Pixelated Microfluidics for Drug Screening on Tumour Spheroids and Ex Vivo Microdissected Tumour Explants
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
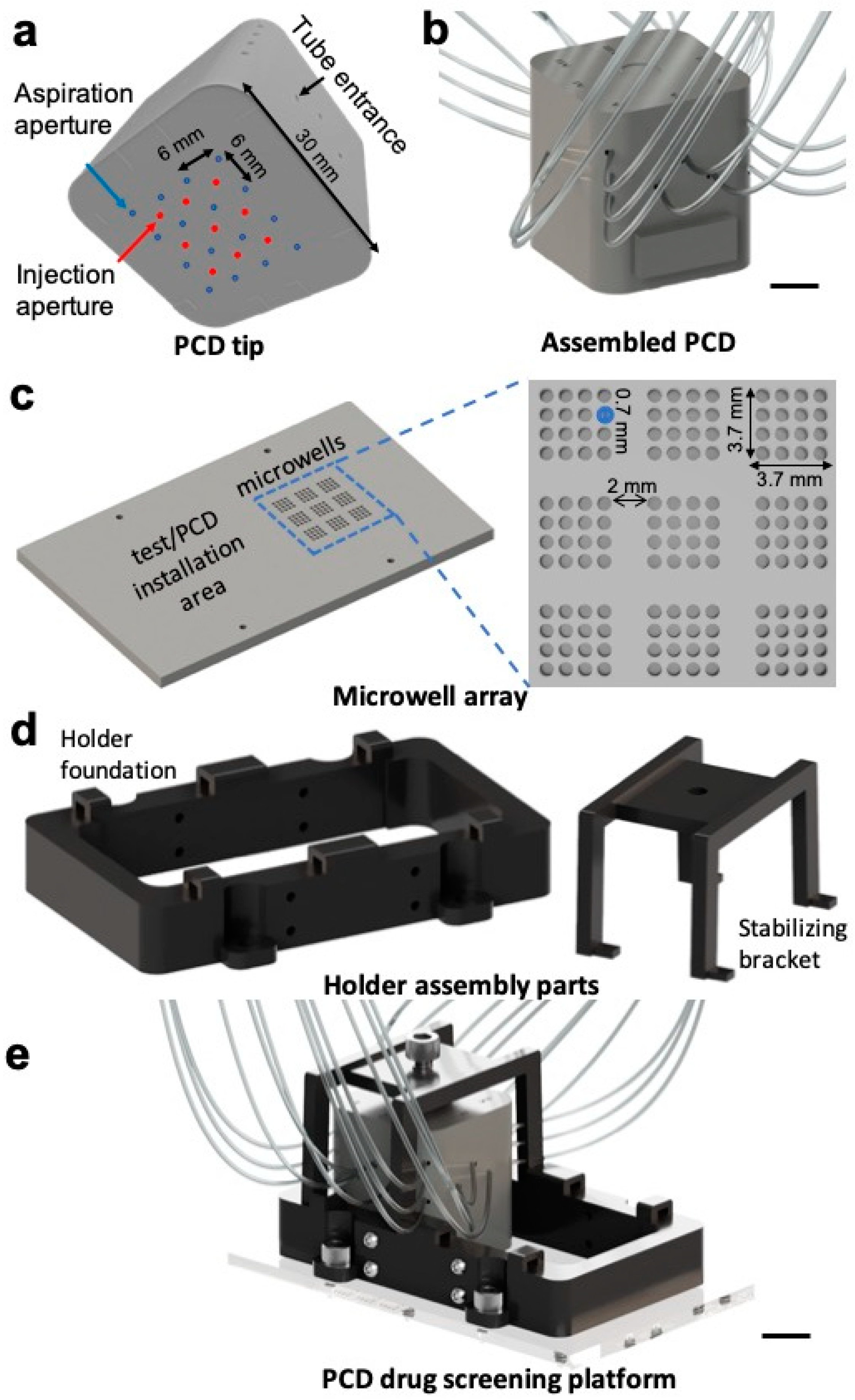
2.1. Design and Fabrication of Parts
2.2. System Operation
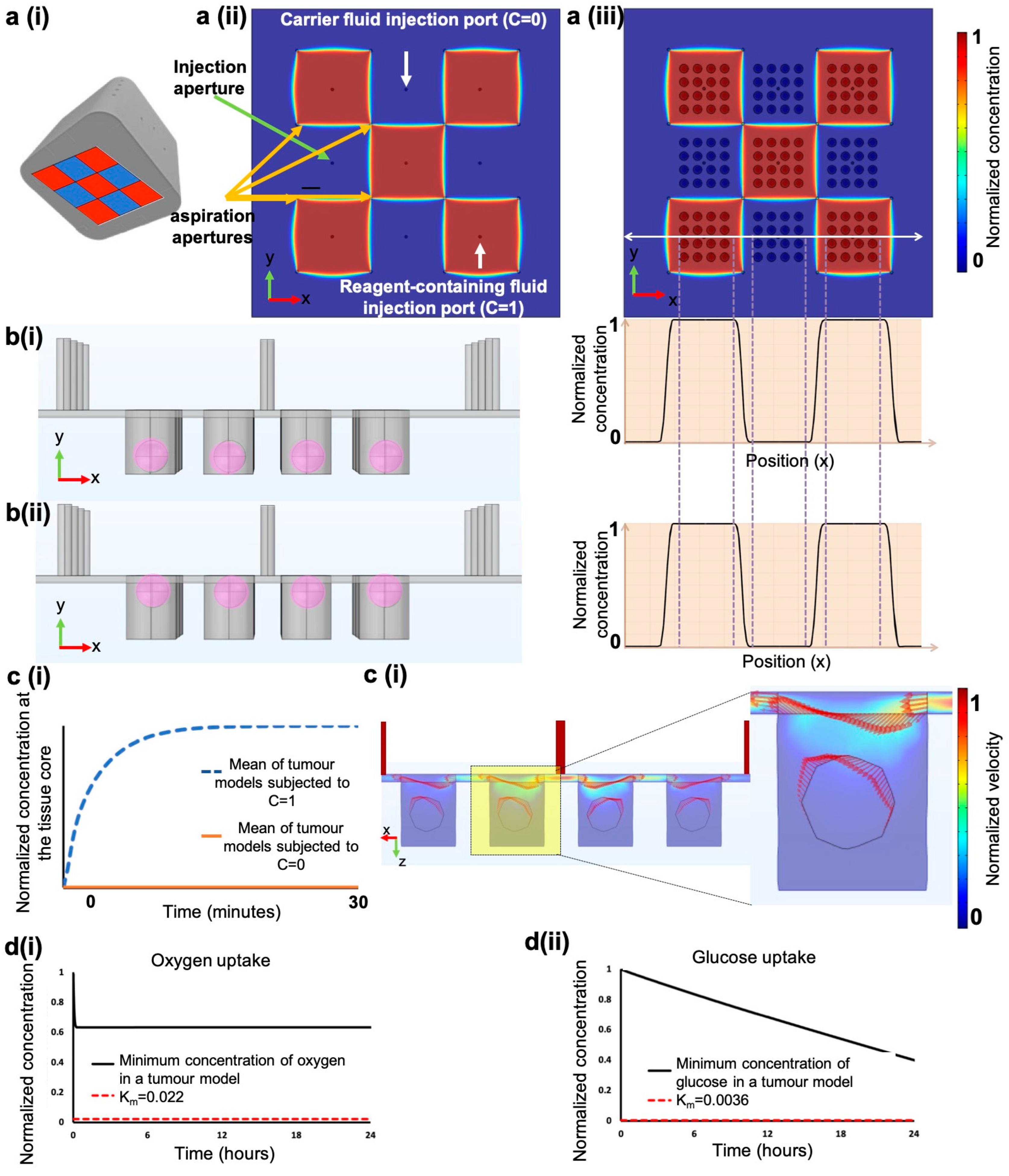
2.3. Finite Element Methodology
2.4. Cancer Cell Lines Xenograft Tumour Production
2.5. MDT Production from Cell Line Xenograft Tumours
2.6. Microwell Preparation and MDT Loading in the Microwell Array
2.7. Spheroid Formation Assay
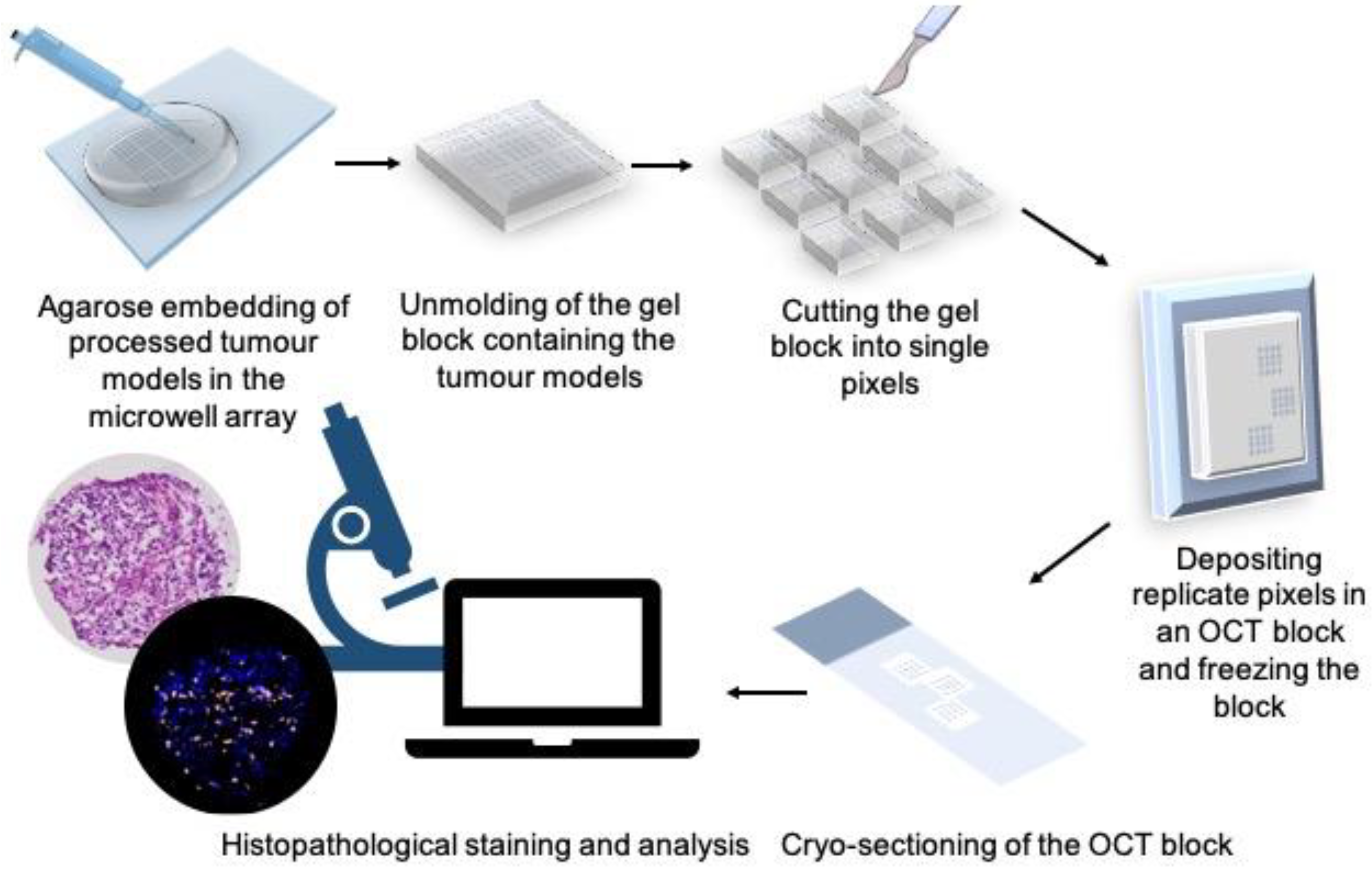
2.8. OCT Embedding Protocol
2.9. Tumour Model Treatment with TNF
2.10. Histopathological Staining
2.11. Quantification of Immunofluorescent Staining
2.12. Statistical Analysis
3. Results
3.1. Design and Fabrication of the Pixelated Chemical Display Drug Screening Platform
3.2. Perifusion vs. Perfusion
3.3. Pressure Pump-Operated Fluidic Lines
3.4. Finite Element Simulations
3.5. High-Throughput Formation of Cancer Cell Line Spheroids Is Possible in the Microwell Array
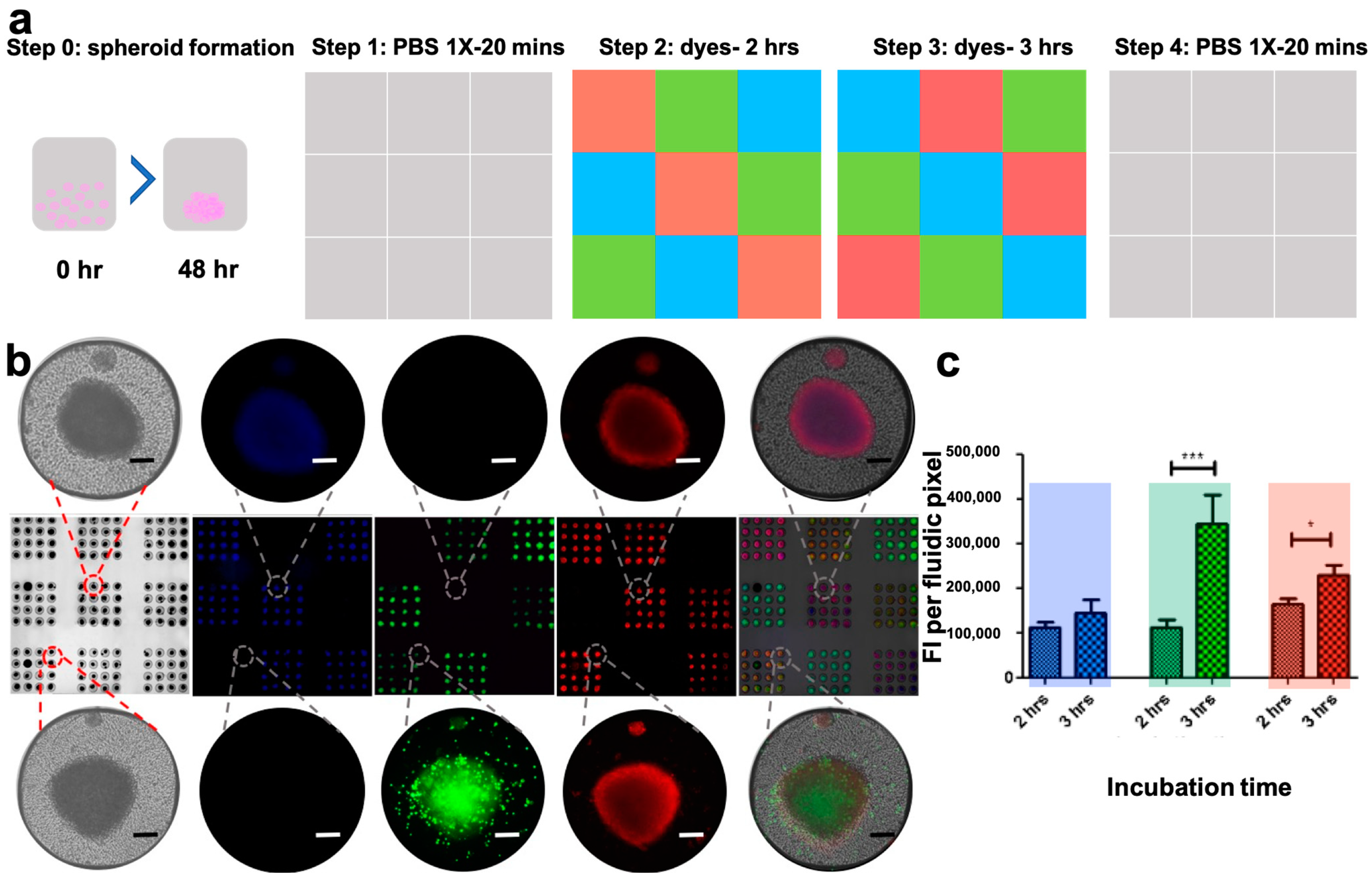
3.6. The PCD Drug Screening Platform Enables Dynamic Multiplexed Staining of Spheroids
3.7. The PCD Drug Screening Platform Can Handle Fragile Ex Vivo Tumour Tissue Explants
3.8. Tumour Tissue Microarray
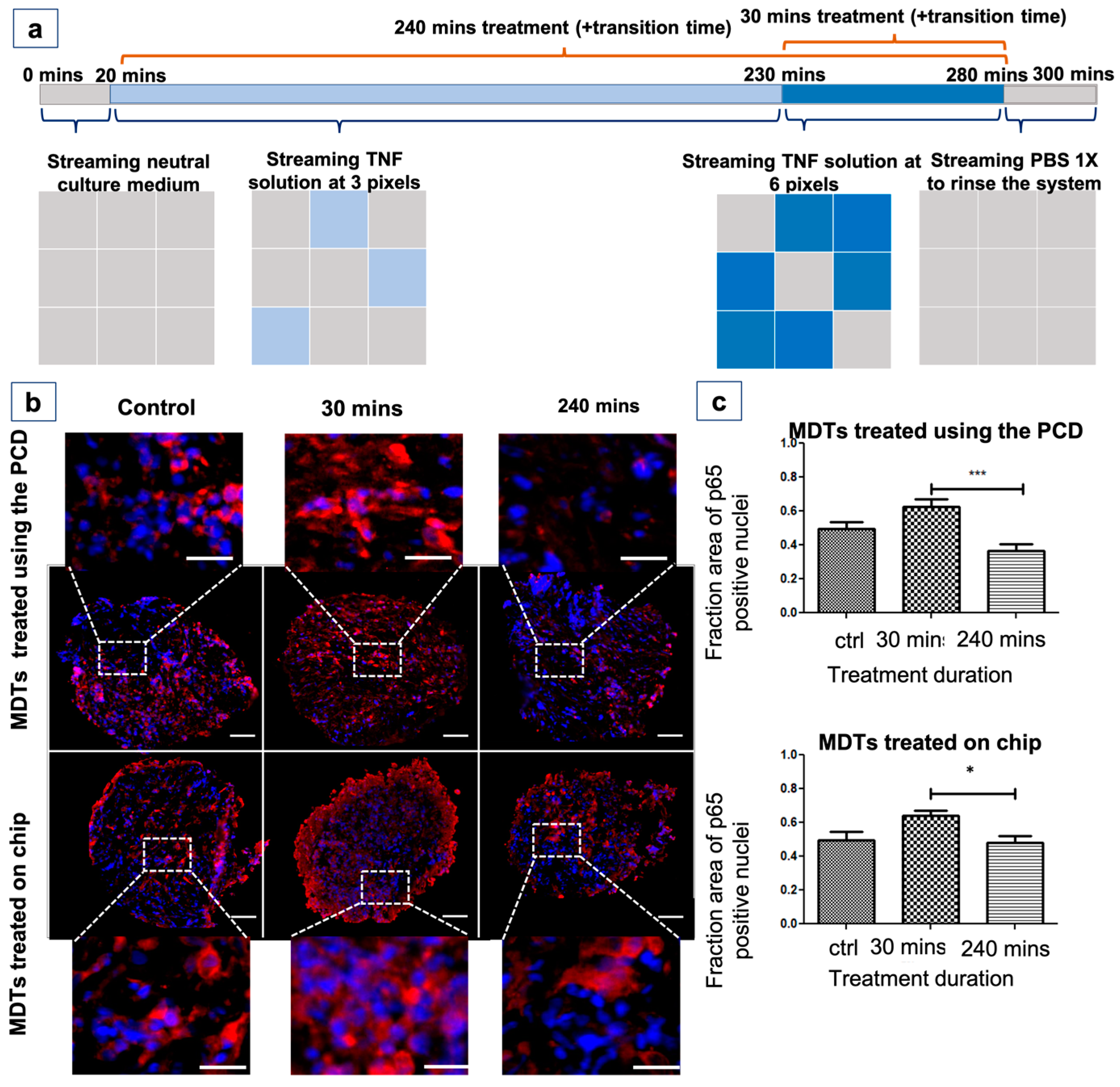
3.9. The PCD Drug Screening Platform Enables the Tracking of Biological Responses in Tumour Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mullard, A. Parsing clinical success rates. Nat. Rev. Drug Discov. 2016, 15, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef]
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2019, 20, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Ibarrola-Villava, M.; Cervantes, A.; Bardelli, A. Preclinical models for precision oncology. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2018, 1870, 239–246. [Google Scholar] [CrossRef]
- Misra, S.; Moro, C.F.; Del Chiaro, M.; Pouso, S.; Sebestyén, A.; Löhr, M.; Björnstedt, M.; Verbeke, C.S. Ex vivo organotypic culture system of precision-cut slices of human pancreatic ductal adenocarcinoma. Sci. Rep. 2019, 9, 2133. [Google Scholar] [CrossRef]
- Lin, R.Z.; Chang, H.Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. Healthc. Nutr. Technol. 2008, 3, 1172–1184. [Google Scholar]
- Kunz-Schughart, L.A.; Freyer, J.P.; Hofstaedter, F.; Ebner, R. The use of 3-D cultures for high-throughput screening: The multicellular spheroid model. J. Biomol. Screen. 2004, 9, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Tuveson, D.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955. [Google Scholar] [CrossRef] [PubMed]
- Powley, I.R.; Patel, M.; Miles, G.; Pringle, H.; Howells, L.; Thomas, A.; Kettleborough, C.; Bryans, J.; Hammonds, T.; MacFarlane, M. Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br. J. Cancer 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, M.N.; Simeone, K.; Leclerc-Deslauniers, K.; Fleury, H.; Carmona, E.; Provencher, D.M.; Mes-Masson, A.-M. Carboplatin response in preclinical models for ovarian cancer: Comparison of 2D monolayers, spheroids, ex vivo tumors and in vivo models. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nyga, A.; Cheema, U.; Loizidou, M. 3D tumour models: Novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 2011, 5, 239. [Google Scholar] [CrossRef] [PubMed]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T. Comparison of 2D-and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Verjans, E.T.; Doijen, J.; Luyten, W.; Landuyt, B.; Schoofs, L. Three-dimensional cell culture models for anticancer drug screening: Worth the effort? J. Cell. Physiol. 2018, 233, 2993–3003. [Google Scholar] [CrossRef]
- Horowitz, L.F.; Rodriguez, A.D.; Au-Yeung, A.; Bishop, K.W.; Barner, L.A.; Mishra, G.; Raman, A.; Delgado, P.; Liu, J.T.; Gujral, T.S. Microdissected “cuboids” for microfluidic drug testing of intact tissues. Lab A Chip 2021, 21, 122–142. [Google Scholar] [CrossRef]
- Helmchen, F.; Denk, W. Deep tissue two-photon microscopy. Nat. Methods 2005, 2, 932–940. [Google Scholar] [CrossRef]
- Gurcan, M.N.; Boucheron, L.E.; Can, A.; Madabhushi, A.; Rajpoot, N.M.; Yener, B. Histopathological image analysis: A review. IEEE Rev. Biomed. Eng. 2009, 2, 147–171. [Google Scholar] [CrossRef]
- Dorrigiv, D.; Simeone, K.; Communal, L.; Kendall-Dupont, J.; St-Georges-Robillard, A.; Péant, B.; Carmona, E.; Mes-Masson, A.-M.; Gervais, T. Microdissected Tissue vs. Tissue Slices—A Comparative Study of Tumor Explant Models Cultured On-Chip and Off-Chip. Cancers 2021, 13, 4208. [Google Scholar] [CrossRef]
- Simeone, K.; Guay-Lord, R.; Lateef, M.A.; Péant, B.; Kendall-Dupont, J.; Orimoto, A.M.; Carmona, E.; Provencher, D.; Saad, F.; Gervais, T. Paraffin-embedding lithography and micro-dissected tissue micro-arrays: Tools for biological and pharmacological analysis of ex vivo solid tumors. Lab A Chip 2019, 19, 693–705. [Google Scholar] [CrossRef]
- Qasaimeh, M.A.; Ricoult, S.G.; Juncker, D. Microfluidic probes for use in life sciences and medicine. Lab A Chip 2013, 13, 40–50. [Google Scholar] [CrossRef]
- Kaigala, G.V.; Lovchik, R.D.; Delamarche, E. Microfluidics in the “open space” for performing localized chemistry on biological interfaces. Angew. Chem. Int. Ed. 2012, 51, 11224–11240. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Kolitz, S.; Lauffenburger, D.A.; Han, J. Microfluidic probe for single-cell analysis in adherent tissue culture. Nat. Commun. 2014, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Brimmo, A.T.; Menachery, A.; Qasaimeh, M.A. Microelectrofluidic probe for sequential cell separation and patterning. Lab A Chip 2019, 19, 4052–4063. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, A.; Autebert, J.; Delamarche, E.; Kaigala, G.V. Selective local lysis and sampling of live cells for nucleic acid analysis using a microfluidic probe. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Queval, A.; Ghattamaneni, N.R.; Perrault, C.M.; Gill, R.; Mirzaei, M.; McKinney, R.A.; Juncker, D. Chamber and microfluidic probe for microperfusion of organotypic brain slices. Lab A Chip 2010, 10, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Lovchik, R.D.; Kaigala, G.V.; Georgiadis, M.; Delamarche, E. Micro-immunohistochemistry using a microfluidic probe. Lab A Chip 2012, 12, 1040–1043. [Google Scholar] [CrossRef]
- Li, X.; Yin, R.; Hu, H.; Li, Y.; Sun, X.; Dey, S.K.; Laskin, J. An integrated microfluidic probe for mass spectrometry imaging of biological samples. Angew. Chem. 2020, 132, 22574–22577. [Google Scholar] [CrossRef]
- Brimmo, A.; Goyette, P.-A.; Alnemari, R.; Gervais, T.; Qasaimeh, M.A. 3D Printed Microfluidic Probes. Sci. Rep. 2018, 8, 10995. [Google Scholar] [CrossRef]
- Qasaimeh, M.A.; Gervais, T.; Juncker, D. Microfluidic quadrupole and floating concentration gradient. Nat. Commun. 2011, 2, 1–8. [Google Scholar] [CrossRef]
- Goyette, P.-A.; Boulais, É.; Tremblay, M.; Gervais, T. Pixel-based open-space microfluidics for versatile surface processing. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Gehrke, S.H.; Fisher, J.P.; Palasis, M.; Lund, M.E. Factors determining hydrogel permeability. Ann. N. Y. Acad. Sci. 1997, 831, 179–207. [Google Scholar] [CrossRef] [PubMed]
- Ramanujan, S.; Pluen, A.; McKee, T.D.; Brown, E.B.; Boucher, Y.; Jain, R.K. Diffusion and convection in collagen gels: Implications for transport in the tumor interstitium. Biophys. J. 2002, 83, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Astolfi, M.; Péant, B.; Lateef, M.; Rousset, N.; Kendall-Dupont, J.; Carmona, E.; Monet, F.; Saad, F.; Provencher, D.; Mes-Masson, A.-M. Micro-dissected tumor tissues on chip: An ex vivo method for drug testing and personalized therapy. Lab A Chip 2016, 16, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Casciari, J.J.; Sotirchos, S.V.; Sutherland, R.M. Variations in tumor cell growth rates and metabolism with oxygen concentration, glucose concentration, and extracellular pH. J. Cell. Physiol. 1992, 151, 386–394. [Google Scholar] [CrossRef]
- Rousset, N.; Monet, F.; Gervais, T. Simulation-assisted design of microfluidic sample traps for optimal trapping and culture of non-adherent single cells, tissues, and spheroids. Sci. Rep. 2017, 7, 245. [Google Scholar] [CrossRef]
- Sorensen, R.; Novak, N. The use of Michaelis-Menten kinetics in cell biology and physiology teaching laboratories. Biochem. Educ. 1996, 24, 26–28. [Google Scholar] [CrossRef]
- Cahuzac, M.; Langlois, P.; Péant, B.; Fleury, H.; Mes-Masson, A.-M.; Saad, F. Pre-activation of autophagy impacts response to olaparib in prostate cancer cells. Commun. Biol. 2022, 5, 1–14. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Labouba, I.; Le Page, C.; Communal, L.; Kristessen, T.; You, X.; Péant, B.; Barrès, V.; Gannon, P.O.; Mes-Masson, A.-M.; Saad, F. Potential Cross-Talk between Alternative and Classical NF-κB Pathways in Prostate Cancer Tissues as Measured by a Multi-Staining Immunofluorescence Co-Localization Assay. PLoS ONE 2015, 10, e0131024. [Google Scholar] [CrossRef]
- Goyette, P.-A.; Boulais, É.; Normandeau, F.; Laberge, G.; Juncker, D.; Gervais, T. Microfluidic multipoles theory and applications. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Hammel, J.H.; Cook, S.R.; Belanger, M.C.; Munson, J.M.; Pompano, R.R. Modeling Immunity In Vitro: Slices, Chips, and Engineered Tissues. Annu. Rev. Biomed. Eng. 2021, 23, 461–491. [Google Scholar] [CrossRef] [PubMed]
- Hattersley, S.M.; Dyer, C.E.; Greenman, J.; Haswell, S.J. Development of a microfluidic device for the maintenance and interrogation of viable tissue biopsies. Lab A Chip 2008, 8, 1842–1846. [Google Scholar] [CrossRef] [PubMed]
- Van Midwoud, P.M.; Groothuis, G.M.; Merema, M.T.; Verpoorte, E. Microfluidic biochip for the perifusion of precision-cut rat liver slices for metabolism and toxicology studies. Biotechnol. Bioeng. 2010, 105, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Dawson, A.; Dyer, C.; Macfie, J.; Davies, J.; Karsai, L.; Greenman, J.; Jacobsen, M. A microfluidic chip based model for the study of full thickness human intestinal tissue using dual flow. Biomicrofluidics 2016, 10, 064101. [Google Scholar] [CrossRef]
- Cheah, L.-T.; Dou, Y.-H.; Seymour, A.-M.L.; Dyer, C.E.; Haswell, S.J.; Wadhawan, J.D.; Greenman, J. Microfluidic perfusion system for maintaining viable heart tissue with real-time electrochemical monitoring of reactive oxygen species. Lab A Chip 2010, 10, 2720–2726. [Google Scholar] [CrossRef]
- Bong, K.W.; Chapin, S.C.; Pregibon, D.C.; Baah, D.; Floyd-Smith, T.M.; Doyle, P.S. Compressed-air flow control system. Lab A Chip 2011, 11, 743–747. [Google Scholar] [CrossRef]
- Zeng, W.; Li, S.; Wang, Z. Characterization of syringe-pump-driven versus pressure-driven microfluidic flows. In Proceedings of the 2015 International Conference on Fluid Power and Mechatronics (FPM), Harbin, China, 5–7 August 2015; pp. 711–715. [Google Scholar]
- Di Carlo, D.; Wu, L.Y.; Lee, L.P. Dynamic single cell culture array. Lab A Chip 2006, 6, 1445–1449. [Google Scholar] [CrossRef]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016, 6, 19103. [Google Scholar] [CrossRef]
- Kwak, B.; Lee, Y.; Lee, J.; Lee, S.; Lim, J. Mass fabrication of uniform sized 3D tumor spheroid using high-throughput microfluidic system. J. Control. Release 2018, 275, 201–207. [Google Scholar] [CrossRef]
- Sakai, Y.; Nakazawa, K. Technique for the control of spheroid diameter using microfabricated chips. Acta Biomater. 2007, 3, 1033–1040. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, M.H.; Tabaei, S.R.; Park, J.H.; Na, K.; Chung, S.; Zhdanov, V.P.; Cho, N.-J. Spheroid formation of hepatocarcinoma cells in microwells: Experiments and Monte Carlo simulations. PloS ONE 2016, 11, e0161915. [Google Scholar] [CrossRef] [PubMed]
- Azizipour, N.; Avazpour, R.; Sawan, M.; Ajji, A.; Rosenzweig, D.H. Surface Optimization and Design Adaptation toward Spheroid Formation On-Chip. Sensors 2022, 22, 3191. [Google Scholar] [CrossRef] [PubMed]
- Majumder, B.; Baraneedharan, U.; Thiyagarajan, S.; Radhakrishnan, P.; Narasimhan, H.; Dhandapani, M.; Brijwani, N.; Pinto, D.D.; Prasath, A.; Shanthappa, B.U. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.V.; Calabresi, P.A. Agar-gelatin for embedding tissues prior to paraffin processing. Biotechniques 2007, 42, 569–570. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, C.M.; Stavnes, H.T.; Kleinberg, L.; Berner, A.; Hernandez, L.F.; Birrer, M.J.; Steinberg, S.M.; Davidson, B.; Kohn, E.C. Nuclear factor κB transcription factors are coexpressed and convey a poor outcome in ovarian cancer. Cancer 2010, 116, 3276–3284. [Google Scholar] [CrossRef]
- Ahn, K.S.; Aggarwal, B.B. Transcription factor NF-κB: A sensor for smoke and stress signals. Ann. N. Y. Acad. Sci. 2005, 1056, 218–233. [Google Scholar] [CrossRef]
- Maguire, O.; Collins, C.; O’Loughlin, K.; Miecznikowski, J.; Minderman, H. Quantifying nuclear p65 as a parameter for NF-κB activation: Correlation between ImageStream cytometry, microscopy, and Western blot. Cytom. Part A 2011, 79, 461–469. [Google Scholar] [CrossRef]
- Badr, C.E.; Niers, J.M.; Tjon-Kon-Fat, L.-A.; Noske, D.P.; Wurdinger, T.; Tannous, B.A. Real-time monitoring of nuclear factor κB activity in cultured cells and in animal models. Mol. Imaging 2009, 8, 7290.2009. 00026. [Google Scholar] [CrossRef]
- Péant, B.; Diallo, J.-S.; Lessard, L.; Delvoye, N.; Le Page, C.; Saad, F.; Mes-Masson, A.-M. Regulation of IκB kinase ε expression by the androgen receptor and the nuclear factor-κB transcription factor in prostate cancer. Mol. Cancer Res. 2007, 5, 87–94. [Google Scholar] [CrossRef]
- Kocbek, V.; Imboden, S.; Nirgianakis, K.; Mueller, M.; McKinnon, B. Dual influence of TNFα on diverse in vitro models of ovarian cancer subtypes. Heliyon 2021, 7, e06099. [Google Scholar] [CrossRef]
- Figenschau, Y.; Sveinbjörnsson, B.; Bertheussen, K. Improvement of a cytokine (TNF-α) bioassay by serum-free target cell (WEHI 164) cultivation. Cytotechnology 1999, 29, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, G.; Zaidi, M.; Iqbal, J. TNF-induced gene expression oscillates in time. Biochem. Biophys. Res. Commun. 2008, 371, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Aulner, N.; Bickle, M.; Davies, A.M.; Nery, E.D.; Ebner, D.; Montoya, M.C.; Östling, P.; Pietiäinen, V.; Price, L.S. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 2016, 15, 751–769. [Google Scholar] [CrossRef] [PubMed]
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114. [Google Scholar] [PubMed]
- Smalley, K.S.; Haass, N.K.; Brafford, P.A.; Lioni, M.; Flaherty, K.T.; Herlyn, M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol. Cancer Ther. 2006, 5, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, N.; Höfig, I.; Rosemann, M.; Szumielewski, J.; Richter, S.; Schorpp, K.; Hadian, K.; Aubele, M.; Atkinson, M.J.; Anastasov, N. Three-dimensional microtissues essentially contribute to preclinical validations of therapeutic targets in breast cancer. Cancer Med. 2016, 5, 703–710. [Google Scholar] [CrossRef]
- Teh, J.L.; Abdul Rahman, S.F.; Domnic, G.; Satiyasilan, L.; Chear, N.J.Y.; Singh, D.; Mohana-Kumaran, N. Rapid spheroid assays in a 3-dimensional cell culture chip. BMC Res. Notes 2021, 14, 1–6. [Google Scholar] [CrossRef]
- Lanz, H.L.; Saleh, A.; Kramer, B.; Cairns, J.; Ng, C.P.; Yu, J.; Trietsch, S.J.; Hankemeier, T.; Joore, J.; Vulto, P. Therapy response testing of breast cancer in a 3D high-throughput perfused microfluidic platform. BMC Cancer 2017, 17, 1–11. [Google Scholar] [CrossRef]
- Place, T.L.; Domann FE Case, A.J. Limitations of oxygen delivery to cells in culture: An underappreciated problem in basic and translational research. Free. Radic. Biol. Med. 2017, 113, 311–322. [Google Scholar] [CrossRef]
- Ariga, O.; Kubo, T.; Sano, Y. Effective diffusivity of glucose in PVA hydrogel. J. Ferment. Bioeng. 1994, 78, 200–201. [Google Scholar]
- Figueiredo, L.; Pace, R.; D’Arros, C.; Réthoré, G.; Guicheux, J.; Le Visage, C.; Weiss, P. Assessing glucose and oxygen diffusion in hydrogels for the rational design of 3D stem cell scaffolds in regenerative medicine. J. Tissue Eng. Regen. Med. 2018, 12, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Markov, D.A.; Lillie, E.M.; Garbett, S.P.; McCawley, L.J. Variation in diffusion of gases through PDMS due to plasma surface treatment and storage conditions. Biomed. Microdevices 2014, 16, 91–96. [Google Scholar] [CrossRef]
- Chowdhury, S.; Bhethanabotla, V.R.; Sen, R. Measurement of oxygen diffusivity and permeability in polymers using fluorescence microscopy. Microsc. Microanal. 2010, 16, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Hicks, K.O.; Ohms, S.J.; van Zijl, P.L.; Denny, W.A.; Hunter, P.J.; Wilson, W.R. An experimental and mathematical model for the extravascular transport of a DNA intercalator in tumours. Br J Cancer. 1997, 76, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Pruijn, F.B.; Patel, K.; Hay, M.P.; Wilson WR Hicks, K.O. Prediction of tumour tissue diffusion coefficients of hypoxia-activated prodrugs from physicochemical parameters. Aust. J. Chem. 2008, 61, 687–693. [Google Scholar] [CrossRef]
- Abaci, H.E.; Truitt, R.; Tan SGerecht, S. Unforeseen decreases in dissolved oxygen levels affect tube formation kinetics in collagen gels. Am. J. Physiol. -Cell Physiol. 2011, 301, C431–C440. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorrigiv, D.; Goyette, P.-A.; St-Georges-Robillard, A.; Mes-Masson, A.-M.; Gervais, T. Pixelated Microfluidics for Drug Screening on Tumour Spheroids and Ex Vivo Microdissected Tumour Explants. Cancers 2023, 15, 1060. https://doi.org/10.3390/cancers15041060
Dorrigiv D, Goyette P-A, St-Georges-Robillard A, Mes-Masson A-M, Gervais T. Pixelated Microfluidics for Drug Screening on Tumour Spheroids and Ex Vivo Microdissected Tumour Explants. Cancers. 2023; 15(4):1060. https://doi.org/10.3390/cancers15041060
Chicago/Turabian StyleDorrigiv, Dina, Pierre-Alexandre Goyette, Amélie St-Georges-Robillard, Anne-Marie Mes-Masson, and Thomas Gervais. 2023. "Pixelated Microfluidics for Drug Screening on Tumour Spheroids and Ex Vivo Microdissected Tumour Explants" Cancers 15, no. 4: 1060. https://doi.org/10.3390/cancers15041060
APA StyleDorrigiv, D., Goyette, P.-A., St-Georges-Robillard, A., Mes-Masson, A.-M., & Gervais, T. (2023). Pixelated Microfluidics for Drug Screening on Tumour Spheroids and Ex Vivo Microdissected Tumour Explants. Cancers, 15(4), 1060. https://doi.org/10.3390/cancers15041060






