Evolving Risk Classifications in AML in a Real-Life Scenario: After Changes upon Changes, Is It More and More Adverse?
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Methods
2.3. Statistical Analyses
3. Results
3.1. Baseline Patient Characteristics
3.2. Mutation Characteristics and Distribution
3.3. Evolving Prognostic Risk Classifications
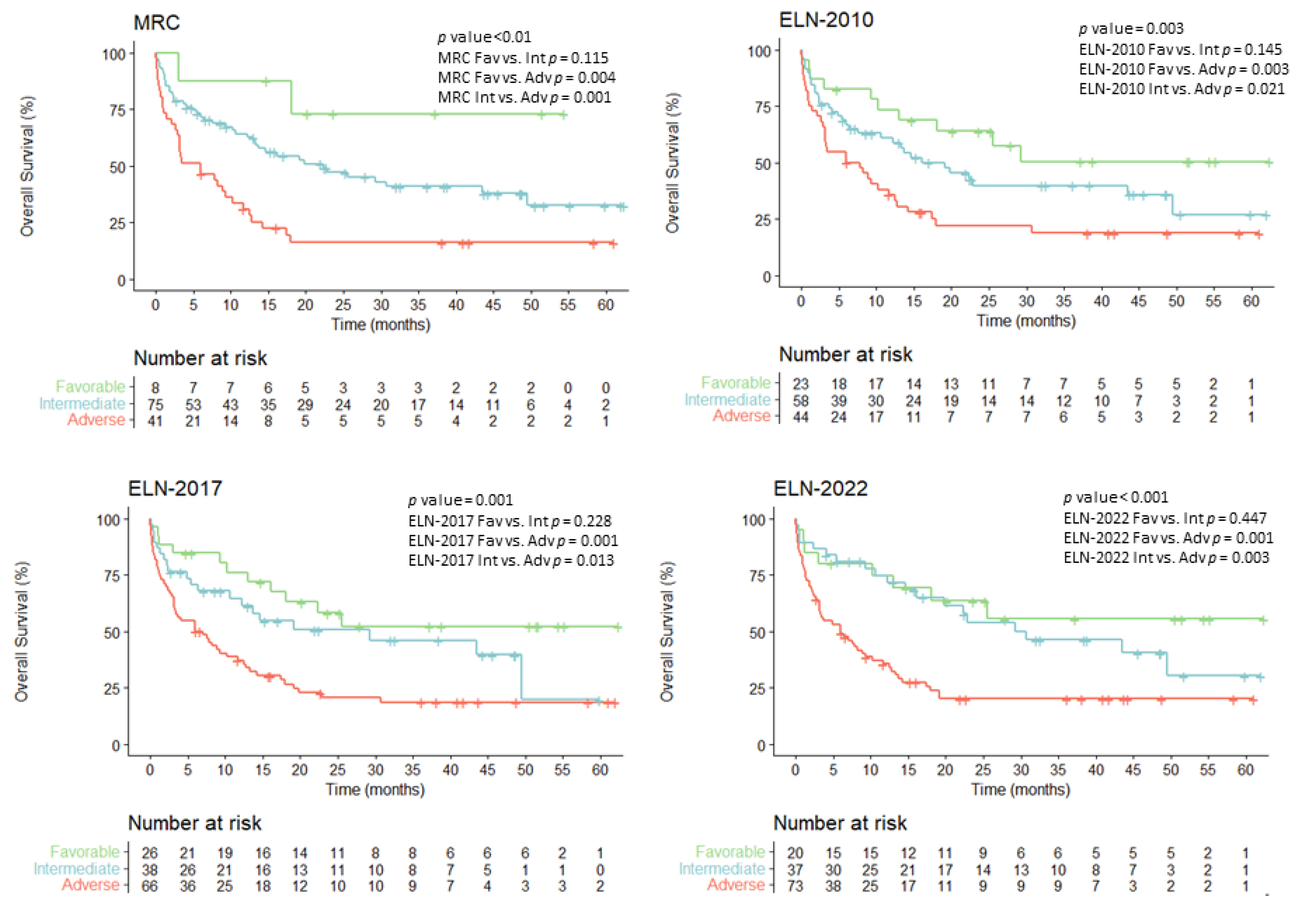
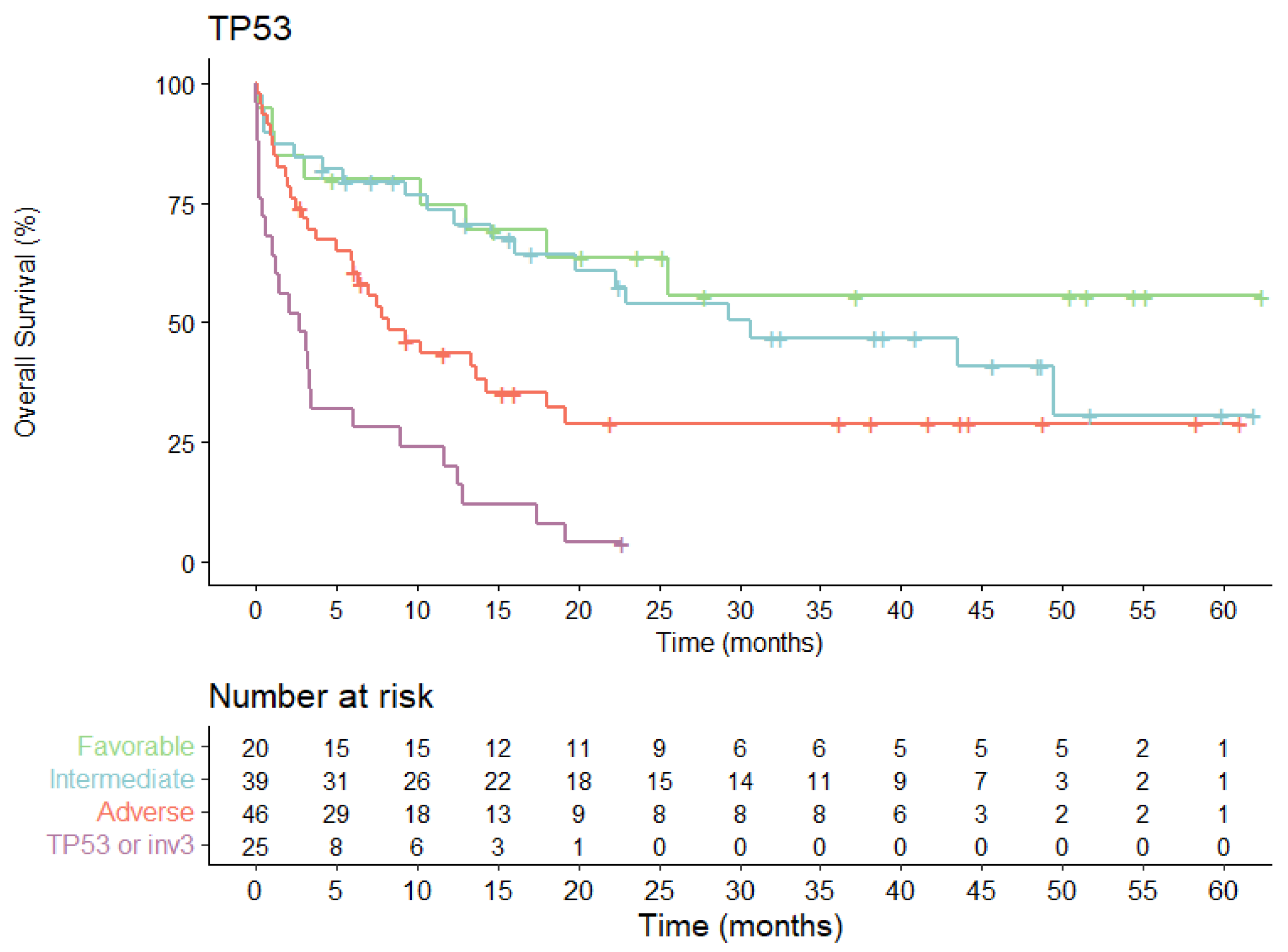
3.4. Overall Survival According to Evolving AML Risk Categories
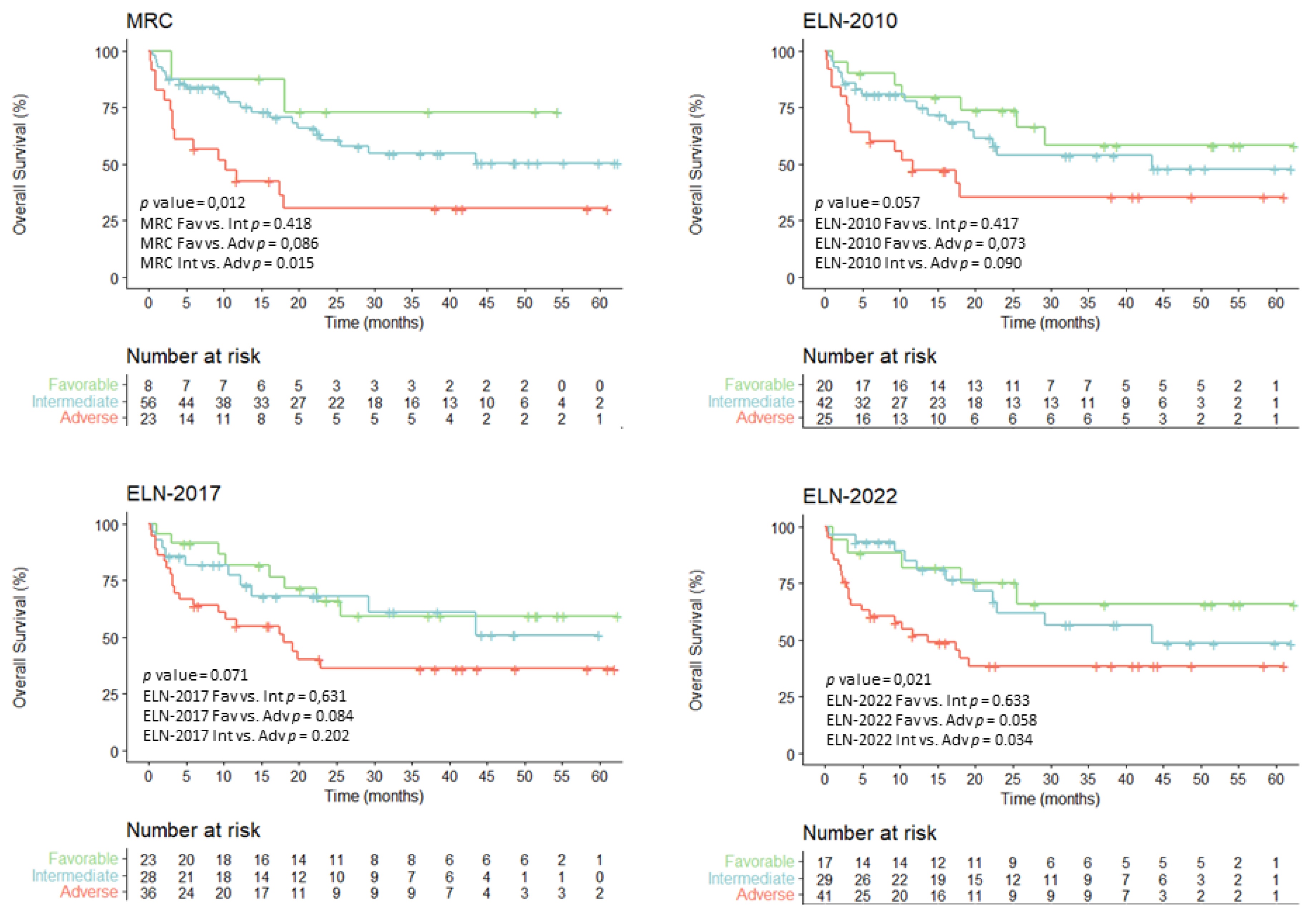
3.5. Overall Survival for Intensively Treated Patients According to Evolving AML Risk Categories
3.6. Univariate and Multivariate Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shallis, R.M.; Wang, R.; Davidoff, A.; Ma, X.; Zeidan, A.M. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 2019, 36, 70–87. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.; Kadia, T.; DiNardo, C.; Daver, N.; Borthakur, G.; Jabbour, E.; Garcia-Manero, G.; Konopleva, M.; Ravandi, F. Acute myeloid leukemia: Current progress and future directions. Blood Cancer J. 2021, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, D.; Hills, R.K.; Moorman, A.V.; Walker, H.; Chatters, S.; Goldstone, A.H.; Wheatley, K.; Harrison, C.J.; Burnett, A.K.; National Cancer Research Institute Adult Leukaemia Working Group. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010, 116, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A.; et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Wouters, B.J.; Löwenberg, B.; Erpelinck-Verschueren, C.A.; van Putten, W.L.; Valk, P.J.; Delwel, R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009, 113, 3088–3091. [Google Scholar] [CrossRef]
- Bataller, A.; Garrido, A.; Guijarro, F.; Oñate, G.; Diaz-Beyá, M.; Arnan, M.; Tormo, M.; Vives, S.; de Llano, M.P.Q.; Coll, R.; et al. European LeukemiaNet 2017 risk stratification for acute myeloid leukemia: Validation in a risk-adapted protocol. Blood Adv. 2022, 6, 1193–1206. [Google Scholar] [CrossRef]
- Herold, T.; Rothenberg-Thurley, M.; Grunwald, V.V.; Janke, H.; Goerlich, D.; Sauerland, M.C.; Konstandin, N.P.; Dufour, A.; Schneider, S.; Neusser, M.; et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia 2020, 34, 3161–3172. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and Management of AML in Adults: 2022 ELN Recommendations from an International Expert Panel. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef]
- Lachowiez, C.A.; Long, N.; Saultz, J.N.; Gandhi, A.; Newell, L.F.; Hayes-Lattin, B.; Maziarz, R.T.; Leonard, J.; Bottomly, D.; McWeeney, S.K.; et al. Comparison and validation of the 2022 European LeukemiaNet guidelines in acute myeloid leukemia. Blood Adv. 2022. [Google Scholar] [CrossRef]
- Sargas, C.; Ayala, R.; Chillón, M.C.; Larráyoz, M.J.; Carrillo-Cruz, E.; Bilbao, C.; Yébenes-Ramírez, M.; Llop, M.; Rapado, I.; García-Sanz, R.; et al. Networking for advanced molecular diagnosis in acute myeloid leukemia patients is possible: The PETHEMA NGS-AML project. Haematologica 2020, 106, 3079–3089. [Google Scholar] [CrossRef]
- Rodríguez-Arbolí, E.; Martínez-Cuadrón, D.; Rodríguez-Veiga, R.; Carrillo-Cruz, E.; Gil-Cortés, C.; Serrano-López, J.; Bernal Del Castillo, T.; Martínez-Sánchez, M.D.P.; Rodríguez-Medina, C.; Vidriales, B.; et al. Long-Term Outcomes After Autologous versus Allogeneic Stem Cell Transplantation in Molecularly-Stratified Patients With Intermediate Cytogenetic Risk Acute Myeloid Leukemia: A PETHEMA Study. Transpl. Cell Ther. 2021, 27, e311. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Cheson, B.D.; Bennett, J.M.; Kopecky, K.J.; Büchner, T.; Willman, C.L.; Estey, E.H.; Schiffer, C.A.; Doehner, H.; Tallman, M.S.; Lister, T.A.; et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J. Clin. Oncol. 2003, 21, 4642–4649. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemia: Integrating Morphological, Clinical, and Genomic Data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Metzeler, K.H.; Herold, T.; Rothenberg-Thurley, M.; Amler, S.; Sauerland, M.C.; Görlich, D.; Schneider, S.; Konstandin, N.P.; Dufour, A.; Bräundl, K.; et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 2016, 128, 686–698. [Google Scholar] [CrossRef]
- Gindin, T.; Hsiao, S.J. Analytical Principles of Cancer Next Generation Sequencing. Clin. Lab. Med. 2022, 42, 395–408. [Google Scholar] [CrossRef]
- Tarlock, K.; Lamble, A.J.; Wang, Y.C.; Gerbing, R.B.; Ries, R.E.; Loken, M.R.; Brodersen, L.E.; Pardo, L.; Leonti, A.; Smith, J.L.; et al. CEBPA-bZip mutations are associated with favorable prognosis in de novo AML: A report from the Children’s Oncology Group. Blood 2021, 138, 1137–1147. [Google Scholar] [CrossRef]
- Taube, F.; Georgi, J.A.; Kramer, M.; Stasik, S.; Middeke, J.M.; Röllig, C.; Krug, U.; Krämer, A.; Scholl, S.; Hochhaus, A.; et al. CEBPA mutations in 4708 patients with acute myeloid leukemia: Differential impact of bZIP and TAD mutations on outcome. Blood 2022, 139, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Ivey, A.; Hills, R.K.; Simpson, M.A.; Jovanovic, J.V.; Gilkes, A.; Grech, A.; Patel, Y.; Bhudia, N.; Farah, H.; Mason, J.; et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N. Engl. J. Med. 2016, 374, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Bornhäuser, M.; Schliemann, C.; Schetelig, J.; Röllig, C.; Kramer, M.; Glass, B.; Platzbecker, U.; Burchert, A.; Hänel, M.; Müller, L.P.; et al. Allogeneic Hematopoietic Cell Transplantation vs Standard Consolidation Chemotherapy in Patients With Intermediate-Risk Acute Myeloid Leukemia: A Randomized Clinical Trial. JAMA Oncol. 2023, e227605. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Y.; Wang, Q.; Chen, L.; Ma, L.; Hao, S. Autologous Stem Cell Transplantation Is a Viable Postremission Therapy for Intermediate-Risk Acute Myeloid Leukemia in First Complete Remission in the Absence of a Matched Identical Sibling: A Meta-Analysis. Acta Haematol. 2019, 141, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Ayala, R.; Carreño-Tarragona, G.; Barragán, E.; Boluda, B.; Larráyoz, M.J.; Chillón, M.C.; Carrillo-Cruz, E.; Bilbao, C.; Sánchez-García, J.; Bernal, T.; et al. Impact of FLT3-ITD Mutation Status and Its Ratio in a Cohort of 2901 Patients Undergoing Upfront Intensive Chemotherapy: A PETHEMA Registry Study. Cancers 2022, 14, 5799. [Google Scholar] [CrossRef]
- Larson, R.A.; Mandrekar, S.J.; Huebner, L.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Döhner, K.; et al. Midostaurin reduces relapse in FLT3-mutant acute myeloid leukemia: The Alliance CALGB 10603/RATIFY trial. Leukemia 2021, 35, 2539–2551. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Döhner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Jentzsch, M.; Bischof, L.; Ussmann, J.; Backhaus, D.; Brauer, D.; Metzeler, K.H.; Merz, M.; Vucinic, V.; Franke, G.N.; Herling, M.; et al. Prognostic impact of the AML ELN2022 risk classification in patients undergoing allogeneic stem cell transplantation. Blood Cancer J. 2022, 12, 170. [Google Scholar] [CrossRef]
- Shahzad, M.; Tariq, E.; Chaudhary, S.G.; Anwar, I.; Iqbal, Q.; Fatima, H.; Abdelhakim, H.; Ahmed, N.; Balusu, R.; Hematti, P.; et al. Outcomes with allogeneic hematopoietic stem cell transplantation in TP53-mutated acute myeloid leukemia: A systematic review and meta-analysis. Leuk Lymphoma 2022, 63, 3409–3417. [Google Scholar] [CrossRef]
- Daver, N.G.; Maiti, A.; Kadia, T.M.; Vyas, P.; Majeti, R.; Wei, A.H.; Garcia-Manero, G.; Craddock, C.; Sallman, D.A.; Kantarjian, H.M. TP53-Mutated Myelodysplastic Syndrome and Acute Myeloid Leukemia: Biology, Current Therapy, and Future Directions. Cancer Discov. 2022, 12, 2516–2529. [Google Scholar] [CrossRef]
- Granowicz, E.M.; Jonas, B.A. Targeting TP53-Mutated Acute Myeloid Leukemia: Research and Clinical Developments. OncoTargets Ther. 2022, 15, 423–436. [Google Scholar] [CrossRef]
- Loschi, M.; Fenaux, P.; Cluzeau, T. How I Treat TP53-Mutated Acute Myeloid Leukemia and Myelodysplastic Syndromes. Cancers 2022, 14, 4519. [Google Scholar] [CrossRef]
- Patel, S.A.; Cerny, J. TP53-mutant myelodysplastic syndrome and acute myeloid leukemia: The black hole of hematology. Blood Adv. 2022, 6, 1917–1918. [Google Scholar] [CrossRef]
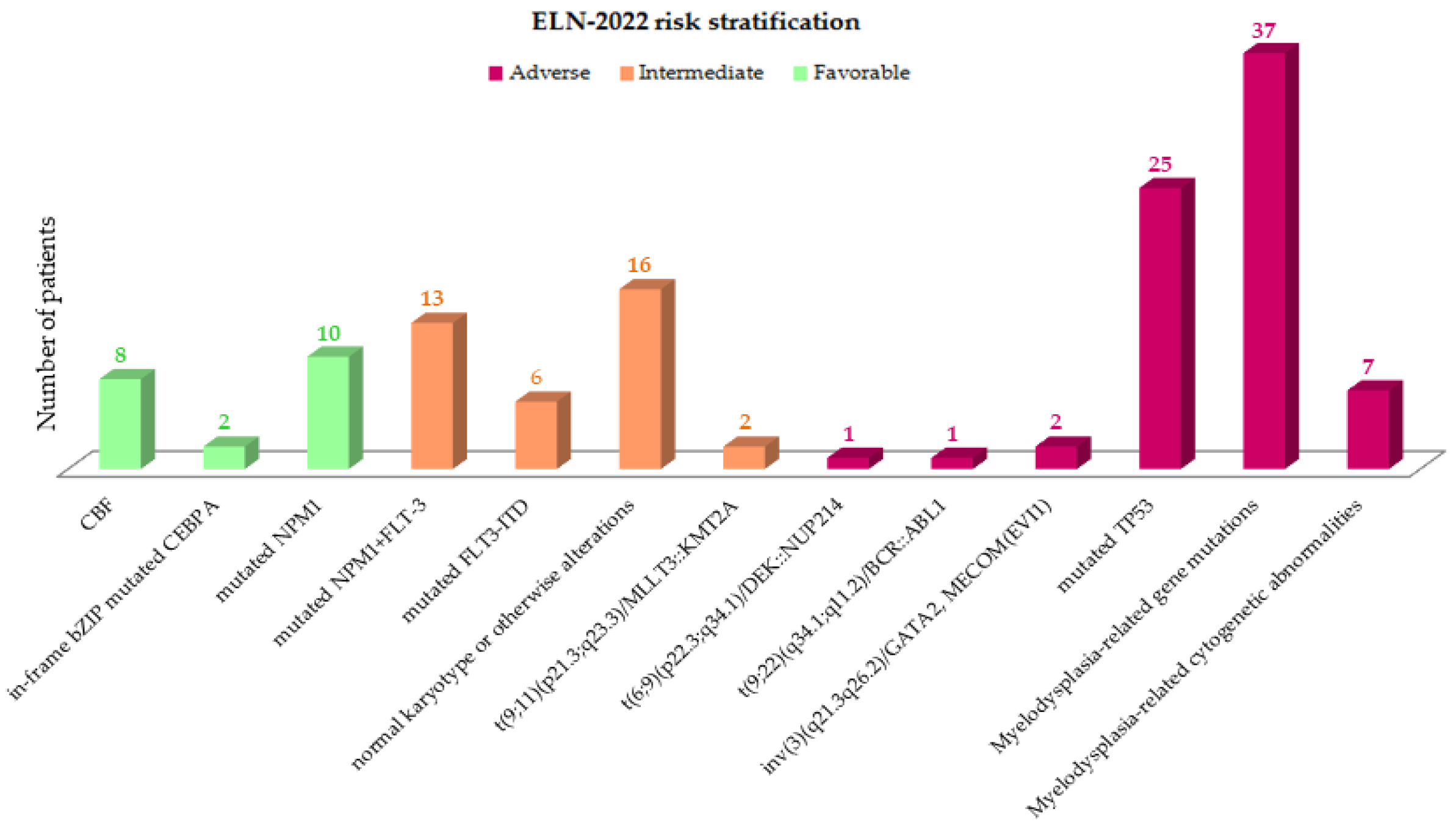
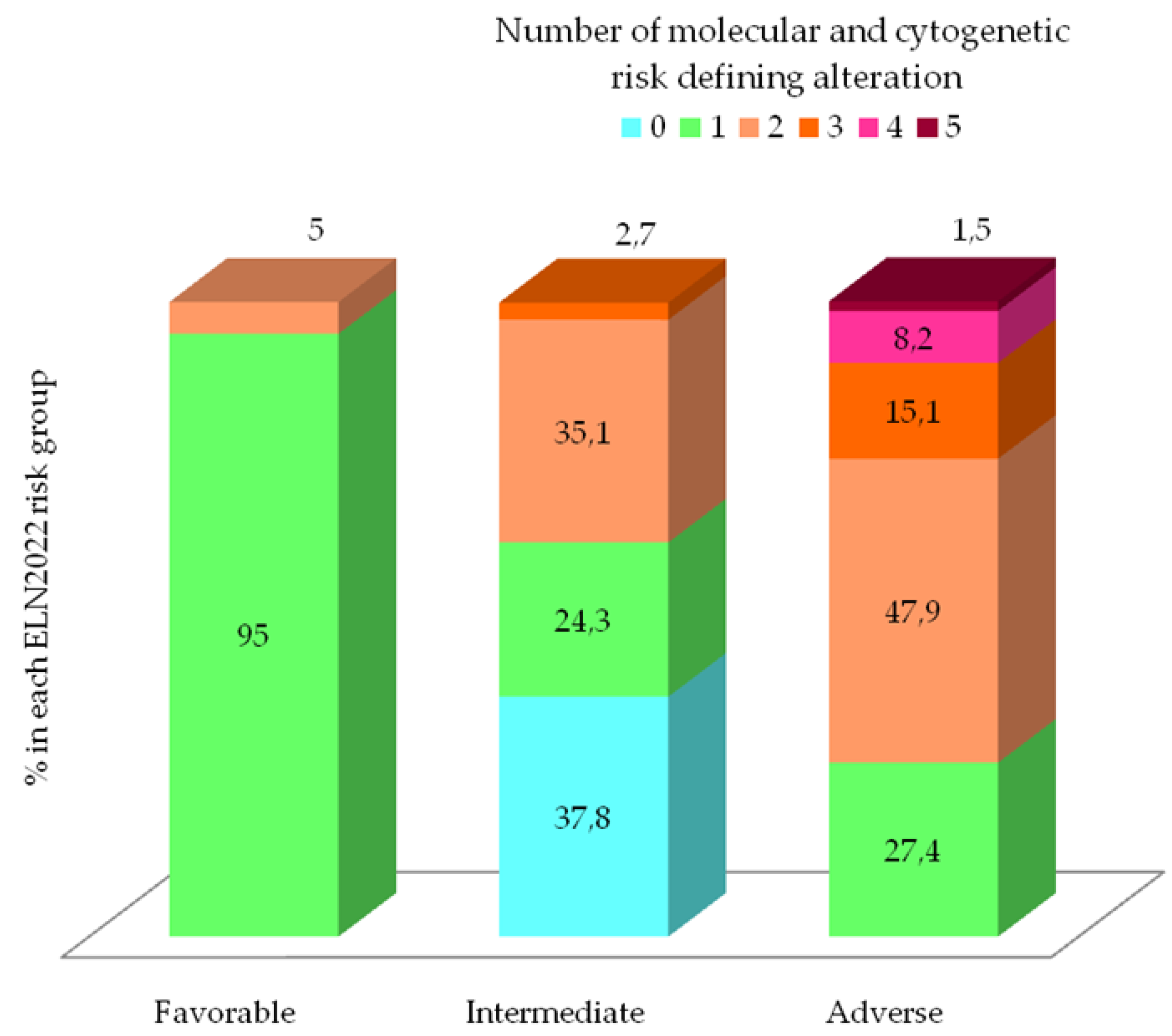

| n (%) | Cohort 130 (100) | ELN2022 Favorable 20 (15.4) | ELN2022 Intermediate 37 (28.5) | ELN2022 Adverse 73 (56.1) | p† |
|---|---|---|---|---|---|
| Age, years median (range) | 65 (18–94) | 50 (25–89) | 55 (18–89) | 69 (30–94) | <0.01 |
| Age ≥ 60 years | 82 (63.1) | 8 (40) | 16 (43.2) | 58 (79.5) | <0.01 |
| Sex, male n (%) | 71 (54.6) | 10 (50) | 13 (35.1) | 48 (65.8) | 0.009 |
| WBC × 109/L, median (range) | 10.4 (0.5–590) | 30 (1.2–158) | 22.4 (0.9–279) | 7.2 (0.5–590) | 0.02 |
| Clinical subtypes n (%) | |||||
| De-novo | 97 (74.6) | 17 (85.0) | 32 (86.5) | 48 (65.8) | 0.01 |
| s-AML | 21 (16.2) | 0 (-) | 2 (5.4) | 19 (26.0) | - |
| t-AML | 12 (9.2) | 3 (15) | 3 (8.1) | 6 (8.2) | - |
| NPM1 n(%) | 23 (17.7) | 10 (50) | 13 (35.1) | 0 (0) | <0.01 |
| FLT3-ITD n (%) | 23 (17.7) | 1 (5) | 16 (43.2) | 6 (8.2) | <0.01 |
| Received treatment n (%) | |||||
| Intensive chemotherapy (IC) | 87 (66.9) | 17 (85) | 29 (78.4) | 41 (56.2) | 0.01 |
| HMA-based/low intensity | 31 (23.9) | 3 (15) | 5 (13.5) | 23 (31.5) | - |
| Supportive care | 12 (9.2) | 0 (0) | 3 (8.1) | 9 (12.3) | - |
| HSCT * | 53 (61) | 6 (35) | 20 (69) | 27 (66) | 0.13 |
| Complete Remission (CR/CRi) * | 58 (66.6) | 16 (94.2) | 22 (75.8) | 20 (48.8) | <0.01 |
| Relapse rate * | 23 (26.4) | 4 (23.5) | 10 (34.5) | 9 (33.3) | 0.78 |
| Exitus rate | 80 (61.5) | 8 (40) | 19 (51.4) | 53 (72.6) | <0.01 |
| Functional Mutations Group n (%) | Mutations | Cohort (n = 130) | <60 Years (n = 49) | ≥60 Years (n = 81) | p |
|---|---|---|---|---|---|
| Signaling pathways | FLT3, KRAS, NRAS, KIT, PTPN | 47 (36.1) | 23 (46.9) | 24 (29.6) | 0.03 |
| Epigenetic modification | |||||
| DNA methylation | DNMT3A, IDH1/2, TET2 | 60 (46.1) | 22 (44.8) | 38 (46.9) | 0.92 |
| Chromatin modifiers | ASXL1, EZH2 y MLL/KMT2A | 19 (14.6) | 3 (6.1) | 16 (19.7) | 0.04 |
| Nucleophosmin | NPM1 | 23 (17.7) | 14 (28.5) | 9 (11.1) | <0.01 |
| Transcription factors | CEBPA, RUNX1 y GATA2 | 24 (18.5) | 7 (14.3) | 17 (21) | 0.37 |
| Tumor Suppressors | TP53 | 25 (19.2) | 5 (10.2) | 20 (24.6) | 0.05 |
| Spliceosome complex | SRSF2, U2AF1, SF3B1 y ZRSR2 | 32 (24.6) | 3 (6.1) | 29 (35.8) | <0.01 |
| Fusiontranscription factors | RUNX1/RUNX1T, MYH11/CBF | 8 (6.1) | 6 (12.3) | 2 (2.5) | 0.02 |
| Variable | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| OR(95% CI) | p | HR(95% CI) | p | ||
| Age (>60 years) | 3.97 (2.31–6.86) | <0.001 | 2.95 (1.65–5.3) | <0.01 | |
| TP53 mutation | 3.71 (2.56–8.8) | <0.001 | 3.17 (1.52–6.6) | 0.001 | |
| ELN-2022 risk stratification | Favorable | reference | - | reference | - |
| Intermediate | 1.36 (0.59–3.1) | 0.47 | 1.24 (0.54–2.86) | 0.613 | |
| Adverse | 3.24 (1.53–6.86) | 0.002 | 1.7 (0.75–3.82) | 0.201 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aparicio-Pérez, C.; Prados de la Torre, E.; Sanchez-Garcia, J.; Martín-Calvo, C.; Martínez-Losada, C.; Casaño-Sanchez, J.; Serrano-López, J.; Serrano, J. Evolving Risk Classifications in AML in a Real-Life Scenario: After Changes upon Changes, Is It More and More Adverse? Cancers 2023, 15, 1425. https://doi.org/10.3390/cancers15051425
Aparicio-Pérez C, Prados de la Torre E, Sanchez-Garcia J, Martín-Calvo C, Martínez-Losada C, Casaño-Sanchez J, Serrano-López J, Serrano J. Evolving Risk Classifications in AML in a Real-Life Scenario: After Changes upon Changes, Is It More and More Adverse? Cancers. 2023; 15(5):1425. https://doi.org/10.3390/cancers15051425
Chicago/Turabian StyleAparicio-Pérez, Clara, Esther Prados de la Torre, Joaquin Sanchez-Garcia, Carmen Martín-Calvo, Carmen Martínez-Losada, Javier Casaño-Sanchez, Juana Serrano-López, and Josefina Serrano. 2023. "Evolving Risk Classifications in AML in a Real-Life Scenario: After Changes upon Changes, Is It More and More Adverse?" Cancers 15, no. 5: 1425. https://doi.org/10.3390/cancers15051425
APA StyleAparicio-Pérez, C., Prados de la Torre, E., Sanchez-Garcia, J., Martín-Calvo, C., Martínez-Losada, C., Casaño-Sanchez, J., Serrano-López, J., & Serrano, J. (2023). Evolving Risk Classifications in AML in a Real-Life Scenario: After Changes upon Changes, Is It More and More Adverse? Cancers, 15(5), 1425. https://doi.org/10.3390/cancers15051425






