Metabolic Reprogramming of Breast Tumor-Educated Macrophages Revealed by NMR Metabolomics
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Collection of Breast Cancer (BC) Cells-Conditioned Medium (CM)
2.3. In Vitro Generation of Tumor-Educated Macrophages (TEMs)
2.4. Samples Collection and Preparation for NMR Metabolomics
2.5. NMR Data Acquisition and Processing
2.6. Multivariate and Qualitative Analysis of Spectral Data
2.7. Quantification of Cytokine Production
2.8. Statistical Analysis
3. Results
3.1. Metabolic Composition of BC Cells-Conditioned Media (CM)
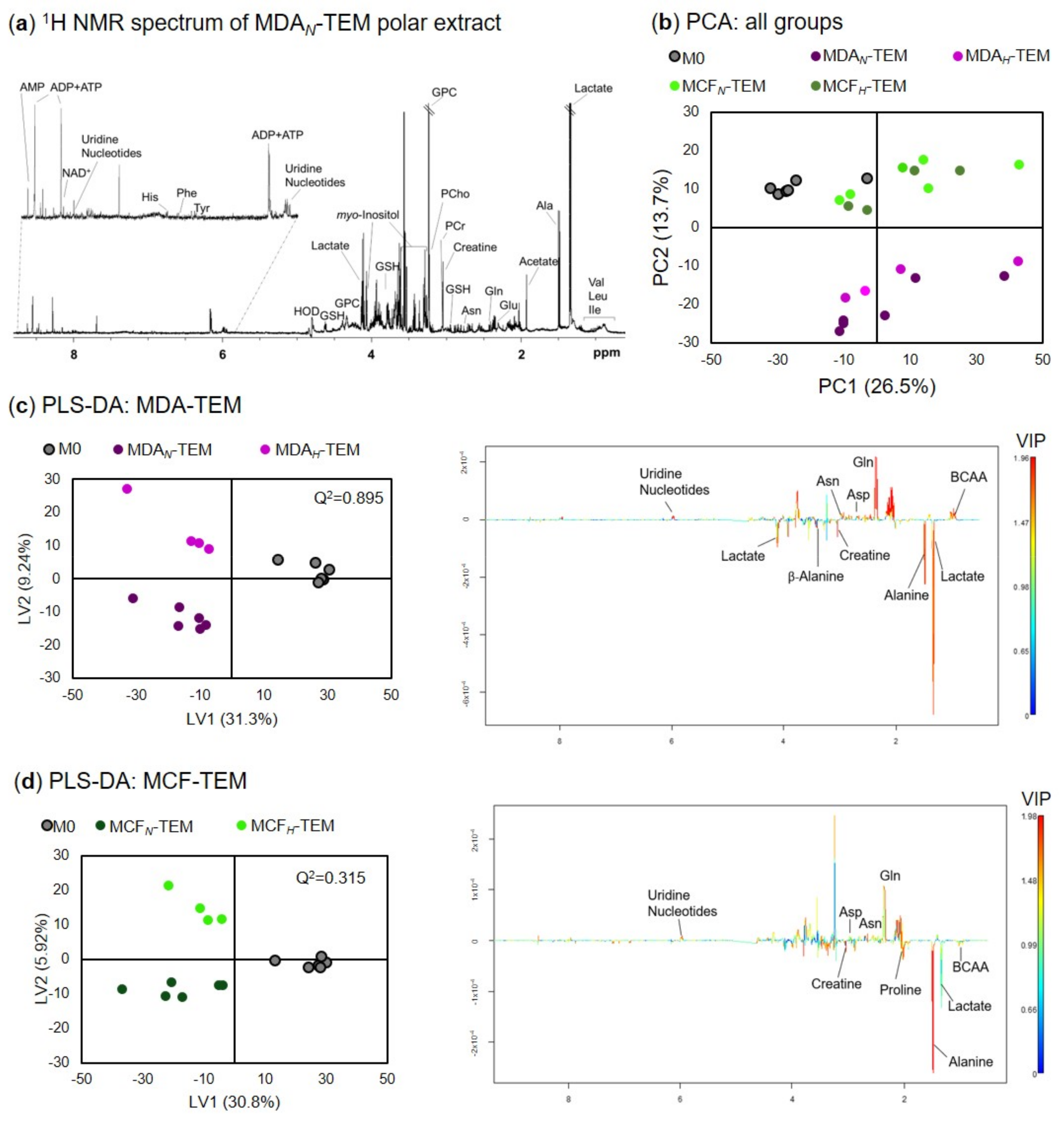
3.2. Exometabolomics of BC TEM
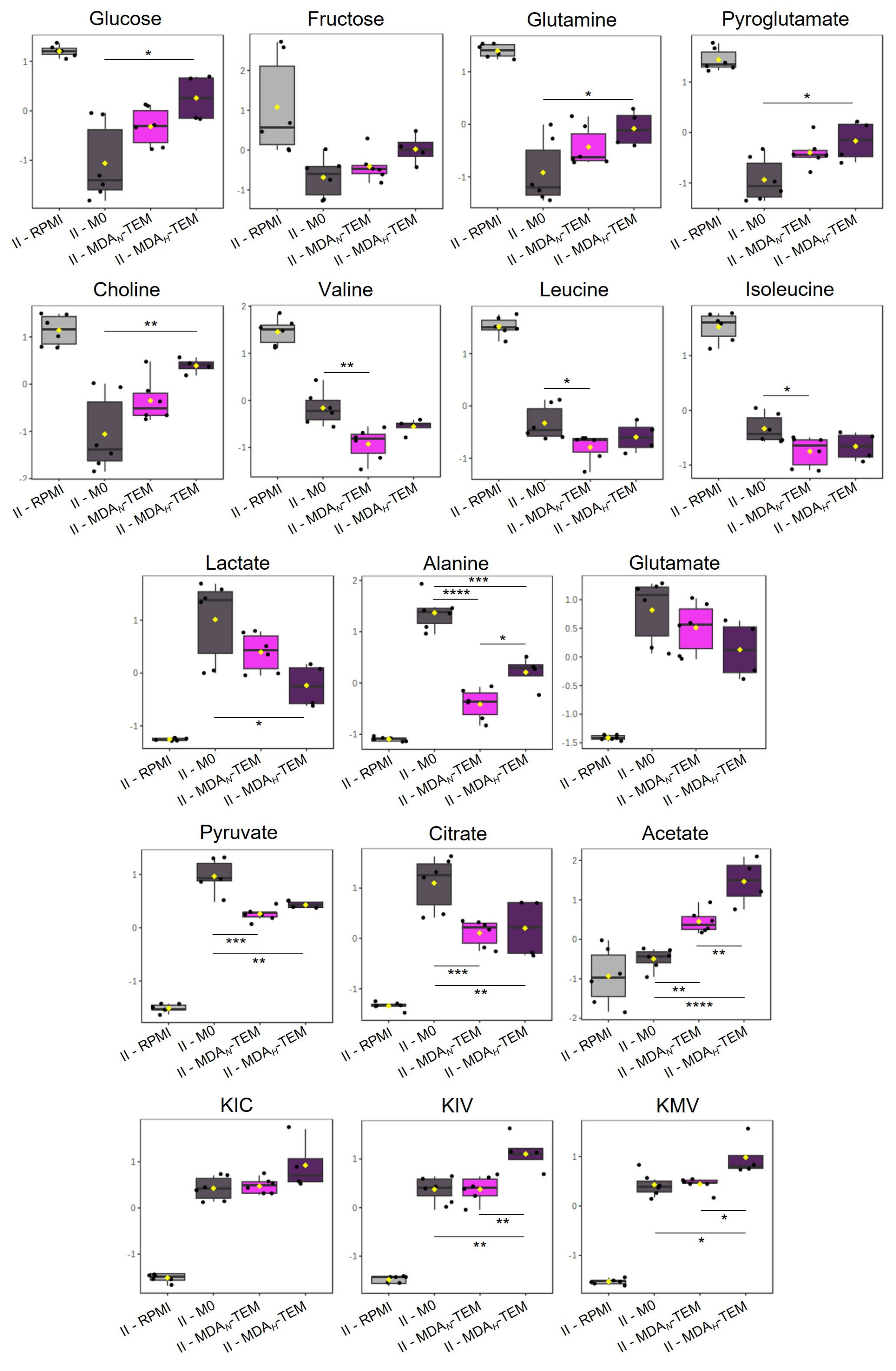
3.3. Endometabolomics of BC TEM
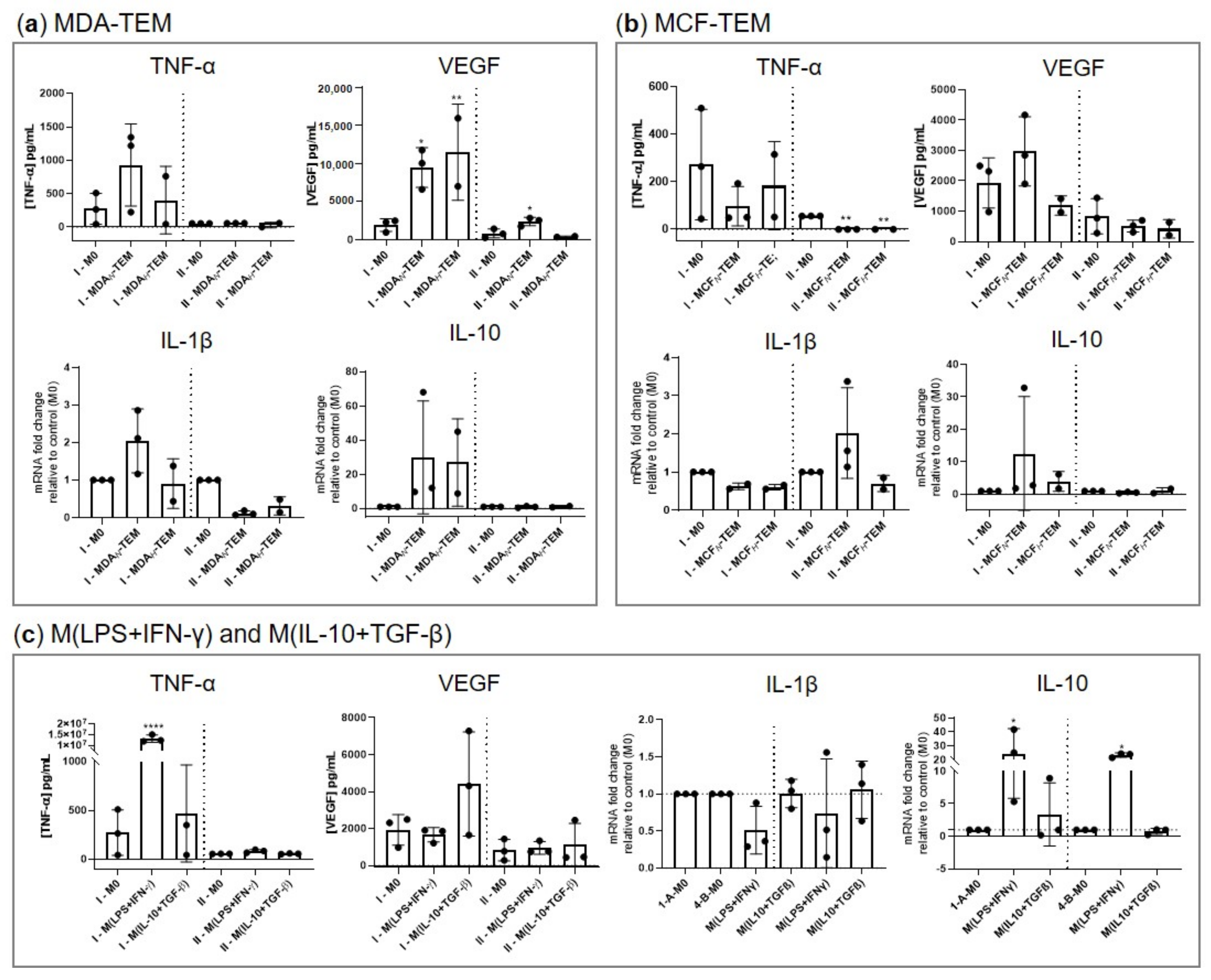
3.4. Cytokine Production by CM-Generated TEM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, S.; Chen, X.; Guo, C.; Wang, W. Tumour-associated macrophages heterogeneity drives resistance to clinical therapy. Expert Rev. Mol. Med. 2022, 24, e17. [Google Scholar] [CrossRef] [PubMed]
- Ruffell, B.; Coussens, L.M. Macrophages and Therapeutic Resistance in Cancer. Cancer Cell 2015, 27, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Guan, R.; Hong, W.; Zhou, Y.; Lin, Y.; Jin, H.; Hou, B.; Jian, Z. Prognostic value of tumor-associated macrophages in pancreatic cancer: A meta-analysis. Cancer Manag. Res. 2019, 11, 4041–4058. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, J.; Chen, S.; Ma, X.; Ying, Y.; Li, J.; Wang, W.; Wang, X.; Xie, L. Prognostic Value of Tumor-Associated Macrophages in Clear Cell Renal Cell Carcinoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2021, 11, 657318. [Google Scholar] [CrossRef]
- Wang, C.; Lin, Y.; Zhu, H.; Zhou, Y.; Mao, F.; Huang, X.; Sun, Q.; Li, C. The Prognostic and Clinical Value of Tumor-Associated Macrophages in Patients with Breast Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12, 905846. [Google Scholar] [CrossRef]
- Larionova, I.; Cherdyntseva, N.; Liu, T.; Patysheva, M.; Rakina, M.; Kzhyshkowska, J. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology 2019, 8, e1596004. [Google Scholar] [CrossRef]
- Medrek, C.; Pontén, F.; Jirström, K.; Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef]
- Tiainen, S.; Masarwah, A.; Oikari, S.; Rilla, K.; Hämäläinen, K.; Sudah, M.; Sutela, A.; Vanninen, R.; Ikonen, J.; Tammi, R.; et al. Tumor microenvironment and breast cancer survival: Combined effects of breast fat, M2 macrophages and hyaluronan create a dismal prognosis. Breast Cancer Res. Treat. 2020, 179, 565–575. [Google Scholar] [CrossRef]
- Jeong, H.; Kim, S.; Hong, B.-J.; Lee, C.-J.; Kim, Y.-E.; Bok, S.; Oh, J.-M.; Gwak, S.-H.; Yoo, M.Y.; Lee, M.S.; et al. Tumor-Associated Macrophages Enhance Tumor Hypoxia and Aerobic Glycolysis. Cancer Res 2019, 79, 795–806. [Google Scholar] [CrossRef]
- Ramos, R.N.; Rodriguez, C.; Hubert, M.; Ardin, M.; Treilleux, I.; Ries, C.H.; Lavergne, E.; Chabaud, S.; Colombe, A.; Trédan, O.; et al. CD163 + tumor-associated macrophage accumulation in breast cancer patients reflects both local differentiation signals and systemic skewing of monocytes. Clin. Transl. Immunol. 2020, 9, e1108. [Google Scholar] [CrossRef]
- Bolli, E.; Movahedi, K.; Laoui, D.; Van Ginderachter, J. Novel insights in the regulation and function of macrophages in the tumor microenvironment. Curr. Opin. Oncol. 2017, 29, 55–61. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; Coussens, L.M.; Kroemer, G.; Galluzzi, L. Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab. 2019, 30, 36–50. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef]
- Dias, A.S.; Almeida, C.R.; Helguero, L.A.; Duarte, I.F. Metabolic crosstalk in the breast cancer microenvironment. Eur. J. Cancer 2019, 121, 154–171. [Google Scholar] [CrossRef]
- Wei, F.; Wang, D.; Wei, J.; Tang, N.; Tang, L.; Xiong, F.; Guo, C.; Zhou, M.; Li, X.; Li, G.; et al. Metabolic crosstalk in the tumor microenvironment regulates antitumor immunosuppression and immunotherapy resisitance. Cell. Mol. Life Sci. 2021, 78, 173–193. [Google Scholar] [CrossRef]
- Colegio, O.R.; Chu, N.-Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef]
- Chen, P.; Zuo, H.; Xiong, H.; Kolar, M.J.; Chu, Q.; Saghatelian, A.; Siegwart, D.J.; Wan, Y. Gpr132 sensing of lactate mediates tumor–macrophage interplay to promote breast cancer metastasis. Proc. Natl. Acad. Sci. USA 2017, 114, 580–585. [Google Scholar] [CrossRef]
- Angelin, A.; Gil-De-Gómez, L.; Dahiya, S.; Jiao, J.; Guo, L.; Levine, M.H.; Wang, Z.; Quinn, W.J., III; Kopinski, P.K.; Wang, L.; et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 2017, 25, 1282–1293.e7. [Google Scholar] [CrossRef]
- Devalaraja, S.; To, T.K.J.; Folkert, I.W.; Natesan, R.; Alam, Z.; Li, M.; Tada, Y.; Budagyan, K.; Dang, M.T.; Zhai, L.; et al. Tumor-Derived Retinoic Acid Regulates Intratumoral Monocyte Differentiation to Promote Immune Suppression. Cell 2020, 180, 1098–1114.e16. [Google Scholar] [CrossRef]
- Di Conza, G.; Tsai, C.H.; Gallart-Ayala, H.; Yu, Y.R.; Franco, F.; Zaffalon, L.; Xie, X.; Li, X.; Xiao, Z.; Raines, L.N.; et al. Tumor-induced reshuffling of lipid composition on the ER membrane sustains macrophage survival and pro-tumorigenic activity. Nat. Immunol. 2021, 22, 1403–1415. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Pontious, C.; Kovalenko, I.; Lapienyte, L.; Dreyer, S.; Lee, H.-J.; Thurston, G.; Zhang, Y.; Lazarus, J.; Sajjakulnukit, P.; et al. Macrophage-Released Pyrimidines Inhibit Gemcitabine Therapy in Pancreatic Cancer. Cell Metab. 2019, 29, 1390–1399.e6. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim. et Biophys. Acta Mol. Cell Res. 2016, 1863, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Kozal, K.; Krześlak, A. The Role of Hypoxia-Inducible Factor Isoforms in Breast Cancer and Perspectives on Their Inhibition in Therapy. Cancers 2022, 14, 4518. [Google Scholar] [CrossRef] [PubMed]
- Carrola, J.; Bastos, V.; de Oliveira, J.M.P.F.; Oliveira, H.; Santos, C.; Gil, A.M.; Duarte, I.F. Insights into the impact of silver nanoparticles on human keratinocytes metabolism through NMR metabolomics. Arch. Biochem. Biophys. 2016, 589, 53–61. [Google Scholar] [CrossRef]
- Berben, L.; Sereika, S.M.; Engberg, S. Effect size estimation: Methods and examples. Int. J. Nurs. Stud. 2012, 49, 1039–1047. [Google Scholar] [CrossRef]
- Mendes, L.F.; Gaspar, V.M.; Conde, T.A.; Mano, J.F.; Duarte, I.F. Flavonoid-mediated immunomodulation of human macrophages involves key metabolites and metabolic pathways. Sci. Rep. 2019, 9, 14906. [Google Scholar] [CrossRef]
- Penny, H.L.; Sieow, J.L.; Adriani, G.; Yeap, W.H.; See, P.; Luis, B.S.; Lee, B.; Lee, T.; Mak, S.Y.; Ho, Y.S.; et al. Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology 2016, 5, e1191731. [Google Scholar] [CrossRef]
- Prasmickaite, L.; Tenstad, E.M.; Pettersen, S.; Jabeen, S.; Egeland, E.V.; Nord, S.; Pandya, A.; Haugen, M.H.; Kristensen, V.N.; Børresen-Dale, A.; et al. Basal-like breast cancer engages tumor-supportive macrophages via secreted factors induced by extracellular S100A4. Mol. Oncol. 2018, 12, 1540–1558. [Google Scholar] [CrossRef]
- Chen, S.-J.; Lian, G.-D.; Li, J.-J.; Zhang, Q.-B.; Zeng, L.-J.; Yang, K.-G.; Huang, C.-M.; Li, Y.-Q.; Chen, Y.-T.; Huang, K.-H. Tumor-driven like macrophages induced by conditioned media from pancreatic ductal adenocarcinoma promote tumor metastasis via secreting IL-8. Cancer Med. 2018, 7, 5679–5690. [Google Scholar] [CrossRef]
- Sawa-Wejksza, K.; Dudek, A.; Lemieszek, M.; Kaławaj, K.; Kandefer-Szerszeń, M. Colon cancer–derived conditioned medium induces differentiation of THP-1 monocytes into a mixed population of M1/M2 cells. Tumor Biol. 2018, 40, 1010428318797880. [Google Scholar] [CrossRef]
- Benner, B.; Scarberry, L.; Suarez-Kelly, L.P.; Duggan, M.C.; Campbell, A.R.; Smith, E.; Lapurga, G.; Jiang, K.; Butchar, J.P.; Tridandapani, S.; et al. Generation of monocyte-derived tumor-associated macrophages using tumor-conditioned media provides a novel method to study tumor-associated macrophages in vitro. J. Immunother. Cancer 2019, 7, 140. [Google Scholar] [CrossRef]
- Boyer, S.; Lee, H.-J.; Steele, N.; Zhang, L.; Sajjakulnukit, P.; Andren, A.; Ward, M.H.; Singh, R.; Basrur, V.; Zhang, Y.; et al. Multiomic characterization of pancreatic cancer-associated macrophage polarization reveals deregulated metabolic programs driven by the GM-CSF–PI3K pathway. Elife 2022, 11, e73796. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Robey, I.F.; Lien, A.; Welsh, S.J.; Baggett, B.K.; Gillies, R.J. Hypoxia-Inducible Factor-1α and the Glycolytic Phenotype in Tumors. Neoplasia 2005, 7, 324–330. [Google Scholar] [CrossRef]
- Sakamoto, T.; Niiya, D.; Seiki, M. Targeting the Warburg Effect That Arises in Tumor Cells Expressing Membrane Type-1 Matrix Metalloproteinase. J. Biol. Chem. 2011, 286, 14691–14704. [Google Scholar] [CrossRef]
- Zare, M.E.; Kansestani, A.N.; Hemmati, S.; Mansouri, K.; Vaisi-Raygani, A. The rate of aerobic glycolysis is a pivotal regulator of tumor progression. J. Diabetes Metab. Disord. 2021, 20, 523–531. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Zhang, B.; Lin, X.; Fu, X.; An, Y.; Zou, Y.; Wang, J.-X.; Wang, Z.; Yu, T. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 2022, 7, 305. [Google Scholar] [CrossRef]
- Liu, N.; Luo, J.; Kuang, D.; Xu, S.; Duan, Y.; Xia, Y.; Wei, Z.; Xie, X.; Yin, B.; Chen, F.; et al. Lactate inhibits ATP6V0d2 expression in tumor-associated macrophages to promote HIF-2α–mediated tumor progression. J. Clin. Investig. 2019, 129, 631–646. [Google Scholar] [CrossRef]
- Geeraerts, X.; Fernández-Garcia, J.; Hartmann, F.J.; de Goede, K.E.; Martens, L.; Elkrim, Y.; Debraekeleer, A.; Stijlemans, B.; Vandekeere, A.; Rinaldi, G.; et al. Macrophages are metabolically heterogeneous within the tumor microenvironment. Cell Rep. 2021, 37, 110171. [Google Scholar] [CrossRef]
- Noel, P.; Hussein, S.; Ng, S.; Antal, C.E.; Lin, W.; Rodela, E.; Delgado, P.; Naveed, S.; Downes, M.; Lin, Y.; et al. Triptolide targets super-enhancer networks in pancreatic cancer cells and cancer-associated fibroblasts. Oncogenesis 2020, 9, 100. [Google Scholar] [CrossRef]
- Liu, D.; Chang, C.; Lu, N.; Wang, X.; Lu, Q.; Ren, X.; Ren, P.; Zhao, D.; Wang, L.; Zhu, Y.; et al. Comprehensive Proteomics Analysis Reveals Metabolic Reprogramming of Tumor-Associated Macrophages Stimulated by the Tumor Microenvironment. J. Proteome Res. 2017, 16, 288–297. [Google Scholar] [CrossRef] [PubMed]
- De-Brito, N.M.; Duncan-Moretti, J.; Da-Costa, H.C.; Saldanha-Gama, R.; Paula-Neto, H.A.; Dorighello, G.; Simões, R.L.; Barja-Fidalgo, C. Aerobic glycolysis is a metabolic requirement to maintain the M2-like polarization of tumor-associated macrophages. Biochim. et Biophys. Acta Mol. Cell Res. 2020, 1867, 118604. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.; Nagy, C.; Knapp, B.; Laengle, J.; Ponweiser, E.; Groeger, M.; Starkl, P.; Bergmann, M.; Wagner, O.; Haschemi, A. Exploring Metabolic Configurations of Single Cells within Complex Tissue Microenvironments. Cell Metab. 2017, 26, 788–800.e6. [Google Scholar] [CrossRef] [PubMed]
- Mycielska, M.E.; Patel, A.; Rizaner, N.; Mazurek, M.P.; Keun, H.; Patel, A.; Ganapathy, V.; Djamgoz, M.B.A. Citrate transport and metabolism in mammalian cells. Bioessays 2009, 31, 10–20. [Google Scholar] [CrossRef]
- Grashei, M.; Biechl, P.; Schilling, F.; Otto, A.M. Conversion of Hyperpolarized [1-13C]Pyruvate in Breast Cancer Cells Depends on Their Malignancy, Metabolic Program and Nutrient Microenvironment. Cancers 2022, 14, 1845. [Google Scholar] [CrossRef]
- Phannasil, P.; Thuwajit, C.; Warnnissorn, M.; Wallace, J.C.; MacDonald, M.J.; Jitrapakdee, S. Pyruvate Carboxylase Is Up-Regulated in Breast Cancer and Essential to Support Growth and Invasion of MDA-MB-231 Cells. PLoS ONE 2015, 10, e0129848. [Google Scholar] [CrossRef]
- Westergaard, N.; Waagepetersen, H.S.; Belhage, B.; Schousboe, A. Citrate, a Ubiquitous Key Metabolite with Regulatory Function in the CNS. Neurochem. Res. 2017, 42, 1583–1588. [Google Scholar] [CrossRef]
- Van Heijster, F.H.A.; Breukels, V.; Jansen, K.F.J.; Schalken, J.A.; Heerschap, A. Carbon sources and pathways for citrate secreted by human prostate cancer cells determined by NMR tracing and metabolic modeling. Proc. Natl. Acad. Sci. USA 2022, 119, e2024357119. [Google Scholar] [CrossRef]
- Peng, M.; Yang, D.; Hou, Y.; Liu, S.; Zhao, M.; Qin, Y.; Chen, R.; Teng, Y.; Liu, M. Intracellular citrate accumulation by oxidized ATM-mediated metabolism reprogramming via PFKP and CS enhances hypoxic breast cancer cell invasion and metastasis. Cell Death Dis. 2019, 10, 228. [Google Scholar] [CrossRef]
- Parkinson, E.K.; Adamski, J.; Zahn, G.; Gaumann, A.; Flores-Borja, F.; Ziegler, C.; Mycielska, M.E. Extracellular citrate and metabolic adaptations of cancer cells. Cancer Metastasis Rev. 2021, 40, 1073–1091. [Google Scholar] [CrossRef]
- Drexler, K.; Schmidt, K.M.; Jordan, K.; Federlin, M.; Milenkovic, V.M.; Liebisch, G.; Artati, A.; Schmidl, C.; Madej, G.; Tokarz, J.; et al. Cancer-associated cells release citrate to support tumour metastatic progression. Life Sci. Alliance 2021, 4, e202000903. [Google Scholar] [CrossRef]
- Sivanand, S.; Vander Heiden, M.G. Emerging roles for branched chain amino acid metabolism in cancer. Cancer Cell 2020, 37, 147–156. [Google Scholar] [CrossRef]
- Zhang, L.; Han, J. Branched-chain amino acid transaminase 1 (BCAT1) promotes the growth of breast cancer cells through improving mTOR-mediated mitochondrial biogenesis and function. Biochem. Biophys. Res. Commun. 2017, 486, 224–231. [Google Scholar] [CrossRef]
- Biswas, D.; Slade, L.; Duffley, L.; Mueller, N.; Dao, K.T.; Mercer, A.; Pakkiriswami, S.; El Hiani, Y.; Kienesberger, P.C.; Pulinilkunnil, T. Inhibiting BCKDK in triple negative breast cancer suppresses protein translation, impairs mitochondrial function, and potentiates doxorubicin cytotoxicity. Cell Death Discov. 2021, 7, 241. [Google Scholar] [CrossRef]
- Mikalayeva, V.; Pankevičiūtė, M.; Žvikas, V.; Skeberdis, V.A.; Bordel, S. Contribution of branched chain amino acids to energy production and mevalonate synthesis in cancer cells. Biochem. Biophys. Res. Commun. 2021, 585, 61–67. [Google Scholar] [CrossRef]
- Lee, J.H.; Cho, Y.-R.; Kim, J.H.; Kim, J.; Nam, H.Y.; Kim, S.W.; Son, J. Branched-chain amino acids sustain pancreatic cancer growth by regulating lipid metabolism. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef]
- Silva, L.S.; Poschet, G.; Nonnenmacher, Y.; Becker, H.M.; Sapcariu, S.; Gaupel, A.-C.; Schlotter, M.; Wu, Y.; Kneisel, N.; Seiffert, M.; et al. Branched-chain ketoacids secreted by glioblastoma cells via MCT1 modulate macrophage phenotype. EMBO Rep. 2017, 18, 2172–2185. [Google Scholar] [CrossRef]
- Cai, Z.; Li, W.; Brenner, M.; Bahiraii, S.; Heiss, E.H.; Weckwerth, W. Branched-chain ketoacids derived from cancer cells modulate macrophage polarization and metabolic reprogramming. Front. Immunol. 2022, 13, 966158. [Google Scholar] [CrossRef]
- Kader, S.A.; Dib, S.; Achkar, I.W.; Thareja, G.; Suhre, K.; Rafii, A.; Halama, A. Defining the landscape of metabolic dysregulations in cancer metastasis. Clin. Exp. Metastasis 2022, 39, 345–362. [Google Scholar] [CrossRef]
- Cruzat, V.; Macedo Rogero, M.; Keane, K.N.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef]
- Choi, J.; Stradmann-Bellinghausen, B.; Yakubov, E.; Savaskan, N.; Régnier-Vigouroux, A. Glioblastoma cells induce differential glutamatergic gene expressions in human tumor-associated microglia/macrophages and monocyte-derived macrophages. Cancer Biol. Ther. 2015, 16, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, E.M.; Menga, A.; Martín-Pérez, R.; Quinto, A.; Riera-Domingo, C.; De Tullio, G.; Hooper, D.C.; Lamers, W.H.; Ghesquière, B.; McVicar, D.W.; et al. Pharmacologic or Genetic Targeting of Glutamine Synthetase Skews Macrophages toward an M1-like Phenotype and Inhibits Tumor Metastasis. Cell Rep. 2017, 20, 1654–1666. [Google Scholar] [CrossRef]
- Li, J.; Meng, Y.; Wu, X.; Sun, Y. Polyamines and related signaling pathways in cancer. Cancer Cell Int. 2020, 20, 539. [Google Scholar] [CrossRef] [PubMed]
- Kieler, M.; Hofmann, M.; Schabbauer, G. More than just protein building blocks: How amino acids and related metabolic pathways fuel macrophage polarization. FEBS J. 2021, 288, 3694–3714. [Google Scholar] [CrossRef] [PubMed]
- Arlauckas, S.P.; Garren, S.B.; Garris, C.S.; Kohler, R.H.; Oh, J.; Pittet, M.J.; Weissleder, R. Arg1 expression defines immunosuppressive subsets of tumor-associated macrophages. Theranostics 2018, 8, 5842–5854. [Google Scholar] [CrossRef]
- LaRue, M.M.; Parker, S.; Puccini, J.; Cammer, M.; Kimmelman, A.C.; Bar-Sagi, D. Metabolic reprogramming of tumor-associated macrophages by collagen turnover promotes fibrosis in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2022, 119, e2119168119. [Google Scholar] [CrossRef]
- Allard, B.; Allard, D.; Buisseret, L.; Stagg, J. The adenosine pathway in immuno-oncology. Nat. Rev. Clin. Oncol. 2020, 17, 611–629. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Yan, S.; Yang, Q.; Zhou, Y.; Zeng, X.; Liu, Z.; An, X.; Toque, H.A.; Dong, Z.; et al. Regulation of endothelial intracellular adenosine via adenosine kinase epigenetically modulates vascular inflammation. Nat. Commun. 2017, 8, 943. [Google Scholar] [CrossRef]
- Zhao, S.; Zhong, Y.; Fu, X.; Wang, Y.; Ye, P.; Cai, J.; Liu, Y.; Sun, J.; Mei, Z.; Jiang, Y.; et al. H3K4 Methylation Regulates LPS-Induced Proinflammatory Cytokine Expression and Release in Macrophages. Shock 2019, 51, 401–406. [Google Scholar] [CrossRef]
- Wu, H.; Han, Y.; Sillke, Y.R.; Deng, H.; Siddiqui, S.; Treese, C.; Schmidt, F.; Friedrich, M.; Keye, J.; Wan, J.; et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol. Med. 2019, 11, e10698. [Google Scholar] [CrossRef]
- Luo, Q.; Zheng, N.; Jiang, L.; Wang, T.; Zhang, P.; Liu, Y.; Zheng, P.; Wang, W.; Xie, G.; Chen, L.; et al. Lipid accumulation in macrophages confers protumorigenic polarization and immunity in gastric cancer. Cancer Sci. 2020, 111, 4000–4011. [Google Scholar] [CrossRef]
- Masetti, M.; Carriero, R.; Portale, F.; Marelli, G.; Morina, N.; Pandini, M.; Iovino, M.; Partini, B.; Erreni, M.; Ponzetta, A.; et al. Lipid-loaded tumor-associated macrophages sustain tumor growth and invasiveness in prostate cancer. J. Exp. Med. 2021, 219, e20210564. [Google Scholar] [CrossRef]
- Goossens, P.; Rodriguez-Vita, J.; Etzerodt, A.; Masse, M.; Rastoin, O.; Gouirand, V.; Ulas, T.; Papantonopoulou, O.; Van Eck, M.; Auphan-Anezin, N.; et al. Membrane Cholesterol Efflux Drives Tumor-Associated Macrophage Reprogramming and Tumor Progression. Cell Metab. 2019, 29, 1376–1389.e4. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias, A.S.; Almeida, C.R.; Helguero, L.A.; Duarte, I.F. Metabolic Reprogramming of Breast Tumor-Educated Macrophages Revealed by NMR Metabolomics. Cancers 2023, 15, 1211. https://doi.org/10.3390/cancers15041211
Dias AS, Almeida CR, Helguero LA, Duarte IF. Metabolic Reprogramming of Breast Tumor-Educated Macrophages Revealed by NMR Metabolomics. Cancers. 2023; 15(4):1211. https://doi.org/10.3390/cancers15041211
Chicago/Turabian StyleDias, Ana S., Catarina R. Almeida, Luisa A. Helguero, and Iola F. Duarte. 2023. "Metabolic Reprogramming of Breast Tumor-Educated Macrophages Revealed by NMR Metabolomics" Cancers 15, no. 4: 1211. https://doi.org/10.3390/cancers15041211
APA StyleDias, A. S., Almeida, C. R., Helguero, L. A., & Duarte, I. F. (2023). Metabolic Reprogramming of Breast Tumor-Educated Macrophages Revealed by NMR Metabolomics. Cancers, 15(4), 1211. https://doi.org/10.3390/cancers15041211






