Influence of Single-Nucleotide Polymorphisms on Clinical Outcomes of Capecitabine-Based Chemotherapy in Colorectal Cancer Patients: A Systematic Review
Abstract
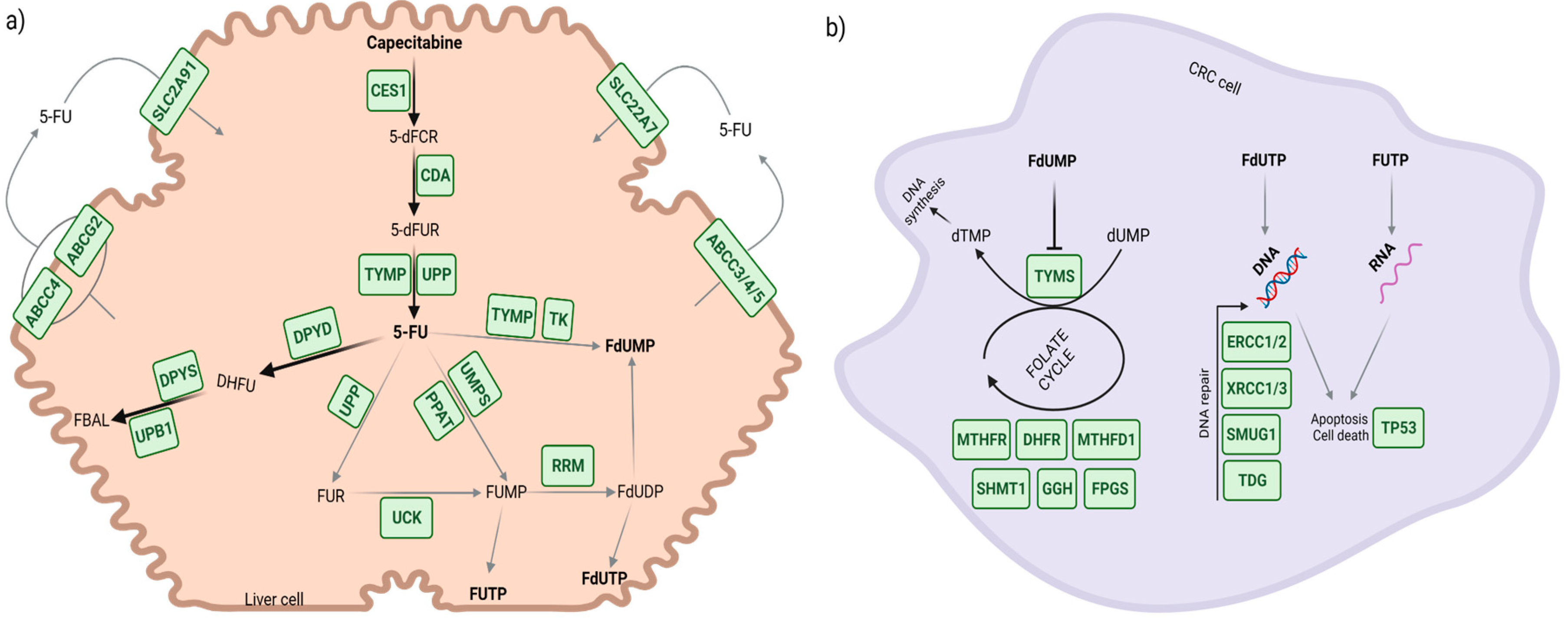
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Data Extraction
2.4. Quality Assessment
3. Results
3.1. Search Results
3.2. Study Characteristics
3.3. SNPs Associated with Capecitabine-Induced Toxicity
3.3.1. Cytidine Deaminase Gene (CDA)
3.3.2. Dihydropyrimidine Dehydrogenase Gene (DPYD)
3.3.3. Uridine Monophosphate Synthetase Gene (UMPS)
3.3.4. Solute Carrier Family 22 Member 7 Gene (SLC22A7)
3.3.5. ATP-Binding Cassette Subfamily B Member 1 Gene (ABCB1)
3.3.6. Thymidylate Synthetase Gene (TYMS) and Enolase Superfamily Member 1 (ENOSF1)
3.3.7. Methylenetetrahydrofolate Reductase Gene (MTHFR)
3.4. Gene Variants Associated with Capecitabine Effectiveness
3.4.1. ATP-Binding Cassette Subfamily B Member 1 Gene (ABCB1)
3.4.2. ERCC Excision Repair 1 (ERCC1)
3.4.3. ERCC Excision Repair 2 (ERCC2)
3.4.4. Methylenetetrahydrofolate Reductase Gene (MTHFR)
3.5. Quality Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- World Health Organization: Regional Office for Europe. World Cancer Report: Cancer Research for Cancer Development; IARC: Lyon, France, 2020; ISBN 978-92-832-0447-3. [Google Scholar]
- European Medicines Agency (EMA). Xeloda® Summary of Product Information. Annex I; CHEPLAPHARM Arzneimittel GmbH: Greifswald, Germany, 2021. [Google Scholar]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised Colon Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D. Metastatic Colorectal Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2014, 25, iii1–iii9. [Google Scholar] [CrossRef]
- Bertholee, D.; Maring, J.G.; van Kuilenburg, A.B.P. Genotypes Affecting the Pharmacokinetics of Anticancer Drugs. Clin. Pharmacokinet. 2017, 56, 317–337. [Google Scholar] [CrossRef]
- Lam, S.W.; Guchelaar, H.J.; Boven, E. The Role of Pharmacogenetics in Capecitabine Efficacy and Toxicity. Cancer Treat. Rev. 2016, 50, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Amstutz, U.; Henricks, L.M.; Offer, S.M.; Barbarino, J.; Schellens, J.H.M.; Swen, J.J.; Klein, T.E.; McLeod, H.L.; Caudle, K.E.; Diasio, R.B.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 210–216. [Google Scholar] [CrossRef]
- Soria-Chacartegui, P.; Villapalos-García, G.; López-Fernández, L.A.; Navares-Gómez, M.; Mejía-Abril, G.; Abad-Santos, F.; Zubiaur, P. Clinical Relevance of Novel Polymorphisms in the Dihydropyrimidine Dehydrogenase (DPYD) Gene in Patients with Severe Fluoropyrimidine Toxicity: A Spanish Case-Control Study. Pharmaceutics 2021, 13, 2036. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.; Kubota, Y.; Ishida, H.; Sekido, M.; Ohkuma, R.; Ishiguro, T.; Hirasawa, Y.; Ariizumi, H.; Tsunoda, T.; Ikusue, T.; et al. Variants of Carboxylesterase 1 Have No Impact on Capecitabine Pharmacokinetics and Toxicity in Capecitabine plus Oxaliplatin Treated-Colorectal Cancer Patients. Cancer Chemother. Pharmacol. 2020, 85, 1119–1128. [Google Scholar] [CrossRef]
- Reigner, B.; Blesch, K.; Weidekamm, E. Clinical Pharmacokinetics of Capecitabine. Clin. Pharmacokinet. 2001, 40, 85–104. [Google Scholar] [CrossRef]
- Thorn, C.F.; Marsh, S.; Carrillo, M.W.; McLeod, H.L.; Klein, T.E.; Altman, R.B. PharmGKB Summary: Fluoropyrimidine Pathways. Pharm. Genom. 2011, 21, 237–242. [Google Scholar] [CrossRef]
- Lage, H. Gene Therapeutic Approaches to Overcome ABCB1-Mediated Drug Resistance. Recent Results Cancer Res. 2016, 209, 87–94. [Google Scholar] [CrossRef]
- Chockalingam, S.; Ghosh, S.S. Amelioration of Cancer Stem Cells in Macrophage Colony Stimulating Factor-Expressing U87MG-Human Glioblastoma upon 5-Fluorouracil Therapy. PLoS ONE 2013, 8, e83877. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Healthcare Interventions: Explanation and Elaboration. BMJ 2009, 339, b2700. [Google Scholar] [CrossRef] [PubMed]
- Little, J.; Higgins, J.P.T.; Ioannidis, J.P.A.; Moher, D.; Gagnon, F.; von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. STrengthening the REporting of Genetic Association Studies (STREGA): An Extension of the STROBE Statement. PLoS Med. 2009, 6, e22. [Google Scholar] [CrossRef]
- Andric, M.; Jacimovic, J.; Jakovljevic, A.; Nikolic, N.; Milasin, J. Gene Polymorphisms in Odontogenic Keratocysts and Ameloblastomas: A Systematic Review. Oral Dis. 2022, 28, 1421–1430. [Google Scholar] [CrossRef]
- Chair, S.Y.; Chan, J.Y.W.; Law, B.M.H.; Waye, M.M.Y.; Chien, W.T. Genetic Susceptibility in Pneumoconiosis in China: A Systematic Review. Int. Arch. Occup. Environ. Health 2022. [Google Scholar] [CrossRef] [PubMed]
- Chair, S.Y.; Law, B.M.H.; Chan, J.Y.W.; So, W.K.W.; Waye, M.M.Y. Association of Genetic Polymorphisms with Psychological Symptoms in Cancer: A Systematic Review. Asia-Pac. J. Oncol. Nurs. 2022, 9, 12–20. [Google Scholar] [CrossRef]
- van Huis-Tanja, L.H.; Gelderblom, H.; Punt, C.J.A.; Guchelaar, H.-J. MTHFR Polymorphisms and Capecitabine-Induced Toxicity in Patients with Metastatic Colorectal Cancer. Pharm. Genom. 2013, 23, 208–218. [Google Scholar] [CrossRef]
- Rosmarin, D.; Palles, C.; Pagnamenta, A.; Kaur, K.; Pita, G.; Martin, M.; Domingo, E.; Jones, A.; Howarth, K.; Freeman-Mills, L.; et al. A Candidate Gene Study of Capecitabine-Related Toxicity in Colorectal Cancer Identifies New Toxicity Variants at DPYD and a Putative Role for ENOSF1 Rather than TYMS. Gut 2015, 64, 111–120. [Google Scholar] [CrossRef] [PubMed]
- García-González, X.; Cortejoso, L.; García, M.I.; García-Alfonso, P.; Robles, L.; Grávalos, C.; González-Haba, E.; Marta, P.; Sanjurjo, M.; López-Fernández, L.A. Variants in CDA and ABCB1 Are Predictors of Capecitabine-Related Adverse Reactions in Colorectal Cancer. Oncotarget 2015, 6, 6422–6430. [Google Scholar] [CrossRef]
- Falvella, F.S.; Cheli, S.; Martinetti, A.; Mazzali, C.; Iacovelli, R.; Maggi, C.; Gariboldi, M.; Pierotti, M.A.; Di Bartolomeo, M.; Sottotetti, E.; et al. DPD and UGT1A1 Deficiency in Colorectal Cancer Patients Receiving Triplet Chemotherapy with Fluoropyrimidines, Oxaliplatin and Irinotecan. Br. J. Clin. Pharmacol. 2015, 80, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Sebio, A.; Salazar, J.; Páez, D.; Berenguer-Llergo, A.; Del Río, E.; Tobeña, M.; Martín-Richard, M.; Sullivan, I.; Targarona, E.; Balart, J.; et al. EGFR Ligands and DNA Repair Genes: Genomic Predictors of Complete Response after Capecitabine-Based Chemoradiotherapy in Locally Advanced Rectal Cancer. Pharm. J. 2015, 15, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, M.; García-González, X.; García, M.I.; Robles, L.; Grávalos, C.; García-Alfonso, P.; Pachón, V.; Longo, F.; Martínez, V.; Blanco, C.; et al. Identification of New SNPs Associated with Severe Toxicity to Capecitabine. Pharmacol. Res. 2017, 120, 133–137. [Google Scholar] [CrossRef]
- Pellicer, M.; García-González, X.; García, M.I.; Blanco, C.; García-Alfonso, P.; Robles, L.; Grávalos, C.; Rueda, D.; Martínez, J.; Pachón, V.; et al. Use of Exome Sequencing to Determine the Full Profile of Genetic Variants in the Fluoropyrimidine Pathway in Colorectal Cancer Patients Affected by Severe Toxicity. Pharmacogenomics 2017, 18, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Matevska-Geshkovska, N.; Staninova-Stojovska, M.; Kapedanovska-Nestorovska, A.; Petrushevska-Angelovska, N.; Panovski, M.; Grozdanovska, B.; Mitreski, N.; Dimovski, A. Influence of MSI and 18q LOH Markers on Capecitabine Adjuvant Monotherapy in Colon Cancer Patients. Pharmgenomics Pers. Med. 2018, 11, 193–203. [Google Scholar] [CrossRef]
- Varma, K.A.; Jayanthi, M.; Dubashi, B.; Shewade, D.G. Influence of DPYD*9A, DPYD*6 and GSTP1 Ile105val Genetic Polymorphisms on Capecitabine and Oxaliplatin (CAPOX) Associated Toxicities in Colorectal Cancer (CRC) Patients. Asian Pac. J. Cancer Prev. 2019, 20, 3093–3100. [Google Scholar] [CrossRef]
- Boige, V.; Mollevi, C.; Gourgou, S.; Azria, D.; Seitz, J.-F.; Vincent, M.; Bigot, L.; Juzyna, B.; Miran, I.; Gerard, J.-P.; et al. Impact of Single-Nucleotide Polymorphisms in DNA Repair Pathway Genes on Response to Chemoradiotherapy in Rectal Cancer Patients: Results from ACCORD-12/PRODIGE-2 Phase III Trial. Int. J. Cancer 2019, 145, 3163–3172. [Google Scholar] [CrossRef]
- Varma, A.; Mathaiyan, J.; Shewade, D.; Dubashi, B.; Sunitha, K. Influence of ABCB-1, ERCC-1 and ERCC-2 Gene Polymorphisms on Response to Capecitabine and Oxaliplatin (CAPOX) Treatment in Colorectal Cancer (CRC) Patients of South India. J. Clin. Pharm. Ther. 2020, 45, 617–627. [Google Scholar] [CrossRef]
- Puerta-García, E.; Urbano-Pérez, D.; Carrasco-Campos, M.I.; Pérez-Ramírez, C.; Segura-Pérez, A.; Calleja-Hernández, M.A.; Cañadas-Garre, M. Effect of DPYD, MTHFR, ABCB1, XRCC1, ERCC1 and GSTP1 on Chemotherapy Related Toxicity in Colorectal Carcinoma. Surg. Oncol. 2020, 35, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.-Q.; Wang, T.-M.; Zhang, J.-B.; He, Y.-Q.; Xue, W.-Q.; Wu, Z.-Y.; Yang, D.-W.; Cao, L.-J.; Huang, J.-W.; Li, X.-Z.; et al. Polymorphisms in TYMS for Prediction of Capecitabine-Induced Hand-Foot Syndrome in Chinese Patients with Colorectal Cancer. Cancer Res. Treat. 2021, 53, 724–732. [Google Scholar] [CrossRef]
- Serdjebi, C.; Milano, G.; Ciccolini, J. Role of Cytidine Deaminase in Toxicity and Efficacy of Nucleosidic Analogs. Expert Opin. Drug Metab. Toxicol. 2015, 11, 665–672. [Google Scholar] [CrossRef]
- Ding, X.; Chen, W.; Fan, H.; Zhu, B. Cytidine Deaminase Polymorphism Predicts Toxicity of Gemcitabine-Based Chemotherapy. Gene 2015, 559, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Q.; Zhuang, M.; Wang, Z.; Huber, R.M. Correlation between Dihydropyrimidine Dehydrogenase and Efficacy and Toxicity of Fluoropyrimidine Drugs. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 2772–2776. [Google Scholar]
- Gilbert, J.A.; Salavaggione, O.E.; Ji, Y.; Pelleymounter, L.L.; Eckloff, B.W.; Wieben, E.D.; Ames, M.M.; Weinshilboum, R.M. Gemcitabine Pharmacogenomics: Cytidine Deaminase and Deoxycytidylate Deaminase Gene Resequencing and Functional Genomics. Clin. Cancer Res. 2006, 12, 1794–1803. [Google Scholar] [CrossRef]
- Tibaldi, C.; Giovannetti, E.; Tiseo, M.; Leon, L.G.; D’Incecco, A.; Loosekoot, N.; Bartolotti, M.; Honeywell, R.; Cappuzzo, F.; Ardizzoni, A.; et al. Correlation of Cytidine Deaminase Polymorphisms and Activity with Clinical Outcome in Gemcitabine-/Platinum-Treated Advanced Non-Small-Cell Lung Cancer Patients. Ann. Oncol. 2012, 23, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, X.; Wang, X. The Impact of CDA A79C Gene Polymorphisms on the Response and Hematologic Toxicity in Gemcitabine-Treated Patients: A Meta-Analysis. Int. J. Biol. Markers 2014, 29, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, X.; Li, X.; Zhang, M.; Zhang, J.; Hou, D.; Tong, Z.; Dong, M. CDA and MTHFR Polymorphisms Are Associated with Clinical Outcomes in Gastroenteric Cancer Patients Treated with Capecitabine-Based Chemotherapy. Cancer Chemother. Pharmacol. 2019, 83, 939–949. [Google Scholar] [CrossRef]
- Meulendijks, D.; Rozeman, E.A.; Cats, A.; Sikorska, K.; Joerger, M.; Deenen, M.J.; Beijnen, J.H.; Schellens, J.H.M. Pharmacogenetic Variants Associated with Outcome in Patients with Advanced Gastric Cancer Treated with Fluoropyrimidine and Platinum-Based Triplet Combinations: A Pooled Analysis of Three Prospective Studies. Pharm. J. 2017, 17, 441–451. [Google Scholar] [CrossRef]
- Hryciuk, B.; Szymanowski, B.; Romanowska, A.; Salt, E.; Wasąg, B.; Grala, B.; Jassem, J.; Duchnowska, R. Severe Acute Toxicity Following Gemcitabine Administration: A Report of Four Cases with Cytidine Deaminase Polymorphisms Evaluation. Oncol. Lett. 2018, 15, 1912–1916. [Google Scholar] [CrossRef]
- Hu, L.; Mao, X.; Gao, C.; Xu, Y.; Li, C.; Wang, T.; Lv, D. Cytidine Deaminase 435C>T Polymorphism Relates to Gemcitabine-Platinum Efficacy and Hematological Toxicity in Chinese Non-Small-Cell Lung Cancer Patients. Neoplasma 2021, 68, 638–644. [Google Scholar] [CrossRef]
- Terrazzino, S.; Cargnin, S.; Del Re, M.; Danesi, R.; Canonico, P.L.; Genazzani, A.A. DPYD IVS14+1G>A and 2846A>T Genotyping for the Prediction of Severe Fluoropyrimidine-Related Toxicity: A Meta-Analysis. Pharmacogenomics 2013, 14, 1255–1272. [Google Scholar] [CrossRef] [PubMed]
- Meulendijks, D.; Henricks, L.M.; Sonke, G.S.; Deenen, M.J.; Froehlich, T.K.; Amstutz, U.; Largiadèr, C.R.; Jennings, B.A.; Marinaki, A.M.; Sanderson, J.D.; et al. Clinical Relevance of DPYD Variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as Predictors of Severe Fluoropyrimidine-Associated Toxicity: A Systematic Review and Meta-Analysis of Individual Patient Data. Lancet Oncol. 2015, 16, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Cho, Y.-A.; Kim, D.-C.; Lee, K.-E. Elevated Risk of Fluoropyrimidine-Associated Toxicity in European Patients with DPYD Genetic Polymorphism: A Systematic Review and Meta-Analysis. J. Pers. Med. 2022, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Božina, N.; Bilić, I.; Ganoci, L.; Šimičević, L.; Pleština, S.; Lešnjaković, L.; Trkulja, V. DPYD Polymorphisms c.496A>G, c.2194G>A and c.85T>C and Risk of Severe Adverse Drug Reactions in Patients Treated with Fluoropyrimidine-Based Protocols. Br. J. Clin. Pharmacol. 2022, 88, 2190–2202. [Google Scholar] [CrossRef]
- Ruzzo, A.; Graziano, F.; Galli, F.; Galli, F.; Rulli, E.; Lonardi, S.; Ronzoni, M.; Massidda, B.; Zagonel, V.; Pella, N.; et al. Dihydropyrimidine Dehydrogenase Pharmacogenetics for Predicting Fluoropyrimidine-Related Toxicity in the Randomised, Phase III Adjuvant TOSCA Trial in High-Risk Colon Cancer Patients. Br. J. Cancer 2017, 117, 1269–1277. [Google Scholar] [CrossRef]
- Del Re, M.; Cinieri, S.; Michelucci, A.; Salvadori, S.; Loupakis, F.; Schirripa, M.; Cremolini, C.; Crucitta, S.; Barbara, C.; Di Leo, A.; et al. DPYD*6 Plays an Important Role in Fluoropyrimidine Toxicity in Addition to DPYD*2A and c.2846A>T: A Comprehensive Analysis in 1254 Patients. Pharm. J. 2019, 19, 556–563. [Google Scholar] [CrossRef]
- Almashagbah, N.A.; Mahasneh, A.A.; Bodoor, K.G. Pharmacogenetic Study of the Dihydropyridine Dehydrogenase Gene in Jordanian Patients with Colorectal Cancer. Asian Pac. J. Cancer Prev. 2022, 23, 3061–3069. [Google Scholar] [CrossRef]
- Cordova-Delgado, M.; Bravo, M.L.; Cumsille, E.; Hill, C.N.; Muñoz-Medel, M.; Pinto, M.P.; Retamal, I.N.; Lavanderos, M.A.; Miquel, J.F.; Rodriguez-Fernandez, M.; et al. A Case-Control Study of a Combination of Single Nucleotide Polymorphisms and Clinical Parameters to Predict Clinically Relevant Toxicity Associated with Fluoropyrimidine and Platinum-Based Chemotherapy in Gastric Cancer. BMC Cancer 2021, 21, 1030. [Google Scholar] [CrossRef]
- Maharjan, A.S.; McMillin, G.A.; Patel, G.K.; Awan, S.; Taylor, W.R.; Pai, S.; Frankel, A.E.; Nelson, C.; Wang, B.; Hosein, P.J.; et al. The Prevalence of DPYD*9A(c.85T>C) Genotype and the Genotype-Phenotype Correlation in Patients with Gastrointestinal Malignancies Treated With Fluoropyrimidines: Updated Analysis. Clin. Color Cancer 2019, 18, e280–e286. [Google Scholar] [CrossRef]
- Joerger, M.; Huitema, A.D.R.; Boot, H.; Cats, A.; Doodeman, V.D.; Smits, P.H.M.; Vainchtein, L.; Rosing, H.; Meijerman, I.; Zueger, M.; et al. Germline TYMS Genotype Is Highly Predictive in Patients with Metastatic Gastrointestinal Malignancies Receiving Capecitabine-Based Chemotherapy. Cancer Chemother. Pharmacol. 2015, 75, 763–772. [Google Scholar] [CrossRef]
- Niu, Y.; Fan, X.; Wang, Y.; Lin, J.; Hua, L.; Li, X.; Qian, R.; Lu, C. Genome-Wide CRISPR Screening Reveals Pyrimidine Metabolic Reprogramming in 5-FU Chronochemotherapy of Colorectal Cancer. Front. Oncol. 2022, 12, 949715. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, X.; Li, X.; Wang, H.; Dong, M. Carboxylesterase 1 Polymorphisms Are Associated with Clinical Outcomes in Gastroenteric Cancer Patients Treated with Capecitabine. Cancer Chemother. Pharmacol. 2021, 87, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Lai, Y.; Rodrigues, A.D. Organic Anion Transporter 2: An Enigmatic Human Solute Carrier. Drug Metab. Dispos. 2017, 45, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Visscher, H.; Rassekh, S.R.; Sandor, G.S.; Caron, H.N.; van Dalen, E.C.; Kremer, L.C.; van der Pal, H.J.; Rogers, P.C.; Rieder, M.J.; Carleton, B.C.; et al. Genetic Variants in SLC22A17 and SLC22A7 Are Associated with Anthracycline-Induced Cardiotoxicity in Children. Pharmacogenomics 2015, 16, 1065–1076. [Google Scholar] [CrossRef]
- Ciszewski, W.M.; Chmielewska-Kassassir, M.; Wozniak, L.A.; Sobierajska, K. Thymidylate Synthase Overexpression Drives the Invasive Phenotype in Colon Cancer Cells. Biomedicines 2022, 10, 1267. [Google Scholar] [CrossRef]
- Wu, Q.; Dolnick, B.J. Detection of Thymidylate Synthase Modulators by a Novel Screening Assay. Mol. Pharmacol. 2003, 63, 167–173. [Google Scholar] [CrossRef]
- Hamzic, S.; Kummer, D.; Froehlich, T.K.; Joerger, M.; Aebi, S.; Palles, C.; Thomlinson, I.; Meulendijks, D.; Schellens, J.H.M.; García-González, X.; et al. Evaluating the Role of ENOSF1 and TYMS Variants as Predictors in Fluoropyrimidine-Related Toxicities: An IPD Meta-Analysis. Pharmacol. Res. 2020, 152, 104594. [Google Scholar] [CrossRef]
- Lin, S.; Yue, J.; Guan, X.; Yuan, P.; Wang, J.; Luo, Y.; Fan, Y.; Cai, R.; Li, Q.; Chen, S.; et al. Polymorphisms of MTHFR and TYMS Predict Capecitabine-Induced Hand-Foot Syndrome in Patients with Metastatic Breast Cancer. Cancer Commun. 2019, 39, 57. [Google Scholar] [CrossRef]
- Zhong, L.; Fu, Q.; Zhou, S.; Chen, L.; Peng, Q. Relevance of MTHFR Polymorphisms with Response to Fluoropyrimidine-Based Chemotherapy in Oesophagogastric Cancer: A Meta-Analysis. BMJ Open 2018, 8, e020767. [Google Scholar] [CrossRef]
- Ramos-Esquivel, A.; Chinchilla, R.; Valle, M. Association of C677T and A1298C MTHFR Polymorphisms and Fluoropyrimidine-Induced Toxicity in Mestizo Patients With Metastatic Colorectal Cancer. Anticancer Res. 2020, 40, 4263–4270. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-Fluorouracil: Mechanisms of Action and Clinical Strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, Y.; Luo, J.; Yuan, H.; Luo, Z. Genetic Polymorphisms and Platinum-Based Chemotherapy-Induced Toxicities in Patients With Lung Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2019, 9, 1573. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Li, H.K.; Shan, T.; Zhang, N. Genetic Variability of DNA Repair Mechanisms in Chemotherapy Treatment Outcome of Gastric Cancer Patients. Genet. Mol. Res. 2015, 14, 17228–17234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ge, J.; Hong, H.; Bi, L.; Sun, Z. Genetic Polymorphisms in ERCC1 and ERCC2 Genes Are Associated with Response to Chemotherapy in Osteosarcoma Patients among Chinese Population: A Meta-Analysis. World J. Surg. Oncol. 2017, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-N.; Liu, G.-F.; Li, X.-F.; Fu, B.-H.; Dong, L.-X.; Zhang, S.-H. Evaluation of Prediction of Polymorphisms of DNA Repair Genes on the Efficacy of Platinum-Based Chemotherapy in Patients With Non-Small Cell Lung Cancer: A Network Meta-Analysis. J. Cell Biochem. 2017, 118, 4782–4791. [Google Scholar] [CrossRef]
- Qian, Y.-Y.; Liu, X.-Y.; Wu, Q.; Song, X.; Chen, X.-F.; Liu, Y.-Q.; Pei, D.; Shen, L.-Z.; Shu, Y.-Q. The ERCC1 C118T Polymorphism Predicts Clinical Outcomes of Colorectal Cancer Patients Receiving Oxaliplatin-Based Chemotherapy: A Meta-Analysis Based on 22 Studies. Asian Pac. J. Cancer Prev. 2014, 15, 8383–8390. [Google Scholar] [CrossRef] [PubMed]
- De Mattia, E.; Cecchin, E.; Toffoli, G. Pharmacogenomics of Intrinsic and Acquired Pharmacoresistance in Colorectal Cancer: Toward Targeted Personalized Therapy. Drug Resist. Updates 2015, 20, 39–70. [Google Scholar] [CrossRef]
- Zeng, H.; Yu, H.; Lu, L.; Jain, D.; Kidd, M.S.; Saif, M.W.; Chanock, S.J.; Hartge, P.; PanScan Consortium; Risch, H.A. Genetic Effects and Modifiers of Radiotherapy and Chemotherapy on Survival in Pancreatic Cancer. Pancreas 2011, 40, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Peethambaram, P.; Fridley, B.L.; Vierkant, R.A.; Larson, M.C.; Kalli, K.R.; Elliott, E.A.; Oberg, A.L.; White, K.L.; Rider, D.N.; Keeney, G.L.; et al. Polymorphisms in ABCB1 and ERCC2 Associated with Ovarian Cancer Outcome. Int. J. Mol. Epidemiol. Genet. 2011, 2, 185–195. [Google Scholar]

| First Author (Year) Country | Study Design | Clinical Data Collection | Ethnicity | N | Median Age (Range) | Women (%) | CRC Stage | Treatment Regimens | Treatment Setting | Genes of Interest Investigated | Object of Study | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Van Huis-Tanja LH (2013) Netherlands | Cohort of an RCT | Prospective | European ancestry | 126 | 61 (27–78) | 49 (39) | IV | CAPE | MX | MTHFR | Effectiveness/Toxicity | [20] |
| Rosmarin D (2015) UK | Cohort of an RCT | Prospective | European ancestry | 940 | 65 (22–85) | 453 (43) | II–III | CAPE, CAPE-B | ADJ | ABCC3, ABCC4, ABCC5, ABCG2, ABCB1, CDA, CES1, CES2, DPYD, DPYS, MTHFR, PPAT, RRM1, RRM2, SLC22A7, SLC29A1, TK1, TYMP, TYMS, UCK1, UCK2, UMPS, UPB1, UPP1, UPP2 | Toxicity | [21] |
| García-González X (2015) Spain | Cohort | Ambispective | NR | 239 | 67 (30–88) | 110 (46) | I–IV | CAPE, CAPOX, CAPIRI, CAPE-AB | ADJ, MX | ABCB1, CDA, ENOSF1, MTHFR, TYMS | Toxicity | [22] |
| Falvella FS (2015) Italy | Cohort of 2 RCTs | Prospective | European ancestry | 64 | 57 (34–73) | 25 (39) | IV | CAPOXIRI-B, CAPOXIRI-CETU | MX | DPYD | Toxicity | [23] |
| Sebio A (2015) Spain | Cohort | NR | European ancestry | 84 | 68 (80–42) | 29 (35) | II–III | CAPE-RT | NEOADJ | ERCC1, ERCC2, TYMS, XRCC1 | Effectiveness | [24] |
| Pellicer M (2017a) Spain | Cross- sectional | Ambispective | European ancestry | 301 | 65 (30–88) | 145 (48) | I–IV | CAPE, CAPOX, CAPIRI, CAPE-AB | NEOADJ, ADJ, MX | CDA, CES1, DPYD, ENOSF1, SLC22A7, TYMP, TYMS, TYMS/ENOSF1, ENOSF1, UMPS | Toxicity | [25] |
| Pellicer M (2017b) Spain | Cohort | NR | European ancestry | 319 | 65 (30–88) | 151 (47) | I–IV | CAPE, CAPOX, CAPIRI, CAPE-AB | NEOADJ, ADJ, MX | ABCC4, DPYD, MTHFR | Toxicity | [26] |
| Matevska-Geshkovska, N (2018) Macedonia | OLCT | Prospective | NR | 126 | 60 (36–81) | 50 (45) | II–III | CAPE | ADJ | MTHFR, TYMS | Effectiveness | [27] |
| Varma A (2019) India | Cohort | Prospective | Asian: Tamilian (76.5%), Andhra (14.4%), N. Indians (9.6%) | 145 | 50 (NR) | 55 (38) | II–IV | CAPOX | NEOADJ, ADJ, P | DPYD | Toxicity | [28] |
| Boige V (2019) France | Cohort of an RCT | Prospective | European ancestry | 316 | 61 (35–79) | 104 (33) | II–III | CAPOX-RT, CAPE-RT | NEOADJ | ERCC1, ERCC2, MTHFR, TYMS, XRCC1, XRCC3 | Effectiveness/Toxicity | [29] |
| Varma A (2020) India | Cohort | Prospective | Asian: Tamilian (76.5%), Andhra (14.4%), Kerala (9.6%) | 145 | 50 (NR) | 55 (38) | II–IV | CAPOX | ADJ | ABCB1, ERCC1, ERCC2 | Effectiveness | [30] |
| Puerta-García E (2020) Spain | Cohort | Retrospective | European ancestry | 84 | 68 (60–72) | 30 (35) | I–IV | CAPE | ADJ | ABCB1, DPYD, MTHFR, ERCC1, XRCC1 | Toxicity | [31] |
| Dong SQ (2021) China | Case-control | Retrospective | Asian | 288 | 59 (27–83) | 108 (38) | I–IV | CAPE, CAPOX, CAPIRI, CAPE-AB | NEOADJ, ADJ, MX | TYMS | Toxicity | [32] |
| Variant rs Number | SNP Position | Variant Type/Consequence | Associated Genotype/Allele vs. Reference | Toxicity | Ref. | ||
|---|---|---|---|---|---|---|---|
| Grade (Type) | p Value a | OR (95% CI) | |||||
| Gene CDA | |||||||
| rs2072671 | c.79A>C | missense | AA vs. AC-CC | 3–4 (diarrhea) 2–4 (HFS) 3–4 (HFS) 3–4 (hematological) 3–4 (asthenia) 3–4 (overall) | 0.157 0.163 0.066 0.531 0.566 0.029 | 1.83 (0.79–4.24) 1.56 (0.83–2.94) 2.89 (0.93–8.98) 1.38 (0.50–3.80) 1.40 (0.44–4.49) 1.84 (1.06–3.18) | [22] |
| AC-CC vs. AA | 3–4 (HFS) 3–4 (overall) | 0.008 0.007 | 0.27 (0.10–0.71) 0.50 (0.30–0.83) | [25] | |||
| rs1048977 | c.435C>T | synonymous | CT-TT vs. CC | 3–4 (hyperbilirubinemia) | 0.044 | 8.62 (1.05–70.24) | [25] |
| DPYD | |||||||
| rs3918290 | c.IVS14+1G>A | splice donor | AG vs. GG | 3–4 (overall) | 0.179 b | 3.02 (0.50–18.15) | [21] |
| rs55886062 | 1679T>G | missense | GT vs. TT | 3–4 (overall) | 0.697 b | 4.02 (0.36–44.47) | [21] |
| rs67376798 | c.2846A>T | missense | AT vs. AA | 3–4 (overall) | 0.001 b | 8.17 (1.73–38.70) | [21] |
| AT vs. AA | 3–4 (overall) 1–4 (GI) 1–4 (cardiovascular) 1–4 (asthenia) 1–4 (cutaneous) 1–4 (respiratory) | 0.287 c 1.000 c 1.000 c 0.250 c 0.412 c 0.125 d | U U 0.00 (0.00–NaN) U U U | [31] | |||
| rs56038477 | c.1236G>A | synonymous | AG vs. GG | 3–4 (overall) | 0.008 b | 2.73 (1.38–5.41) | [21] |
| rs1801160 | c.2194G>A | missense | AG-AA vs. GG | 3–4 (overall) | 0.827 b | 1.16 (0.69–1.96) | [21] |
| AG-AA vs. GG | 2–4 (overall) | 0.029 | 2.11 (1.08–4.13) | [26] | |||
| AG-AA vs. GG | 1–4 (anemia) 1–4 (thrombocytopenia) 1–4 (neutropenia) 1–4 (vomiting) 1–4 (diarrhea) 1–4 (HFS) 1–4 (PN) | 0.800 e 0.600 e 0.200 e 0.900 e 0.100 e 0.300 e 0.700 e | 1.90 (0.40–2.60) 1.20 (0.50–3.10) 1.70 (0.60–1.70) 1.00 (0.40–2.00) 1.80 (0.70–2.00) 0.60 (0.20–1.00) 1.10 (0.40–2.00) | [28] | |||
| rs1801265 | c.85T>C | missense | CT-CC vs. TT | 3–4 (overall) | 0.078 b | 0.76 (0.58–1.01) | [21] |
| CT-CC vs. TT | 1–4 (anemia) 1–4 (thrombocytopenia) 1–4 (neutropenia) 1–4 (vomiting) 1–4 (diarrhea) 1–4 (HFS) 1–4 (PN) | 0.800 e 0.010 e 0.500 e 0.060 e 0.040 e 0.020 e 0.900 e | 0.90 (0.40–1.80) 2.40 (1.18–5.10) 1.24 (0.50–2.90) 1.00 (0.90–4.00) 2.70 (1.80–4.00) 2.30 (1.80–4.00) 0.90 (0.40–2.00) | [28] | |||
| rs1801158 | c.1601G>A | missense | AG vs. GG | 3–4 (overall) | 0.368 b | 1.38 (0.73–2.59) | [21] |
| rs1801159 | c.1627A>G | missense | AG-GG vs. AA | 3–4 (overall) | 0.560 b | 1.03 (0.77–1.36) | [21] |
| rs2297595 | c.496A>G | missense | AG-GG vs. AA | 3–4 (overall) | 0.415 b | 0.92 (0.64–1.31) | [21] |
| AG vs. AA | 3–4 (overall) | 0.022 | 5.94 (1.29–27.22) | [23] | |||
| rs12022243 | c.1906-14763G>A | intron | A vs. G | 3–4 (overall) 3–4 (HFS) 3–4 (diarrhea) | 2.55 × 10−5 b 0.009 b 9.86 × 10−6 b | 1.69 (1.45–1.94) 1.43 (1.16–1.70) 1.79 (1.54–2.05) | [21] |
| rs7548189 | C.1906-19696G>T | intron | T vs. G | 3–4 (overall) 3–4 (HFS) 3–4 (diarrhea) 2–4 (diarrhea) | 3.79 × 10−5 b 0.011 b 0.001 b 1.72 × 10−5 b | 1.67 (1.43–1.91) 1.42 (1.15–1.69) 1.21 (0.84–1.58) 1.76 (1.50–2.02) | [21] |
| rs45589337 | c.775A>G | missense | AG vs. AA | 3–4 (overall) | 0.723 b | 0.80 (0.25–2.56) | [21] |
| rs76387818 | g.97539400G>A | – | A vs. G | 3–4 (overall) 3–4 (HFS) 3–4 (diarrhea) | 2.11 × 10−6 b 1.75 × 10−8 b 0.071 b | 4.05 (3.47–4.62) 6.44 (5.79–7.09) 0.44 (0.00–1.33) | [21] |
| rs12132152 | g.97523004G>A | – | A vs. G | 3–4 (overall) 3–4 (HFS) 3–4 (diarrhea) | 4.31 × 10−6 b 3.29 × 10−8 b 0.065 b | 3.83 (3.26–4.40) 6.12 (5.48–6.76) 0.44 (0.00–1.32) | [21] |
| rs17376848 | 1896T>C | synonymous | CT vs. TT | 3–4 (overall) | 0.027 | 14.53 (1.36–155.20) | [23] |
| CT vs. TT | 3–4 (overall) 1–4 (GI) 1–4 (cardiovascular) 1–4 (asthenia) 1–4 (skin) 1–4 (respiratory) | 0.494c 1.000 c 1.000 c 1.000 c 1.000 c 1.000 c | 2.51 (0.03–202.96) U 0.00 (0.00–NaN) 0.00 (0.00–NaN) 1.43 (0.02–115.15) 0.00 (0.00–NaN) | [31] | |||
| rs12119882 | c.680+2545T>C | intron | CT-CC vs. TT | 3–4 (hyperbilirubinemia) | 0.031 | 4.86 (1.16–20.38) | [25] |
| UMPS | |||||||
| rs4678145 | c.156+607G>C | intron | CG-CC vs. GG | 3–4 (asthenia) | 0.006 | 4.54 (1.55–13.24) | [25] |
| rs2279199 | c.-67T>C | 2KB upstream | CT-CC vs. TT | 3–4 (nausea and vomiting) | 0.036 | 0.21 (0.04–0.90) | [25] |
| SLC22A7 | |||||||
| rs2270860 | 1269C>T | synonymous | TT vs. CT-CC | 3–4 (skin) | 0.016 | 17.08 (1.71–170.26) | [25] |
| rs4149178 | 1592+206A>G | intron | AG-GG vs. AA | 3–4 (diarrhea) | 0.034 | 0.34 (0.12–0.92) | [25] |
| ABCB1 | |||||||
| rs1128503 | c.1236T>C | synonymous | CC vs. CT-TT | 3–4 (overall) 1–4 (GI) 1–4 (cardiovascular) 1–4 (asthenia) 1–4 (skin) 1–4 (respiratory) | 0.044 d 0.643 d 0.562 d 0.372 d 0.402 d 1.000 c | 0.22 (0.02–1.11) 0.77 (0.24–2.72) 1.63 (0.03–33.00) 0.49 (0.08–2.04) 0.66 (0.22–1.92) 0.88 (0.13–4.30) | [31] |
| TYMS | |||||||
| rs2853741 | c.-391T>C | 2KB upstream | CC vs. CT-TT | 3–4 (diarrhea) | 0.008 | 0.31 (0.13–0.74) | [25] |
| rs3786362 | c.381A>G | synonymous | AA vs. AG vs. GG | 2–3 (HFS) | 1.89 × 10−3 | 0.38 (0.21–0.70) | [32] |
| TYMS/ENOSF1 | |||||||
| rs699517 | c.*19C>T | 3′UTR/noncoding transcript | TT vs. CT-CC | 3–4 (nausea and vomiting) 3–4 (anorexia) | 0.014 0.006 | 7.93 (1.51–41.63) 128.82 (4.16–3988.96) | [25] |
| CT-TT vs. CC | 3–4 (asthenia) | 0.021 | 0.24 (0.07–0.81) | [25] | |||
| CC vs. CT vs. TT | 2–3 (HFS) | 4.62 × 10−4 | 2.12 (1.39–3.24) | [32] | |||
| rs2790 | c.*89A>G | 3′UTR/intron | AA vs. AG vs. GG | 2–3 (HFS) | 8.80 × 10−3 | 0.58 (0.39–0.87) | [32] |
| ENOSF1 | |||||||
| rs2612091 | c.496-227G>A | intron | G vs. A | 3–4 (overall) 3–4 (HFS) 3–4 (diarrhea) | 5.28 × 10−6 b 2.94 × 10−6 b 0.290 b | 1.59 (1.39–1.79) 1.57 (–) 1.18 (0.55–1.15) | [21] |
| GG vs. GA-AA | 3–4 (diarrhea) 2–4 (HFS) 3–4 (HFS) 3–4 (hematological) 3–4 (asthenia) 3–4 (overall) | 0.431 0.027 0.114 0.541 0.063 0.789 | 0.60 (0.17–2.12) 2.28 (1.10–4.76) 2.53 (0.80–8.02) 0.62 (0.14–2.84) 3.15 (0.94–10.57) 0.91 (0.45–1.82) | [22] | |||
| rs2741171 | c.63+5783A>G | intron | A vs. G | 3–4 (overall) 3–4 (HFS) 3–4 (diarrhea) | 6.64 × 10−6 b 1.64 × 10−6 b 0.920 b | 1.60 (1.39–1.80) 1.74 (1.51–1.97) 1.01 (0.70–1.32) | [21] |
| MTHFR | |||||||
| rs1801131 | c.1286A>C | missense | CC vs. AC-AA | 3–4 (overall) 3–4 (diarrhea) 3–4 (HFS) | 0.355 f 0.041 f 0.406 f | 1.85 (0.55–6.11) 6.00 (1.28–28.09) 1.90 (0.47–7.75) | [20] |
| CC vs. AC-AA | 3–4 (overall) 1–4 (GI) 1–4 (cardiovascular) 1–4 (asthenia) 1–4 (skin) 1–4 (respiratory) | 0.529 d 1.000 c 1.000 c 0.741 c 0.464 d 0.682 c | 1.47 (0.34–5.73) 1.24 (0.31–6.07) – 1.24 (0.25–5.12) 1.52 (0.40–5.79) 0.49 (0.01–4.13) | [31] | |||
| rs1801133 | c.665C>T | missense | TT vs. CT-CC | 3–4 (overall) 3–4 (diarrhea) 3–4 (HFS) | 0.770 f 0.596 f 0.237 f | 1.35 (0.44–4.17) 0.00 (0.00–NaN) 2.40 (0.67–8.59) | [20] |
| TT vs. CT-CC | 3–4 (overall) 1–4 (GI) 1–4 (cardiovascular) 1–4 (asthenia) 1–4 (skin) 1–4 (respiratory) | 0.403 c 0.676 c 1.000 c 0.009 c 0.693 c 0.209 c | 1.95 (0.26–12.79) 0.61 (0.09–4.56) – 9.30 (1.36–106.8) 0.55 (0.05–3.61) 3.18 (0.26–23.9) | [31] | |||
| Variant rs Number | SNP Position | Variant Type/Consequence | Associated Genotype/Allele vs. Reference | Effectiveness Outcomes | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| PFS | Response | OS | ||||||||
| p Value a | HR (95% CI) | p Value c | OR (95% CI) | p Value | HR (95% CI) | |||||
| Gene | ||||||||||
| ABCB1 | ||||||||||
| rs1128503 | c.1236T>C | synonymous | CT-CC vs. TT | - | – | 0.040 d | 3.70 (0.70–19.00) | – | – | [30] |
| rs1045642 | c.3435T>C | synonymous | CT-CC vs. TT | - | – | 0.050 d | 3.10 (0.80–13.00) | – | – | [30] |
| ERCC1 | ||||||||||
| rs11615 | c.354T>C | synonymous | CC vs. CT-TT | - | – | 0.023 | NE | – | – | [24] |
| CT-CC vs. TT | - | – | 0.300 d | 0.50 (0.10–2.00) | – | – | [30] | |||
| rs10412761 | g.45908461A>G | – | AG-GG vs. AA | - | – | 0.042 | 0.57 (0.34–0.98) | 0.160 | 1.47 (0.85–2.56) | [29] |
| ERCC2 | ||||||||||
| rs13181 | c.2251A>C | stop gained | AC-CC vs. AA | - | – | – | – | 0.235 | 0.73 (0.43–1.22) | [29] |
| AC-CC vs. AA | - | – | 0.500 d | 0.80 (0.10–4.00) | – | – | [30] | |||
| rs1799787 | c.1832-70C>T | intron | CT-TT vs. CC | - | – | 0.027 | 0.55 (0.33–0.93) | 0.276 | 0.75 (0.45–1.25) | [29] |
| MTHFR | ||||||||||
| rs1801131 | 1286A>C | missense | CC vs. AC-AA | 0.904 b | – | 0.691 | – | 0.758 | – | [20] |
| rs1801133 | 665C>T | missense | TT vs. CT-TT | 0.807 b | – | 0.127 | – | 0.270 | – | [20] |
| TT vs. CT-TT | 0.225 | 0.29 (0.04–2.13) | – | – | – | – | [27] | |||
| rs7553194 | c.-578C>T | noncoding transcript | CT-TT vs. CC | - | – | – | – | 0.108 | 0.49 (0.20–1.26) | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cura, Y.; Pérez-Ramírez, C.; Sánchez-Martín, A.; Membrive-Jimenez, C.; Valverde-Merino, M.I.; González-Flores, E.; Morales, A.J. Influence of Single-Nucleotide Polymorphisms on Clinical Outcomes of Capecitabine-Based Chemotherapy in Colorectal Cancer Patients: A Systematic Review. Cancers 2023, 15, 1821. https://doi.org/10.3390/cancers15061821
Cura Y, Pérez-Ramírez C, Sánchez-Martín A, Membrive-Jimenez C, Valverde-Merino MI, González-Flores E, Morales AJ. Influence of Single-Nucleotide Polymorphisms on Clinical Outcomes of Capecitabine-Based Chemotherapy in Colorectal Cancer Patients: A Systematic Review. Cancers. 2023; 15(6):1821. https://doi.org/10.3390/cancers15061821
Chicago/Turabian StyleCura, Yasmin, Cristina Pérez-Ramírez, Almudena Sánchez-Martín, Cristina Membrive-Jimenez, María Isabel Valverde-Merino, Encarnación González-Flores, and Alberto Jiménez Morales. 2023. "Influence of Single-Nucleotide Polymorphisms on Clinical Outcomes of Capecitabine-Based Chemotherapy in Colorectal Cancer Patients: A Systematic Review" Cancers 15, no. 6: 1821. https://doi.org/10.3390/cancers15061821
APA StyleCura, Y., Pérez-Ramírez, C., Sánchez-Martín, A., Membrive-Jimenez, C., Valverde-Merino, M. I., González-Flores, E., & Morales, A. J. (2023). Influence of Single-Nucleotide Polymorphisms on Clinical Outcomes of Capecitabine-Based Chemotherapy in Colorectal Cancer Patients: A Systematic Review. Cancers, 15(6), 1821. https://doi.org/10.3390/cancers15061821








