Associations of T-Cell Receptor Repertoire Diversity with L-Asparaginase Allergy in Childhood Acute Lymphoblastic Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. L-Asparaginase Therapy and Measurement of L-asp Activity
2.3. Phenotyping of Clinical Allergy to Asparaginase
2.4. T-Cell Receptor Sequencing and Generation of TCR Repertoires
2.5. Analysis of T-Cell Receptor Repertoire Diversity and Statistical Analysis
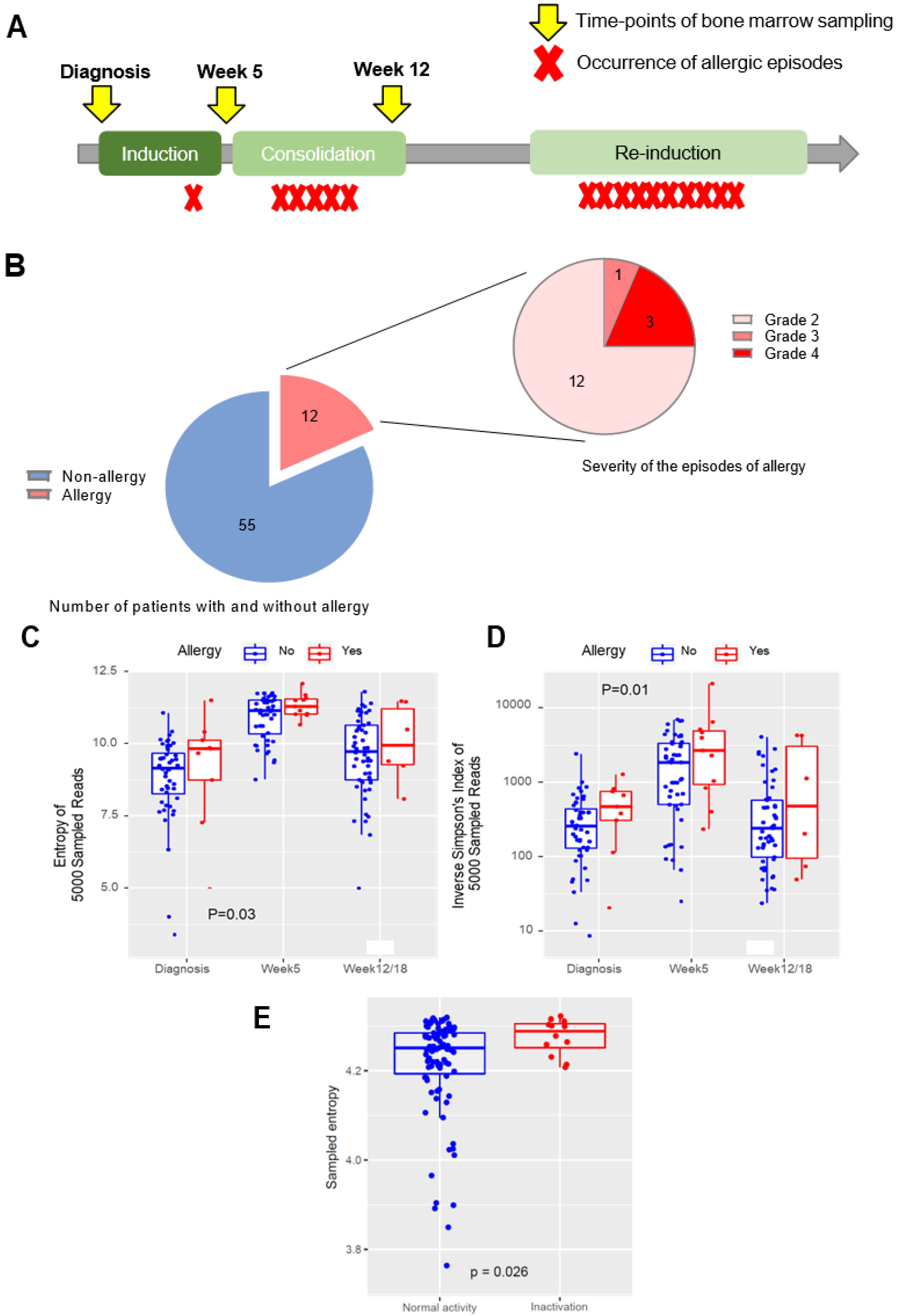
3. Results
3.1. Clinical Characteristics of L-asp Hypersensitivity
3.2. Higher TCR-Beta Diversity at All Earlier Timepoints Is Associated with the Occurrence of Future Allergy and L-Asparaginase Inactivation
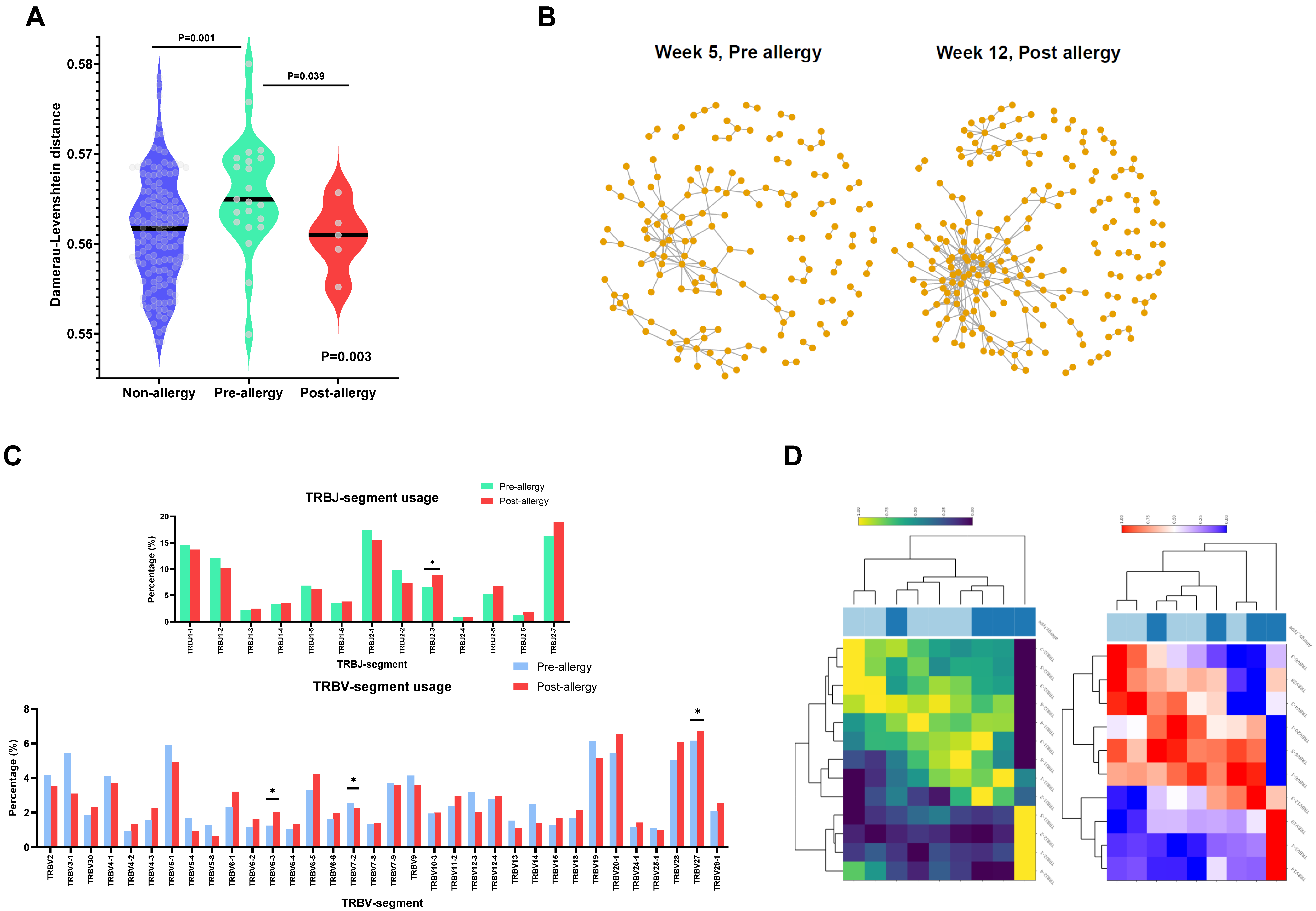
3.3. Higher TCR-Beta Diversity Is Characterized by Infrequent, Dynamically Changing, and Less Shared Clonotypes
3.4. Occurrence of L-Asparaginase Allergy Is Associated with Convergent TCR Response to Common Antigens and Shaping of TCR Gene Usage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pui, C.H.; Liu, Y.; Relling, M.V. How to solve the problem of hypersensitivity to asparaginase? Pediatr. Blood Cancer 2018, 65, e26884. [Google Scholar] [CrossRef] [PubMed]
- van der Sluis, I.M.; Vrooman, L.M.; Pieters, R.; Baruchel, A.; Escherich, G.; Goulden, N.; Mondelaers, V.; Sanchez de Toledo, J.; Rizzari, C.; Silverman, L.B.; et al. Consensus expert recommendations for identification and management of asparaginase hypersensitivity and silent inactivation. Haematologica 2016, 101, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Oettgen, H.F.; Stephenson, P.A.; Schwartz, M.K.; Leeper, R.D.; Tallal, L.; Tan, C.C.; Clarkson, B.D.; Golbey, R.B.; Krakoff, I.H.; Karnofsky, D.A.; et al. Toxicity of E. coli L- asparaginase in man. Cancer 1970, 25, 253–278. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kawedia, J.D.; Cheng, C.; Pei, D.; Fernandez, C.A.; Cai, X.; Crews, K.R.; Kaste, S.C.; Panetta, J.C.; Bowman, W.P.; et al. Clinical utility and implications of asparaginase antibodies in acute lymphoblastic leukemia. Leukemia 2012, 26, 2303–2309. [Google Scholar] [CrossRef]
- Vrooman, L.M.; Stevenson, K.E.; Supko, J.G.; O'Brien, J.; Dahlberg, S.E.; Asselin, B.L.; Athale, U.H.; Clavell, L.A.; Kelly, K.M.; Kutok, J.L.; et al. Postinduction dexamethasone and individualized dosing of Escherichia Coli L-asparaginase each improve outcome of children and adolescents with newly diagnosed acute lymphoblastic leukemia: Results from a randomized study--Dana-Farber Cancer Institute ALL Consortium Protocol 00-01. J. Clin. Oncol. 2013, 31, 1202–1210. [Google Scholar]
- Silverman, L.B.; Gelber, R.D.; Dalton, V.K.; Asselin, B.L.; Barr, R.D.; Clavell, L.A.; Hurwitz, C.A.; Moghrabi, A.; Samson, Y.; Schorin, M.A.; et al. Improved outcome for children with acute lymphoblastic leukemia: Results of Dana-Farber Consortium Protocol 91-01. Blood 2001, 97, 1211–1218. [Google Scholar] [CrossRef]
- Yeoh, A.E.J.; Lu, Y.; Chin, W.H.N.; Chiew, E.K.H.; Lim, E.H.; Li, Z.; Kham, S.K.Y.; Chan, Y.H.; Abdullah, W.A.; Lin, H.P.; et al. Intensifying Treatment of Childhood B- Lymphoblastic Leukemia With IKZF1 Deletion Reduces Relapse and Improves Overall Survival: Results of Malaysia-Singapore ALL 2010 Study. J. Clin. Oncol. 2018, 36, 2726–2735. [Google Scholar] [CrossRef]
- Liu, H.C.; Yeh, T.C.; Hou, J.Y.; Chen, K.H.; Huang, T.H.; Chang, C.Y.; Liang, D.C. Triple intrathecal therapy alone with omission of cranial radiation in children with acute lymphoblastic leukemia. J. Clin. Oncol. 2014, 32, 1825–1829. [Google Scholar] [CrossRef]
- Antillon, F.G.; Blanco, J.G.; Valverde, P.D.; Castellanos, M.; Garrido, C.P.; Giron, V.; Letona, T.R.; Osorio, E.J.; Borrayo, D.A.; Mack, R.A.; et al. The treatment of childhood acute lymphoblastic leukemia in Guatemala: Biologic features, treatment hurdles, and results. Cancer 2017, 123, 436–448. [Google Scholar] [CrossRef]
- Navarrete, M.; Rossi, E.; Brivio, E.; Carrillo, J.M.; Bonilla, M.; Vasquez, R.; Pena, A.; Fu, L.; Martinez, R.; Espinoza, C.M.; et al. Treatment of childhood acute lymphoblastic leukemia in central America: A lower-middle income countries experience. Pediatr. Blood Cancer 2014, 61, 803–809. [Google Scholar] [CrossRef]
- Liu, Y.; Smith, C.A.; Panetta, J.C.; Yang, W.; Thompson, L.E.; Counts, J.P.; Molinelli, A.R.; Pei, D.; Kornegay, N.M.; Crews, K.R.; et al. Antibodies Predict Pegaspargase Allergic Reactions and Failure of Rechallenge. J. Clin. Oncol. 2019, 37, 2051–2061. [Google Scholar] [CrossRef] [PubMed]
- Rathod, S.; Ramsey, M.; Finkelman, F.D.; Fernandez, C.A. Genetic inhibition of NFATC2 attenuates asparaginase hypersensitivity in mice. Blood Adv. 2020, 4, 4406–4416. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.A.; Smith, C.; Yang, W.; Mullighan, C.G.; Qu, C.; Larsen, E.; Bowman, W.P.; Liu, C.; Ramsey, L.B.; Chang, T.; et al. Genome-wide analysis links NFATC2 with asparaginase hypersensitivity. Blood 2015, 126, 69–75. [Google Scholar] [CrossRef]
- Rathod, S.; Ramsey, M.; DiGiorgio, D.; Berrios, R.; Finkelman, F.D.; Fernandez, C.A. Asparaginase immune complexes induce Fc-gammaRIII-dependent hypersensitivity in naive mice. FASEB J. 2019, 33, 10996–11005. [Google Scholar] [CrossRef]
- Tong, W.H.; Pieters, R.; Kaspers, G.J.; te Loo, D.M.; Bierings, M.B.; van den Bos, C.; Kollen, W.J.; Hop, W.C.; Lanvers-Kaminsky, C.; Relling, M.V.; et al. A prospective study on drug monitoring of PEGasparaginase and Erwinia asparaginase and asparaginase antibodies in pediatric acute lymphoblastic leukemia. Blood 2014, 123, 2026–2033. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.A.; Smith, C.; Yang, W.; Date, M.; Bashford, D.; Larsen, E.; Bowman, W.P.; Liu, C.; Ramsey, L.B.; Chang, T.; et al. HLA-DRB1*07:01 is associated with a higher risk of asparaginase allergies. Blood 2014, 124, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Chua, G.T.; Rosa Duque, J.S.; Cheuk, D.K.L.; Leung, A.W.K.; Wong, W.H.S.; Liu, A.P.Y.; Lee, P.P.W.; Ha, S.Y.; Chiang, A.K.S.; Ho, M.H.K.; et al. HLA alleles associated with asparaginase hypersensitivity in Chinese children. J. Hematol. Oncol. 2021, 14, 182. [Google Scholar] [CrossRef]
- Liu, S.; Gao, C.; Wu, Y.; Lin, W.; Li, J.; Xue, T.; Wang, L.; Zheng, H.; Zhang, R. HLA-DRB1*16:02 is associated with PEG- asparaginase hypersensitivity. Pharmacogenomics 2021, 22, 1135–1142. [Google Scholar] [CrossRef]
- Hojfeldt, S.G.; Wolthers, B.O.; Tulstrup, M.; Abrahamsson, J.; Gupta, R.; Harila-Saari, A.; Heyman, M.; Henriksen, L.T.; Jonsson, O.G.; Lahteenmaki, P.M.; et al. Genetic predisposition to PEG- asparaginase hypersensitivity in children treated according to NOPHO ALL2008. Br. J. Haematol. 2019, 184, 405–417. [Google Scholar] [CrossRef]
- Bradley, P.; Thomas, P.G. Using T Cell Receptor Repertoires to Understand the Principles of Adaptive Immune Recognition. Annu. Rev. Immunol. 2019, 37, 547–570. [Google Scholar] [CrossRef]
- Nikolich-Zugich, J.; Slifka, M.K.; Messaoudi, I. The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 2004, 4, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Pavlos, R.; Mallal, S.; Ostrov, D.; Buus, S.; Metushi, I.; Peters, B.; Phillips, E. T cell-mediated hypersensitivity reactions to drugs. Annu. Rev. Med. 2015, 66, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Wu, J.; Li, X.; Xie, H.; Tang, C.; Zhao, X.; Wang, S.; Chen, L.; Zhang, W.; An, Y.; et al. T-cell receptor repertoire data provides new evidence for hygiene hypothesis of allergic diseases. Allergy 2020, 75, 681–683. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.H.; Stadinski, B.D.; Henderson, L.A.; Ott de Bruin, L.; Delmonte, O.; Lee, Y.N.; de la Morena, M.T.; Goyal, R.K.; Hayward, A.; Huang, C.H.; et al. Abnormalities of T-cell receptor repertoire in CD4+ regulatory and conventional T cells in patients with RAG mutations: Implications for autoimmunity. J. Allergy Clin. Immunol. 2017, 140, 1739–1743.e7. [Google Scholar] [CrossRef]
- Bechara, R.; Feray, A.; Pallardy, M. Drug and Chemical Allergy: A Role for a Specific Naive T-Cell Repertoire? Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Pan, R.Y.; Chu, M.T.; Wang, C.W.; Lee, Y.S.; Lemonnier, F.; Michels, A.W.; Schutte, R.; Ostrov, D.A.; Chen, C.B.; Phillips, E.J.; et al. Identification of drug-specific public TCR driving severe cutaneous adverse reactions. Nat. Commun. 2019, 10, 3569. [Google Scholar] [CrossRef]
- Yeoh, A.E.; Ariffin, H.; Chai, E.L.; Kwok, C.S.; Chan, Y.H.; Ponnudurai, K.; Campana, D.; Tan, P.L.; Chan, M.Y.; Kham, S.K.; et al. Minimal residual disease-guided treatment deintensification for children with acute lymphoblastic leukemia: Results from the Malaysia- Singapore acute lymphoblastic leukemia 2003 study. J. Clin. Oncol. 2012, 30, 2384–2392. [Google Scholar] [CrossRef]
- Zalewska-Szewczyk, B.; Andrzejewski, W.; Mlynarski, W.; Jedrychowska-Danska, K.; Witas, H.; Bodalski, J. The anti- asparagines antibodies correlate with L-asparagines activity and may affect clinical outcome of childhood acute lymphoblastic leukemia. Leuk Lymphoma 2007, 48, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lee, S.H.R.; Chin, W.H.N.; Lu, Y.; Jiang, N.; Lim, E.H.; Coustan-Smith, E.; Chiew, K.H.; Oh, B.L.Z.; Koh, G.S.; et al. Distinct clinical characteristics of DUX4- and PAX5-altered childhood B-lymphoblastic leukemia. Blood Adv. 2021, 5, 5226–5238. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Venturi, V.; Kedzierska, K.; Turner, S.J.; Doherty, P.C.; Davenport, M.P. Methods for comparing the diversity of samples of the T cell receptor repertoire. J. Immunol. Methods 2007, 321, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Keylock, C.J. Simpson diversity and the Shannon-Wiener index as special cases of a generalized entropy. Oikos 2005, 109, 203–207. [Google Scholar] [CrossRef]
- Stewart, J.J.; Lee, C.Y.; Ibrahim, S.; Watts, P.; Shlomchik, M.; Weigert, M.; Litwin, S. A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol. Immunol. 1997, 34, 1067–1082. [Google Scholar] [CrossRef] [PubMed]
- Shugay, M.; Bagaev, D.V.; Zvyagin, I.V.; Vroomans, R.M.; Crawford, J.C.; Dolton, G.; Komech, E.A.; Sycheva, A.L.; Koneva, A.E.; Egorov, E.S.; et al. VDJdb: A curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res 2018, 46, D419–D427. [Google Scholar] [CrossRef]
- Chen, S.H.; Pei, D.; Yang, W.; Cheng, C.; Jeha, S.; Cox, N.J.; Evans, W.E.; Pui, C.H.; Relling, M.V. Genetic variations in GRIA1 on chromosome 5q33 related to asparaginase hypersensitivity. Clin. Pharmacol. Ther. 2010, 88, 191–196. [Google Scholar] [CrossRef]
- Lambrecht, B.N.; Hammad, H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat. Immunol. 2017, 18, 1076–1083. [Google Scholar] [CrossRef]
- Hosie, L.; Pachnio, A.; Zuo, J.; Pearce, H.; Riddell, S.; Moss, P. Cytomegalovirus-Specific T Cells Restricted by HLA-Cw*0702 Increase Markedly with Age and Dominate the CD8+ T-Cell Repertoire in Older People. Front. Immunol. 2017, 8, 1776. [Google Scholar] [CrossRef] [PubMed]
- Zamora, A.E.; Crawford, J.C.; Thomas, P.G. Hitting the Target: How T Cells Detect and Eliminate Tumors. J. Immunol. 2018, 200, 392–399. [Google Scholar] [CrossRef]
- Esser, P.R.; Kimber, I.; Martin, S.F. Correlation of contact sensitizer potency with T cell frequency and TCR repertoire diversity. Exp. Suppl. 2014, 104, 101–114. [Google Scholar]
- Ko, T.M.; Chung, W.H.; Wei, C.Y.; Shih, H.Y.; Chen, J.K.; Lin, C.H.; Chen, Y.T.; Hung, S.I. Shared and restricted T-cell receptor use is crucial for carbamazepine-induced Stevens-Johnson syndrome. J. Allergy Clin. Immunol. 2011, 128, 1266–1276.e11. [Google Scholar] [CrossRef]
- Yun, J.; Marcaida, M.J.; Eriksson, K.K.; Jamin, H.; Fontana, S.; Pichler, W.J.; Yerly, D. Oxypurinol directly and immediately activates the drug-specific T cells via the preferential use of HLA-B*58:01. J. Immunol. 2014, 192, 2984–2993. [Google Scholar] [CrossRef] [PubMed]
- Reuben, A.; Gittelman, R.; Gao, J.; Zhang, J.; Yusko, E.C.; Wu, C.J.; Emerson, R.; Zhang, J.; Tipton, C.; Li, J.; et al. TCR Repertoire Intratumor Heterogeneity in Localized Lung Adenocarcinomas: An Association with Predicted Neoantigen Heterogeneity and Postsurgical Recurrence. Cancer Discov 2017, 7, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Keane, C.; Gould, C.; Jones, K.; Hamm, D.; Talaulikar, D.; Ellis, J.; Vari, F.; Birch, S.; Han, E.; Wood, P.; et al. The T-cell Receptor Repertoire Influences the Tumor Microenvironment and Is Associated with Survival in Aggressive B-cell Lymphoma. Clin. Cancer Res. 2017, 23, 1820–1828. [Google Scholar] [CrossRef] [PubMed]
- Pokrovsky, V.S.; Kazanov, M.D.; Dyakov, I.N.; Pokrovskaya, M.V.; Aleksandrova, S.S. Comparative immunogenicity and structural analysis of epitopes of different bacterial L-asparaginases. BMC Cancer 2016, 16, 89. [Google Scholar] [CrossRef]
- Okada, H.; Kuhn, C.; Feillet, H.; Bach, J.F. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: An update. Clin. Exp. Immunol. 2010, 160, 1–9. [Google Scholar] [CrossRef]

| Patient | Age | Gender | All Subtype | CNS Status | WBC at Diagnosis (× 109/L) | Day of Therapy of Hypersensitivity Occurrence (Phase of Therapy) | Grade of Asparaginase Hypersensitivity | Date of Rechallenge | Grade of Hypersensitivity to Rechallenge | Subsequent Treatment |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 8 | F | B Other | CNS I | 3.7 | 161 (Reinduction #1) | 2 | - | - | Switched to Erwinase |
| 2 | 5 | M | ETV6- RUNX1 | CNS I | 8.9 | 48 (Consolidation) | 2 | 3 days after first allergy | 2 | Switched to Erwinase after 2nd episode |
| 3 | 2 | F | B Other | CNS I | 57.0 | 353 (Reinduction #3) | 4 | - | - | Switched to Erwinase |
| 4 | 1 | F | KMT2A | CNS I | 5.1 | 391 (Reinduction #3) | 2 | - | - | Switched to Erwinase |
| 5 | 6 | M | ETV6- RUNX1 | CNS I | 8.0 | 165 (Reinduction #1) | 2 | - | - | Switched to Erwinase |
| 6 | 3 | M | Hyperdiploid | CNS I | 8.4 | 174 (Reinduction #1) | 2 | - | - | Omitted asparaginase |
| 7 | 5 | F | ETV6- RUNX1 | CNS I | 1.2 | 246 (Reinduction #2) | 2 | - | - | Omitted asparaginase |
| 8 * | 1 | F | KMT2A | CNS I | 120.7 | 255 (Reinduction #2) | 2 | 2 weeks after first allergy | 2 | Switched to Erwinase after 3rd episode |
| 9 | 2 | F | Hyperdiploid | CNS I | 3.3 | 49 (Consolidation) | 4 | - | - | Switched to Erwinase |
| 10 | 11 | M | B Other | CNS II | 11.0 | 25 (Induction) | 2 | 3 weeks after first allergy | 4 | Switched to Erwinase after 2nd episode |
| 11 | 1 | M | B Other | CNS I | 23.0 | 147 (Reinduction #1) | 2 | - | - | Omitted asparaginase |
| 12 | 5 | M | Hyperdiploid | CNS I | 2.0 | 49 (Consolidation) | 2 | - | - | Switched to Erwinase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.H.R.; Li, Z.; Lim, E.H.Z.; Chin, W.H.N.; Jiang, N.; Chiew, K.H.; Chen, Z.; Oh, B.L.Z.; Tan, A.M.; Ariffin, H.; et al. Associations of T-Cell Receptor Repertoire Diversity with L-Asparaginase Allergy in Childhood Acute Lymphoblastic Leukemia. Cancers 2023, 15, 1829. https://doi.org/10.3390/cancers15061829
Lee SHR, Li Z, Lim EHZ, Chin WHN, Jiang N, Chiew KH, Chen Z, Oh BLZ, Tan AM, Ariffin H, et al. Associations of T-Cell Receptor Repertoire Diversity with L-Asparaginase Allergy in Childhood Acute Lymphoblastic Leukemia. Cancers. 2023; 15(6):1829. https://doi.org/10.3390/cancers15061829
Chicago/Turabian StyleLee, Shawn H. R., Zhenhua Li, Evelyn H. Z. Lim, Winnie H. N. Chin, Nan Jiang, Kean Hui Chiew, Zhiwei Chen, Bernice L. Z. Oh, Ah Moy Tan, Hany Ariffin, and et al. 2023. "Associations of T-Cell Receptor Repertoire Diversity with L-Asparaginase Allergy in Childhood Acute Lymphoblastic Leukemia" Cancers 15, no. 6: 1829. https://doi.org/10.3390/cancers15061829
APA StyleLee, S. H. R., Li, Z., Lim, E. H. Z., Chin, W. H. N., Jiang, N., Chiew, K. H., Chen, Z., Oh, B. L. Z., Tan, A. M., Ariffin, H., Yang, J. J., & Yeoh, A. E. J. (2023). Associations of T-Cell Receptor Repertoire Diversity with L-Asparaginase Allergy in Childhood Acute Lymphoblastic Leukemia. Cancers, 15(6), 1829. https://doi.org/10.3390/cancers15061829





