S.; Madabhushi, A. Quantitative nuclear histomorphometric features are predictive of Oncotype DX risk categories in ductal carcinoma in situ: Preliminary findings. Breast Cancer Res. 2019, 21, 114. [Google Scholar] [CrossRef]
- Toss, M.S.; Abidi, A.; Lesche, D.; Joseph, C.; Mahale, S.; Saunders, H.; Kader, T.; Miligy, I.M.; Green, A.R.; Gorringe, K.L.; et al. The prognostic significance of immune microenvironment in breast ductal carcinoma in situ. Br. J. Cancer 2020, 122, 1496–1506. [Google Scholar] [CrossRef]
- Salgado, R.; Fineberg, S.; De Caluwe, A.; de Azambuja, E. Tumour infiltrating lymphocytes and ductal carcinoma in situ: The art of thinking counterintuitively. Eur. J. Cancer 2022, 168, 138–140. [Google Scholar] [CrossRef]
- Komforti, M.; Badve, S.S.; Harmon, B.; Lo, Y.; Fineberg, S. Tumour-infiltrating lymphocytes in ductal carcinoma in situ (DCIS)-assessment with three different methodologies and correlation with Oncotype DX DCIS Score. Histopathology 2020, 77, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Badve, S.S.; Cho, S.; Lu, X.; Cao, S.; Ghose, S.; Thike, A.A.; Tan, P.H.; Ocal, I.T.; Generali, D.; Zanconati, F.; et al. Tumor Infiltrating Lymphocytes in Multi-National Cohorts of Ductal Carcinoma In Situ (DCIS) of Breast. Cancers 2022, 14, 3916. [Google Scholar] [CrossRef]
- Shmatko, A.; Ghaffari Laleh, N.; Gerstung, M.; Kather, J.N. Artificial intelligence in histopathology: Enhancing cancer research and clinical oncology. Nat. Cancer 2022, 3, 1026–1038. [Google Scholar] [CrossRef]
- Fu, Y.; Jung, A.W.; Torne, R.V.; Gonzalez, S.; Vohringer, H.; Shmatko, A.; Yates, L.R.; Jimenez-Linan, M.; Moore, L.; Gerstung, M. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 2020, 1, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Rong, R.; Wang, S.; Zhang, X.; Wen, Z.; Cheng, X.; Jia, L.; Yang, D.M.; Xie, Y.; Zhan, X.; Xiao, G. Enhanced Pathology Image Quality with Restore-Generative Adversarial Network. Am. J. Pathol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.; Liu, S.; Russo, C.; Nadort, A.; Di Ieva, A. Generative Adversarial Networks in Digital Pathology and Histopathological Image Processing: A Review. J. Pathol. Inform. 2021, 12, 43. [Google Scholar] [CrossRef]
- Tschuchnig, M.E.; Oostingh, G.J.; Gadermayr, M. Generative Adversarial Networks in Digital Pathology: A Survey on Trends and Future Potential. Patterns 2020, 1, 100089. [Google Scholar] [CrossRef]
- Levine, A.B.; Peng, J.; Farnell, D.; Nursey, M.; Wang, Y.; Naso, J.R.; Ren, H.; Farahani, H.; Chen, C.; Chiu, D.; et al. Synthesis of diagnostic quality cancer pathology images by generative adversarial networks. J. Pathol. 2020, 252, 178–188. [Google Scholar] [CrossRef]
- Liu, S.; Shah, Z.; Sav, A.; Russo, C.; Berkovsky, S.; Qian, Y.; Coiera, E.; Di Ieva, A. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci. Rep. 2020, 10, 7733. [Google Scholar] [CrossRef]
- Quiros, A.C.; Murray-Smith, R.; Yuan, K. PathologyGAN: Learning deep representations of cancer tissue. arXiv 2019, arXiv:1907.02644. [Google Scholar]
- Liu, D.; Chen, X.; Peng, D. Cosine Similarity Measure between Hybrid Intuitionistic Fuzzy Sets and Its Application in Medical Diagnosis. Comput. Math. Methods Med. 2018, 2018, 3146873. [Google Scholar] [CrossRef] [PubMed]
- Scutari, M.; Denis, J.-B. Bayesian Networks: With Examples in R; Chapman and Hall/CRC: New York, NY, USA, 2021. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the 31st AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Kalinsky, K.; Barlow, W.E.; Gralow, J.R.; Meric-Bernstam, F.; Albain, K.S.; Hayes, D.F.; Lin, N.U.; Perez, E.A.; Goldstein, L.J.; Chia, S.K.L.; et al. 21-Gene Assay to Inform Chemotherapy Benefit in Node-Positive Breast Cancer. N. Engl. J. Med. 2021, 385, 2336–2347. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; Olson, J.A., Jr.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef] [PubMed]


| Oxford (N = 67) | Singapore (N = 66) | |
|---|---|---|
| Follow up time | ||
| Mean (SD) | 7.74 (4.36) | 5.1 (2.17) |
| Range (BCE) | 0.83–15.17 | 0.96–13.71 |
| Range (Non-BCE) | 4.9–17.08 | 3.89–6.16 |
| Breast Cancer Event | ||
| N | 40 (59.7%) | 48 (72.7%) |
| Y | 27 (40.3%) | 18 (27.3%) |
| Age | ||
| N-Miss | 1 | 0 |
| ≤50 | 19 (28.8%) | 23 (34.8%) |
| >50 | 47 (71.2%) | 43 (65.2%) |
| Size | ||
| N-Miss | 39 | 0 |
| ≤20 | 18 (64.3%) | 34 (51.5%) |
| >20 | 10 (35.7%) | 32 (48.5%) |
| Grade | ||
| N-Miss | 8 | 0 |
| Low | 8 (13.6%) | 8 (12.1%) |
| Intermediate | 17 (28.8%) | 22 (33.3%) |
| High | 34 (57.6%) | 36 (54.5%) |
| Lymphocyte | ||
| 0–5% | 27 (40.3%) | 22 (33.3%) |
| >5% | 40 (59.7%) | 44 (66.7%) |
| Touching TILs | ||
| 0 | 52 (77.6%) | 52 (78.8%) |
| >0 | 15 (22.4%) | 14 (21.2%) |
| Circumferential TILs | ||
| No | 51 (76.1%) | 38 (57.6%) |
| Yes | 16 (23.9%) | 28 (42.4%) |
| Hotspot | ||
| No | 40 (59.7%) | 30 (45.5%) |
| dense | 27 (40.3%) | 36 (54.5%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghose, S.; Cho, S.; Ginty, F.; McDonough, E.; Davis, C.; Zhang, Z.; Mitra, J.; Harris, A.L.; Thike, A.A.; Tan, P.H.; et al. Predicting Breast Cancer Events in Ductal Carcinoma In Situ (DCIS) Using Generative Adversarial Network Augmented Deep Learning Model. Cancers 2023, 15, 1922. https://doi.org/10.3390/cancers15071922
Ghose S, Cho S, Ginty F, McDonough E, Davis C, Zhang Z, Mitra J, Harris AL, Thike AA, Tan PH, et al. Predicting Breast Cancer Events in Ductal Carcinoma In Situ (DCIS) Using Generative Adversarial Network Augmented Deep Learning Model. Cancers. 2023; 15(7):1922. https://doi.org/10.3390/cancers15071922
Chicago/Turabian StyleGhose, Soumya, Sanghee Cho, Fiona Ginty, Elizabeth McDonough, Cynthia Davis, Zhanpan Zhang, Jhimli Mitra, Adrian L. Harris, Aye Aye Thike, Puay Hoon Tan, and et al. 2023. "Predicting Breast Cancer Events in Ductal Carcinoma In Situ (DCIS) Using Generative Adversarial Network Augmented Deep Learning Model" Cancers 15, no. 7: 1922. https://doi.org/10.3390/cancers15071922
APA StyleGhose, S., Cho, S., Ginty, F., McDonough, E., Davis, C., Zhang, Z., Mitra, J., Harris, A. L., Thike, A. A., Tan, P. H., Gökmen-Polar, Y., & Badve, S. S. (2023). Predicting Breast Cancer Events in Ductal Carcinoma In Situ (DCIS) Using Generative Adversarial Network Augmented Deep Learning Model. Cancers, 15(7), 1922. https://doi.org/10.3390/cancers15071922









