Potential Role of the Fragile Histidine Triad in Cancer Evo-Dev
Abstract
Simple Summary
Abstract
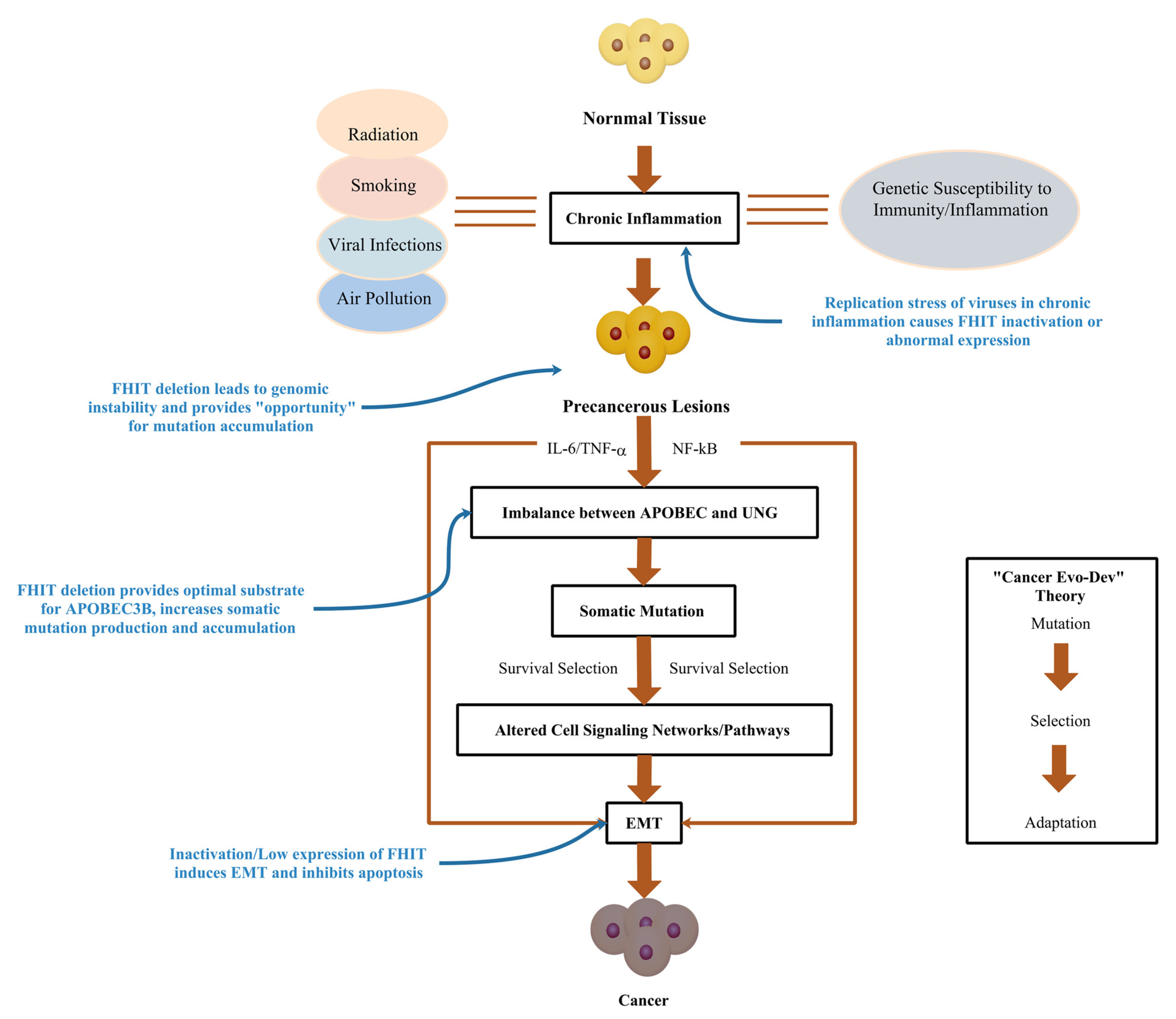
1. Introduction
2. Inactivation/Low Expression of the FHIT Gene
2.1. The FHIT/FRA3B Is Sensitive to Replication Pressure
2.2. Repeated Breakages and Repairs Cause Loss of Heterozygosity in the FHIT
2.3. CpG Methylation at the FHIT Promoter Region
3. Aberrant Expression of the FHIT Contributes to the Genome Chaos
3.1. Decreased Expression of the FHIT Induces Replication Fork Stagnation
3.2. Replication Stress Causes Chromosome Instability
3.3. Decreased Expression of the FHIT Stops Cellular Checkpoints
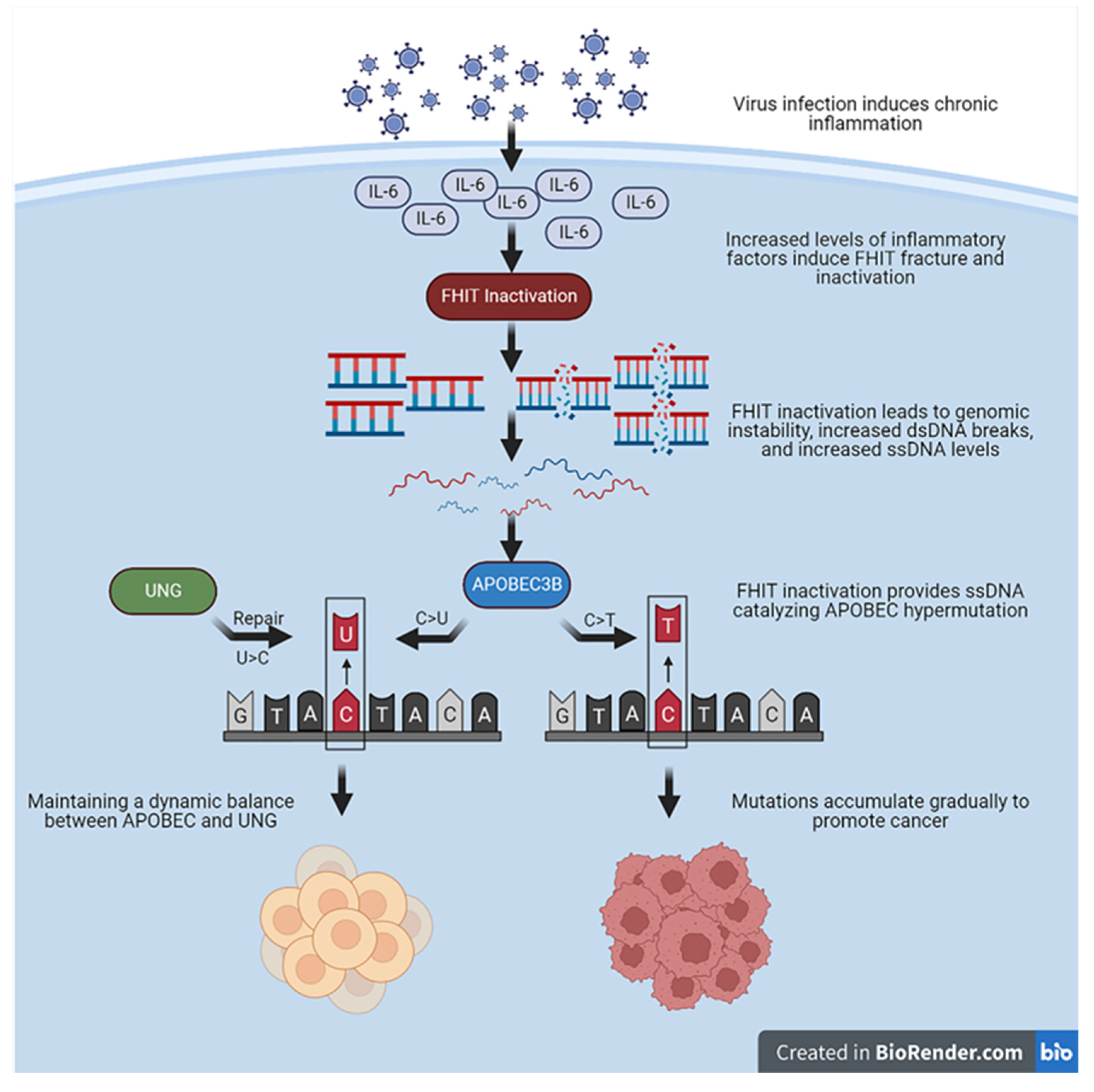
4. Decreased Expression of the FHIT Catalyzes APOBEC3B Hypermutation
4.1. APOBEC3B Requires Co-Factors to Promote Hypermutation
4.2. Mutation Characteristics Caused by Decreased Expression of FHIT Are Similar to APOBEC Signature
4.3. FHIT Provides Optimal Substrate for APOBEC3B Hypermutation
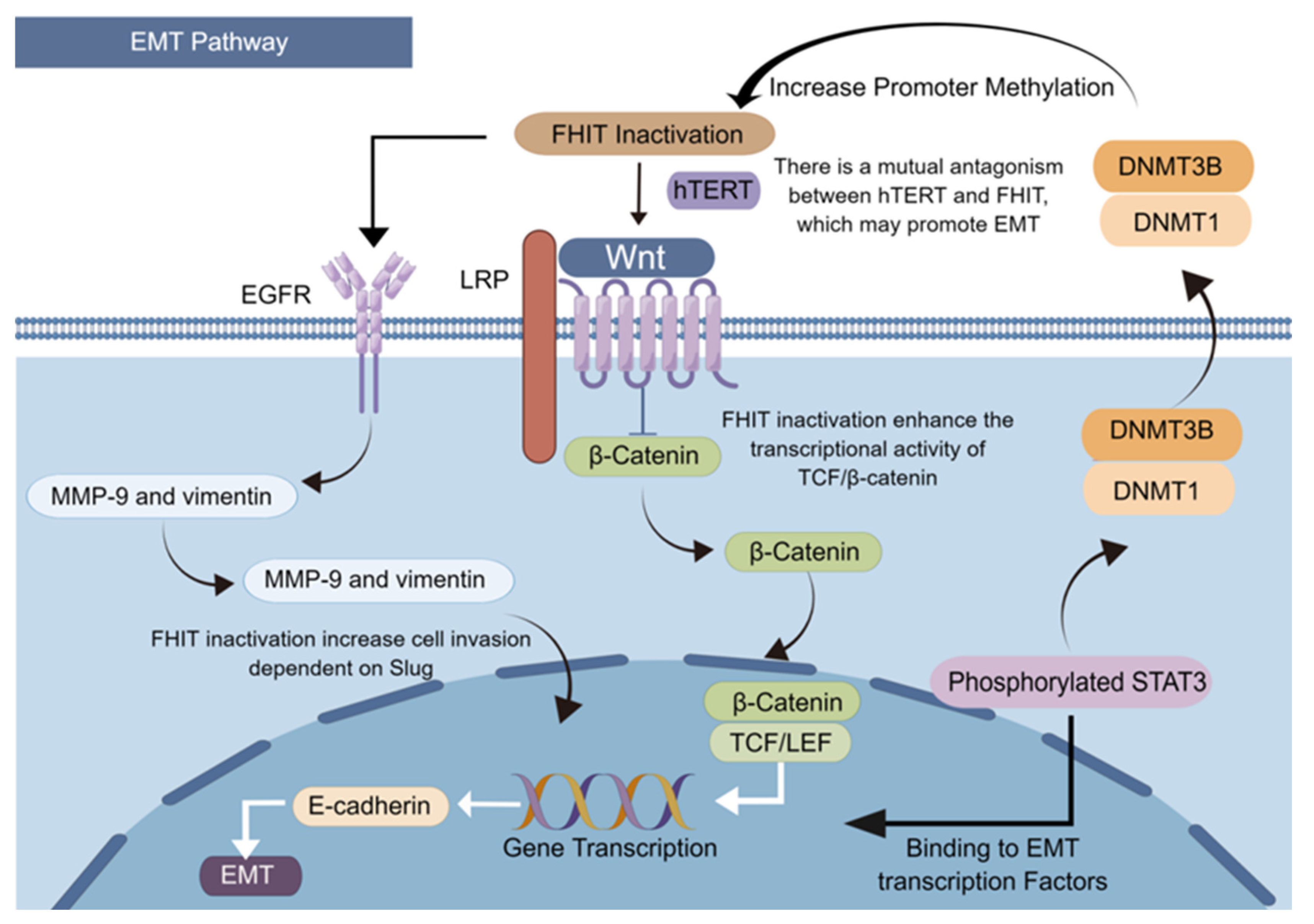
5. Abnormal Expression of FHIT Induces EMT and Inhibits Apoptosis
5.1. FHIT Deficiency Affects the Mitochondria-Mediated Apoptosis Pathway
5.2. Decreased Expression of the FHIT Promotes Reverse Differentiation of Cancer Cells
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Somarelli, J.A.; DeGregori, J.; Gerlinger, M.; Heng, H.H.; Marusyk, A.; Welch, D.R.; Laukien, F.H. Questions to guide cancer evolution as a framework for furthering progress in cancer research and sustainable patient outcomes. Med. Oncol. 2022, 39, 137. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Zhang, Q.; Ji, X.; Hou, X.; Lu, F.; Du, Y.; Yin, J.; Sun, X.; Deng, Y.; Zhao, J.; Han, X.; et al. Effect of functional nuclear factor-kappaB genetic polymorphisms on hepatitis B virus persistence and their interactions with viral mutations on the risk of hepatocellular carcinoma. Ann. Oncol. 2014, 25, 2413–2419. [Google Scholar] [CrossRef]
- Cao, G.-W. Cancer Evo-Dev, a novel hypothesis derived from studies on hepatitis B virus-induced carcinogenesis. Hepatoma Res. 2017, 3, 241–259. [Google Scholar] [CrossRef]
- Paul, D. Cancer as a form of life: Musings of the cancer and evolution symposium. Prog. Biophys. Mol. Biol. 2021, 165, 120–139. [Google Scholar] [CrossRef]
- Heng, H.H.Q.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, K.J.; Reddy, P.-V.; Wu, G.S.; Wang, Y.A.; Tainsky, M.A.; Ye, C.J. Stochastic cancer progression driven by non-clonal chromosome aberrations. J. Cell. Physiol. 2006, 208, 461–472. [Google Scholar] [CrossRef]
- Heng, J.; Heng, H.H. Genome chaos: Creating new genomic information essential for cancer macroevolution. Semin. Cancer Biol. 2022, 81, 160–175. [Google Scholar] [CrossRef]
- Li, X.; Zhong, Y.; Zhang, X.; Sood, A.K.; Liu, J. Spatiotemporal view of malignant histogenesis and macroevolution via formation of polyploid giant cancer cells. Oncogene 2023, 1–14. [Google Scholar] [CrossRef]
- Liu, W.; Deng, Y.; Li, Z.; Chen, Y.; Zhu, X.; Tan, X.; Cao, G. Cancer Evo–Dev: A Theory of Inflammation-Induced Oncogenesis. Front. Immunol. 2021, 12, 768098. [Google Scholar] [CrossRef]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Petljak, M.; Dananberg, A.; Chu, K.; Bergstrom, E.N.; Striepen, J.; von Morgen, P.; Chen, Y.; Shah, H.; Sale, J.E.; Alexandrov, L.B.; et al. Mechanisms of APOBEC3 mutagenesis in human cancer cells. Nature 2022, 607, 799–807. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Ng, A.W.T.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]
- Pu, R.; Liu, W.; Zhou, X.; Chen, X.; Hou, X.; Cai, S.; Chen, L.; Wu, J.; Yang, F.; Tan, X.; et al. The Effects and Underlying Mechanisms of Hepatitis B Virus X Gene Mutants on the Development of Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 836517. [Google Scholar] [CrossRef]
- Liu, W.; Cai, S.; Pu, R.; Li, Z.; Liu, D.; Zhou, X.; Yin, J.; Chen, X.; Chen, L.; Wu, J.; et al. HBV preS Mutations Promote Hepatocarcinogenesis by Inducing Endoplasmic Reticulum Stress and Upregulating Inflammatory Signaling. Cancers 2022, 14, 3274. [Google Scholar] [CrossRef]
- Martincorena, I.; Raine, K.M.; Gerstung, M.; Dawson, K.J.; Haase, K.; Van Loo, P.; Davies, H.; Stratton, M.R.; Campbell, P.J. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 2017, 171, 1029–1041.e21. [Google Scholar] [CrossRef]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of non-coding somatic drivers in 2658 cancer whole genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef]
- Suarez-Carmona, M.; Lesage, J.; Cataldo, D.; Gilles, C. EMT and inflammation: Inseparable actors of cancer progression. Mol. Oncol. 2017, 11, 805–823. [Google Scholar] [CrossRef]
- Babaei, G.; Aziz, S.G.-G.; Jaghi, N.Z.Z. EMT, cancer stem cells and autophagy; The three main axes of metastasis. Biomed. Pharmacother. 2020, 133, 110909. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, M.; Ebrahimi, D.; Temiz, N.A.; Harris, R.S. Mutation Signatures Including APOBEC in Cancer Cell Lines. JNCI Cancer Spectr. 2018, 2, pky002. [Google Scholar] [CrossRef]
- Venkatesan, S.; Angelova, M.; Puttick, C.; Zhai, H.; Caswell, D.R.; Lu, W.-T.; Dietzen, M.; Galanos, P.; Evangelou, K.; Bellelli, R.; et al. Induction of APOBEC3 Exacerbates DNA Replication Stress and Chromosomal Instability in Early Breast and Lung Cancer Evolution. Cancer Discov. 2021, 11, 2456–2473. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ji, H.; Zhao, J.; Song, J.; Zheng, S.; Chen, L.; Li, P.; Tan, X.; Ding, Y.; Pu, R.; et al. Transcriptional repression and apoptosis influence the effect of APOBEC3A / 3B functional polymorphisms on biliary tract cancer risk. Int. J. Cancer 2022, 150, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.L.; Collins, C.D.; Thompson, S.; Coxon, M.; Mertz, T.M.; Roberts, S.A. Single-stranded DNA binding proteins influence APOBEC3A substrate preference. Sci. Rep. 2021, 11, 21008. [Google Scholar] [CrossRef] [PubMed]
- Salter, J.D.; Smith, H.C. Modeling the Embrace of a Mutator: APOBEC Selection of Nucleic Acid Ligands. Trends Biochem. Sci. 2018, 43, 606–622. [Google Scholar] [CrossRef]
- Waters, C.E.; Saldivar, J.C.; Hosseini, S.A.; Huebner, K. The FHIT gene product: Tumor suppressor and genome “caretaker”. Cell. Mol. Life Sci. 2014, 71, 4577–4587. [Google Scholar] [CrossRef]
- Ohta, M.; Inoue, H.; Cotticelli, M.G.; Kastury, K.; Baffa, R.; Palazzo, J.; Siprashvili, Z.; Mori, M.; McCue, P.; Druck, T.; et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 1996, 84, 587–597. [Google Scholar] [CrossRef]
- Palumbo, E.; Tosoni, E.; Matricardi, L.; Russo, A. Genetic instability of the tumor suppressor gene FHIT in normal human cells. Genes Chromosomes Cancer 2013, 52, 832–844. [Google Scholar] [CrossRef]
- Prosseda, S.D.; Tian, X.; Kuramoto, K.; Boehm, M.; Sudheendra, D.; Miyagawa, K.; Zhang, F.; Solow-Cordero, D.; Saldivar, J.C.; Austin, E.D.; et al. FHIT, a Novel Modifier Gene in Pulmonary Arterial Hypertension. Am. J. Respir. Crit. Care Med. 2019, 199, 83–98. [Google Scholar] [CrossRef]
- Bai, Y.; Shen, Y.; Yuan, Q.; Lv, C.; Xing, Q. Evaluation of Relationship between Occurrence of Liver Cancer and Methylation of Fragile Histidine Triad (FHIT) and P16 Genes. Experiment 2019, 25, 1301–1306. [Google Scholar] [CrossRef]
- Rabelo, R.A.S.; Antunes, L.; Etchebehere, R.M.; Nomelini, R.S.; Nascentes, G.A.N.; Murta, E.F.C.; Pedrosa, A.L. Loss of heterozygosity in the fragile histidine triad (FHIT) locus and expression analysis of FHIT protein in patients with breast disorders. Clin. Exp. Obstet. Gynecol. 2013, 40, 89–94. [Google Scholar]
- Fassan, M.; Rusev, B.; Corbo, V.; Gasparini, P.; Luchini, C.; Vicentini, C.; Mafficini, A.; Paiella, S.; Salvia, R.; Cataldo, I.; et al. Fhit down-regulation is an early event in pancreatic carcinogenesis. Virchows Arch. 2017, 470, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, J.L.; Komissarova, E.V.; Kongkarnka, S.; Friedman, R.A.; Davison, J.M.; Levy, B.; Bryk, D.; Jobanputra, V.; Del Portillo, A.; Falk, G.W.; et al. High-resolution genomic alterations in Barrett’s metaplasia of patients who progress to esophageal dysplasia and adenocarcinoma. Int. J. Cancer 2019, 145, 2754–2766. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Smith, D.I. Very large common fragile site genes and their potential role in cancer development. Cell. Mol. Life Sci. 2014, 71, 4601–4615. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.-W.; Hsu, N.-Y.; Wang, Y.-C.; Lee, M.-C.; Cheng, Y.-W.; Chen, C.-Y.; Lee, H. c-Myc suppresses microRNA-29b to promote tumor aggressiveness and poor outcomes in non-small cell lung cancer by targeting FHIT. Oncogene 2015, 34, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.; Jouida, A.; Ancel, J.; Dalstein, V.; Routhier, J.; Delepine, G.; Cutrona, J.; Jonquet, A.; Dewolf, M.; Birembaut, P.; et al. FHIT(low) /pHER2(high) signature in non-small cell lung cancer is predictive of anti-HER2 molecule efficacy. J. Pathol. 2020, 51, 187–199. [Google Scholar] [CrossRef]
- Guadarrama-Ponce, R.; Aranda-Anzaldo, A. The epicenter of chromosomal fragility of Fra14A2, the mouse ortholog of human FRA3B common fragile site, is largely attached to the nuclear matrix in lymphocytes but not in other cell types that do not express such a fragility. J. Cell. Biochem. 2020, 121, 2209–2224. [Google Scholar] [CrossRef]
- Sarni, D.; Sasaki, T.; Tur-Sinai, M.I.; Miron, K.; Rivera-Mulia, J.C.; Magnuson, B.; Ljungman, M.; Gilbert, D.M.; Kerem, B. 3D genome organization contributes to genome instability at fragile sites. Nat. Commun. 2020, 11, 3613. [Google Scholar] [CrossRef]
- Ji, F.; Liao, H.; Pan, S.; Ouyang, L.; Fu, Z.; Zhang, F.; Geng, X.; Wang, X.; Li, T.; Liu, S.; et al. Genome-wide high-resolution mapping of mitotic DNA synthesis sites and common fragile sites by direct sequencing. Cell Res. 2020, 30, 1009–1023. [Google Scholar] [CrossRef]
- Epum, E.A.; Haber, J.E. DNA replication: The recombination connection. Trends Cell Biol. 2022, 32, 45–57. [Google Scholar] [CrossRef]
- Park, S.H.; Bennett-Baker, P.; Ahmed, S.; Arlt, M.F.; Ljungman, M.; Glover, T.W.; Wilson, T.E. Locus-specific transcription silencing at the FHIT gene suppresses replication stress-induced copy number variant formation and associated replication delay. Nucleic Acids Res. 2021, 49, 7507–7524. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Minocherhomji, S.; Ying, S.; Bjerregaard, V.A.; Bursomanno, S.; Aleliunaite, A.; Wu, W.; Mankouri, H.W.; Shen, H.; Liu, Y.; Hickson, I.D. Replication stress activates DNA repair synthesis in mitosis. Nature 2015, 528, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xu, D. Repair pathway choice for double-strand breaks. Essays Biochem. 2020, 64, 765–777. [Google Scholar] [CrossRef]
- Yoshioka, K.-I.; Kusumoto-Matsuo, R.; Matsuno, Y.; Ishiai, M. Genomic Instability and Cancer Risk Associated with Erroneous DNA Repair. Int. J. Mol. Sci. 2021, 22, 12254. [Google Scholar] [CrossRef] [PubMed]
- Wali, A.; Srinivasan, R.; Shabnam, M.S.; Majumdar, S.; Joshi, K.; Behera, D. Loss of Fragile Histidine Triad Gene Expression in Advanced Lung Cancer Is Consequent to Allelic Loss at 3p14 Locus and Promoter Methylation. Mol. Cancer Res. 2006, 4, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Saldivar, J.C.; Miuma, S.; Bene, J.; Hosseini, S.A.; Shibata, H.; Sun, J.; Wheeler, L.J.; Mathews, C.K.; Huebner, K. Initiation of Genome Instability and Preneoplastic Processes through Loss of Fhit Expression. PLoS Genet. 2012, 8, e1003077. [Google Scholar] [CrossRef]
- Cao, J.; Li, W.; Xie, J.; Du, H.; Tang, W.; Wang, H.; Chen, X.; Xiao, W.; Li, Y. Down-regulation of FHIT inhibits apoptosis of colorectal cancer: Mechanism and clinical implication. Surg. Oncol. 2006, 15, 223–233. [Google Scholar] [CrossRef]
- Hassan, I.; Naiyer, A.; Ahmad, F. Fragile histidine triad protein: Structure, function, and its association with tumorogenesis. J. Cancer Res. Clin. Oncol. 2010, 136, 333–350. [Google Scholar] [CrossRef]
- Czarnecka, K.H.; Migdalska-Sęk, M.; Domańska, D.; Pastuszak-Lewandoska, D.; Dutkowska, A.; Kordiak, J.; Nawrot, E.; Kiszałkiewicz, J.; Antczak, A.; Brzeziańska-Lasota, E. FHIT promoter methylation status, low protein and high mRNA levels in patients with non-small cell lung cancer. Int. J. Oncol. 2016, 49, 1175–1184. [Google Scholar] [CrossRef]
- Brisebarre, A.; Ancel, J.; Ponchel, T.; Loeffler, E.; Germain, A.; Dalstein, V.; Dormoy, V.; Durlach, A.; Delepine, G.; Deslée, G.; et al. Transcriptomic FHITlow/pHER2high signature as a predictive factor of outcome and immunotherapy response in non-small cell lung cancer. Front. Immunol. 2022, 13, 1058531. [Google Scholar] [CrossRef] [PubMed]
- Ismail, H.M.S.; Medhat, A.M.; Karim, A.M.; Zakhary, N.I. FHIT gene and flanking region on chromosome 3p are subjected to extensive allelic loss in Egyptian breast cancer patients. Mol. Carcinog. 2011, 50, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Callahan, C.L.; Bonner, M.R.; Nie, J.; Han, D.; Wang, Y.; Tao, M.-H.; Shields, P.G.; Marian, C.; Eng, K.H.; Trevisan, M.; et al. Lifetime exposure to ambient air pollution and methylation of tumor suppressor genes in breast tumors. Environ. Res. 2017, 161, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Cao, B.; Guo, M. The detective, prognostic, and predictive value of DNA methylation in human esophageal squamous cell carcinoma. Clin. Epigenetics 2016, 8, 43. [Google Scholar] [CrossRef]
- Heng, H.; Liu, G.; Stevens, J.; Abdallah, B.; Horne, S.; Ye, K.; Bremer, S.; Chowdhury, S.; Ye, C. Karyotype Heterogeneity and Unclassified Chromosomal Abnormalities. Cytogenet. Genome Res. 2013, 139, 144–157. [Google Scholar] [CrossRef]
- Heng, H.H.; Bremer, S.W.; Stevens, J.B.; Horne, S.D.; Liu, G.; Abdallah, B.Y.; Ye, K.J.; Ye, C.J. Chromosomal instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 2013, 32, 325–340. [Google Scholar] [CrossRef]
- Heng, H.H. The genome-centric concept: Resynthesis of evolutionary theory. Bioessays 2009, 31, 512–525. [Google Scholar] [CrossRef]
- Bissell, M.J.; Hines, W.C. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011, 17, 320–329. [Google Scholar] [CrossRef]
- Ye, C.J.; Regan, S.; Liu, G.; Alemara, S.; Heng, H.H. Understanding aneuploidy in cancer through the lens of system inheritance, fuzzy inheritance and emergence of new genome systems. Mol. Cytogenet. 2018, 11, 31. [Google Scholar] [CrossRef]
- Aguilera, A.; Gómez-González, B. Genome instability: A mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008, 9, 204–217. [Google Scholar] [CrossRef]
- Gatt, M.E.; Goldschmidt, N.; Kalichman, I.; Friedman, M.; Arronson, A.C.; Barak, V. Thymidine kinase levels correlate with prognosis in aggressive lymphoma and can discriminate patients with a clinical suspicion of indolent to aggressive transformation. Anticancer. Res. 2015, 35, 3019–3026. [Google Scholar] [PubMed]
- Hu, C.M.; Chang, Z.F. Mitotic control of dTTP pool: A necessity or coincidence? J. Biomed. Sci. 2007, 14, 491–497. [Google Scholar] [CrossRef] [PubMed]
- McAllister, K.A.; Yasseen, A.A.; McKerr, G.; Downes, C.S.; McKelvey-Martin, V.J. FISH comets show that the salvage enzyme TK1 contributes to gene-specific DNA repair. Front. Genet. 2014, 5, 233. [Google Scholar] [CrossRef] [PubMed]
- Bester, A.C.; Roniger, M.; Oren, Y.S.; Im, M.M.; Sarni, D.; Chaoat, M.; Bensimon, A.; Zamir, G.; Shewach, D.S.; Kerem, B. Nucleotide Deficiency Promotes Genomic Instability in Early Stages of Cancer Development. Cell 2011, 145, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Karras, J.R.; Paisie, C.A.; Huebner, K. Replicative Stress and the FHIT Gene: Roles in Tumor Suppression, Genome Stability and Prevention of Carcinogenesis. Cancers 2014, 6, 1208–1219. [Google Scholar] [CrossRef]
- Técher, H.; Pasero, P. The Replication Stress Response on a Narrow Path Between Genomic Instability and Inflammation. Front. Cell Dev. Biol. 2021, 9, 702584. [Google Scholar] [CrossRef]
- Hosseini, S.A.; Horton, S.; Saldivar, J.C.; Miuma, S.; Stampfer, M.R.; Heerema, N.A.; Huebner, K. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes Chromosom. Cancer 2013, 52, 1017–1029. [Google Scholar] [CrossRef]
- Saldivar, J.C.; Park, D. Mechanisms shaping the mutational landscape of the FRA3B/FHIT -deficient cancer genome. Genes Chromosom. Cancer 2019, 58, 317–323. [Google Scholar] [CrossRef]
- Deem, A.; Keszthelyi, A.; Blackgrove, T.; Vayl, A.; Coffey, B.; Mathur, R.; Chabes, A.; Malkova, A. Break-Induced Replication Is Highly Inaccurate. PLoS Biol. 2011, 9, e1000594. [Google Scholar] [CrossRef]
- Pilié, P.G.; Tang, C.; Mills, G.B.; Yap, T.A. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 2019, 16, 81–104. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Cantley, L.C. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef]
- Smith, J.; Tho, L.M.; Xu, N.; Gillespie, D.A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010, 108, 73–112. [Google Scholar] [PubMed]
- Wenzel, E.S.; Singh, A.T.K. Cell-cycle Checkpoints and Aneuploidy on the Path to Cancer. In Vivo 2018, 32, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef] [PubMed]
- Mertz, T.M.; Collins, C.D.; Dennis, M.; Coxon, M.; Roberts, S.A. APOBEC-Induced Mutagenesis in Cancer. Annu. Rev. Genet. 2022, 56, 229–252. [Google Scholar] [CrossRef] [PubMed]
- Bergstrom, E.N.; Luebeck, J.; Petljak, M.; Khandekar, A.; Barnes, M.; Zhang, T.; Steele, C.D.; Pillay, N.; Landi, M.T.; Bafna, V.; et al. Mapping clustered mutations in cancer reveals APOBEC3 mutagenesis of ecDNA. Nature 2022, 602, 510–517. [Google Scholar] [CrossRef]
- Boichard, A.; Tsigelny, I.F.; Kurzrock, R. High expression of PD-1 ligands is associated with kataegis mutational signature and APOBEC3 alterations. Oncoimmunology 2017, 6, e1284719. [Google Scholar] [CrossRef]
- Chan, K.; Roberts, S.A.; Klimczak, L.J.; Sterling, J.F.; Saini, N.; Malc, E.; Kim, J.; Kwiatkowski, D.J.; Fargo, D.C.; Mieczkowski, P.; et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015, 47, 1067–1072. [Google Scholar] [CrossRef]
- Zhao, D.; Li, J.; Li, S.; Xin, X.; Hu, M.; Price, M.A.; Rosser, S.J.; Bi, C.; Zhang, X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021, 39, 35–40. [Google Scholar] [CrossRef]
- Liu, W.; Wu, J.; Yang, F.; Ma, L.; Ni, C.; Hou, X.; Wang, L.; Xu, A.; Song, J.; Deng, Y.; et al. Genetic Polymorphisms Predisposing the Interleukin 6–Induced APOBEC3B-UNG Imbalance Increase HCC Risk via Promoting the Generation of APOBEC-Signature HBV Mutations. Clin. Cancer Res. 2019, 25, 5525–5536. [Google Scholar] [CrossRef]
- Serebrenik, A.A.; Starrett, G.J.; Leenen, S.; Jarvis, M.C.; Shaban, N.M.; Salamango, D.J.; Nilsen, H.; Brown, W.L.; Harris, R.S. The deaminase APOBEC3B triggers the death of cells lacking uracil DNA glycosylase. Proc. Natl. Acad. Sci. USA 2019, 116, 22158–22163. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Du, Y.; Zhang, Q.; Han, X.; Cao, G. Human cytidine deaminases facilitate hepatitis B virus evolution and link inflammation and hepatocellular carcinoma. Cancer Lett. 2014, 343, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Vural, S.; Simon, R.; Krushkal, J. Correlation of gene expression and associated mutation profiles of APOBEC3A, APOBEC3B, REV1, UNG, and FHIT with chemosensitivity of cancer cell lines to drug treatment. Hum. Genom. 2018, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Karras, J.R.; Schrock, M.S.; Batar, B.; Huebner, K. Fragile Genes That Are Frequently Altered in Cancer: Players Not Passengers. Cytogenet. Genome Res. 2016, 150, 208–216. [Google Scholar] [CrossRef]
- Paisie, C.A.; Schrock, M.S.; Karras, J.R.; Zhang, J.; Miuma, S.; Ouda, I.M.; Waters, C.E.; Saldivar, J.C.; Druck, T.; Huebner, K. Exome-wide single-base substitutions in tissues and derived cell lines of the constitutive Fhit knockout mouse. Cancer Sci. 2016, 107, 528–535. [Google Scholar] [CrossRef]
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N.; Nikas, J.B.; et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366–370. [Google Scholar] [CrossRef]
- Waters, C.E.; Saldivar, J.C.; Amin, Z.A.; Schrock, M.S.; Huebner, K. FHIT loss-induced DNA damage creates optimal APOBEC substrates: Insights into APOBEC-mediated mutagenesis. Oncotarget 2014, 6, 3409–3419. [Google Scholar] [CrossRef]
- Guo, R.; Wang, Y.; Shi, W.-Y.; Liu, B.; Hou, S.-Q.; Liu, L. MicroRNA miR-491-5p Targeting both TP53 and Bcl-XL Induces Cell Apoptosis in SW1990 Pancreatic Cancer Cells through Mitochondria Mediated Pathway. Molecules 2012, 17, 14733–14747. [Google Scholar] [CrossRef]
- Su, C.M.; Weng, Y.S.; Kuan, L.Y.; Chen, J.H.; Hsu, F.T. Suppression of PKCδ/NF-κB Signaling and Apoptosis Induction through Extrinsic/Intrinsic Pathways Are Associated Magnolol-Inhibited Tumor Progression in Colorectal Cancer In Vitro and In Vivo. Int. J. Mol. Sci. 2020, 21, 3527. [Google Scholar] [CrossRef]
- Rimessi, A.; Marchi, S.; Fotino, C.; Romagnoli, A.; Huebner, K.; Croce, C.M.; Pinton, P.; Rizzuto, R. Intramitochondrial calcium regulation by the FHIT gene product sensitizes to apoptosis. Proc. Natl. Acad. Sci. USA 2009, 106, 12753–12758. [Google Scholar] [CrossRef]
- Silveira Zavalhia, L.; Weber Medeiros, A.; Oliveira Silva, A.; Vial Roehe, A. Do FHIT gene alterations play a role in human solid tumors? Asia Pac. J. Clin. Oncol. 2018, 14, e214–e223. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.-S.; Yoo, J.Y.; Cui, R.; Kaur, B.; Huebner, K.; Lee, T.-K.; Aqeilan, R.I.; Croce, C.M. FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs. PLoS Genet. 2014, 10, e1004652. [Google Scholar] [CrossRef] [PubMed]
- Joannes, A.; Grelet, S.; Duca, L.; Gilles, C.; Kileztky, C.; Dalstein, V.; Birembaut, P.; Polette, M.; Nawrocki-Raby, B. Fhit Regulates EMT Targets through an EGFR/Src/ERK/Slug Signaling Axis in Human Bronchial Cells. Mol. Cancer Res. 2014, 12, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Tian, W.; Jiang, Z.; Huang, T.; Ge, C.; Liu, T.; Zhao, F.; Chen, T.; Cui, Y.; Li, H.; et al. A Positive Feedback Loop of AKR1C3-Mediated Activation of NF-κB and STAT3 Facilitates Proliferation and Metastasis in Hepatocellular Carcinoma. Cancer Res. 2021, 81, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-H.; Chang, Y.-W.; Hong, M.-X.; Hsu, T.-C.; Lee, K.-C.; Lin, C.; Lee, J.-L. STAT3 phosphorylation at Ser727 and Tyr705 differentially regulates the EMT–MET switch and cancer metastasis. Oncogene 2020, 40, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.L.; Klement, J.D.; Lu, C.; Redd, P.S.; Xiao, W.; Yang, D.; Browning, D.D.; Savage, N.M.; Buckhaults, P.J.; Morse, H.C., 3rd; et al. Myeloid-Derived Suppressor Cells Produce IL-10 to Elicit DNMT3b-Dependent IRF8 Silencing to Promote Colitis-Associated Colon Tumorigenesis. Cell Rep. 2018, 25, 3036–3046.e6. [Google Scholar] [CrossRef]
- Leão, R.; Apolónio, J.D.; Lee, D.; Figueiredo, A.; Tabori, U.; Castelo-Branco, P. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J. Biomed. Sci. 2018, 25, 22. [Google Scholar] [CrossRef]
- Zhang, K.; Guo, Y.; Wang, X.; Zhao, H.; Ji, Z.; Cheng, C.; Li, L.; Fang, Y.; Xu, D.; Zhu, H.; et al. WNT/β-Catenin Directs Self-Renewal Symmetric Cell Division of hTERT(high) Prostate Cancer Stem Cells. Cancer Res. 2017, 77, 2534–2547. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Z.; Jiang, D.; Shen, J.; Liu, W.; Tan, X.; Cao, G. Potential Role of the Fragile Histidine Triad in Cancer Evo-Dev. Cancers 2023, 15, 1144. https://doi.org/10.3390/cancers15041144
Niu Z, Jiang D, Shen J, Liu W, Tan X, Cao G. Potential Role of the Fragile Histidine Triad in Cancer Evo-Dev. Cancers. 2023; 15(4):1144. https://doi.org/10.3390/cancers15041144
Chicago/Turabian StyleNiu, Zheyun, Dongming Jiang, Jiaying Shen, Wenbin Liu, Xiaojie Tan, and Guangwen Cao. 2023. "Potential Role of the Fragile Histidine Triad in Cancer Evo-Dev" Cancers 15, no. 4: 1144. https://doi.org/10.3390/cancers15041144
APA StyleNiu, Z., Jiang, D., Shen, J., Liu, W., Tan, X., & Cao, G. (2023). Potential Role of the Fragile Histidine Triad in Cancer Evo-Dev. Cancers, 15(4), 1144. https://doi.org/10.3390/cancers15041144





