The Receptor for Advanced Glycation Endproducts (RAGE) and Its Ligands S100A8/A9 and High Mobility Group Box Protein 1 (HMGB1) Are Key Regulators of Myeloid-Derived Suppressor Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. S100A8 and A9 Are Pro-inflammatory Proteins
3. S100A8 and A9 Are Widely Expressed in Cancer and Strongly Associated with Tumor Progression
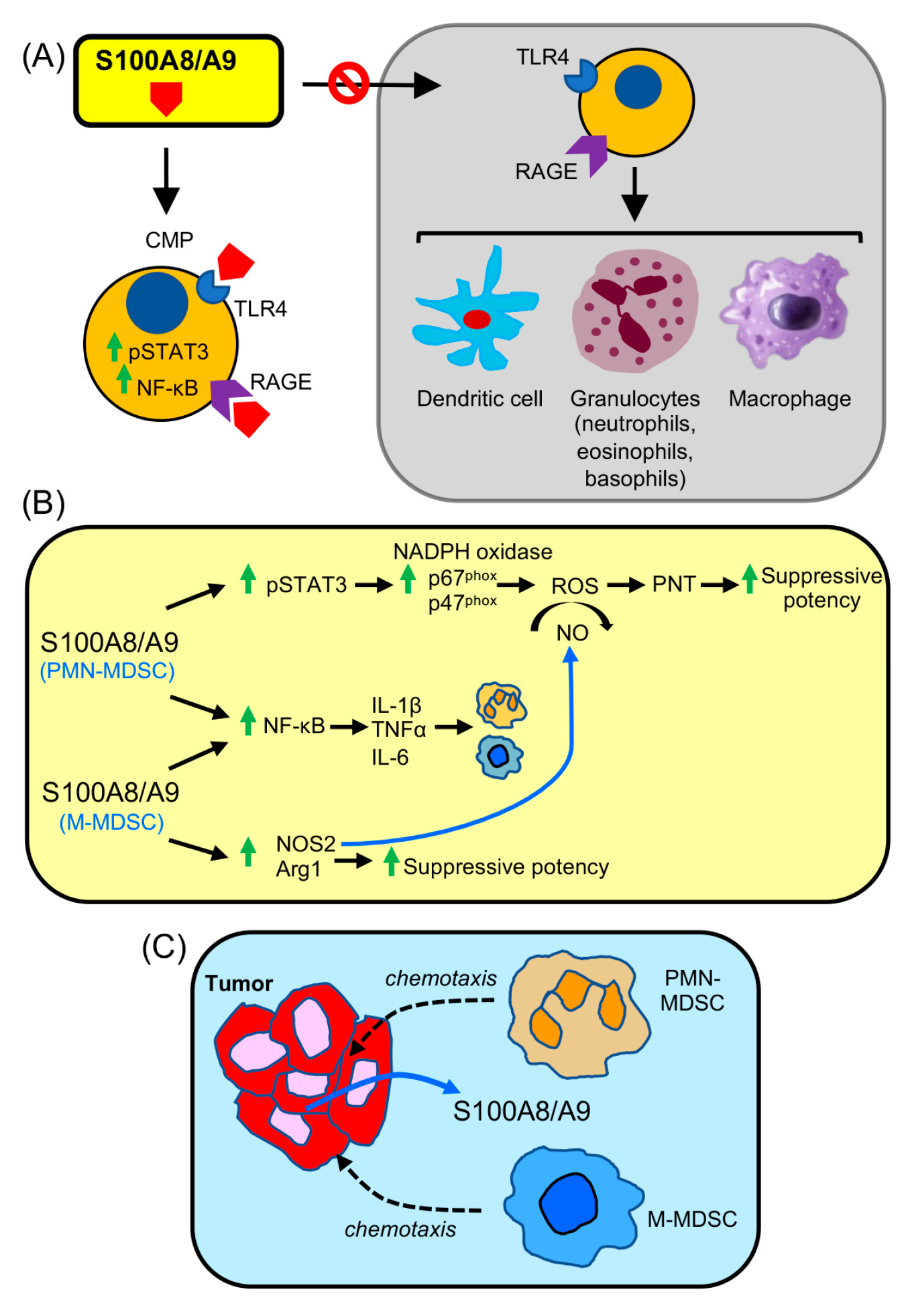
4. S100A8/A9 Drives the Accumulation and Potency of MDSCs in Cancer
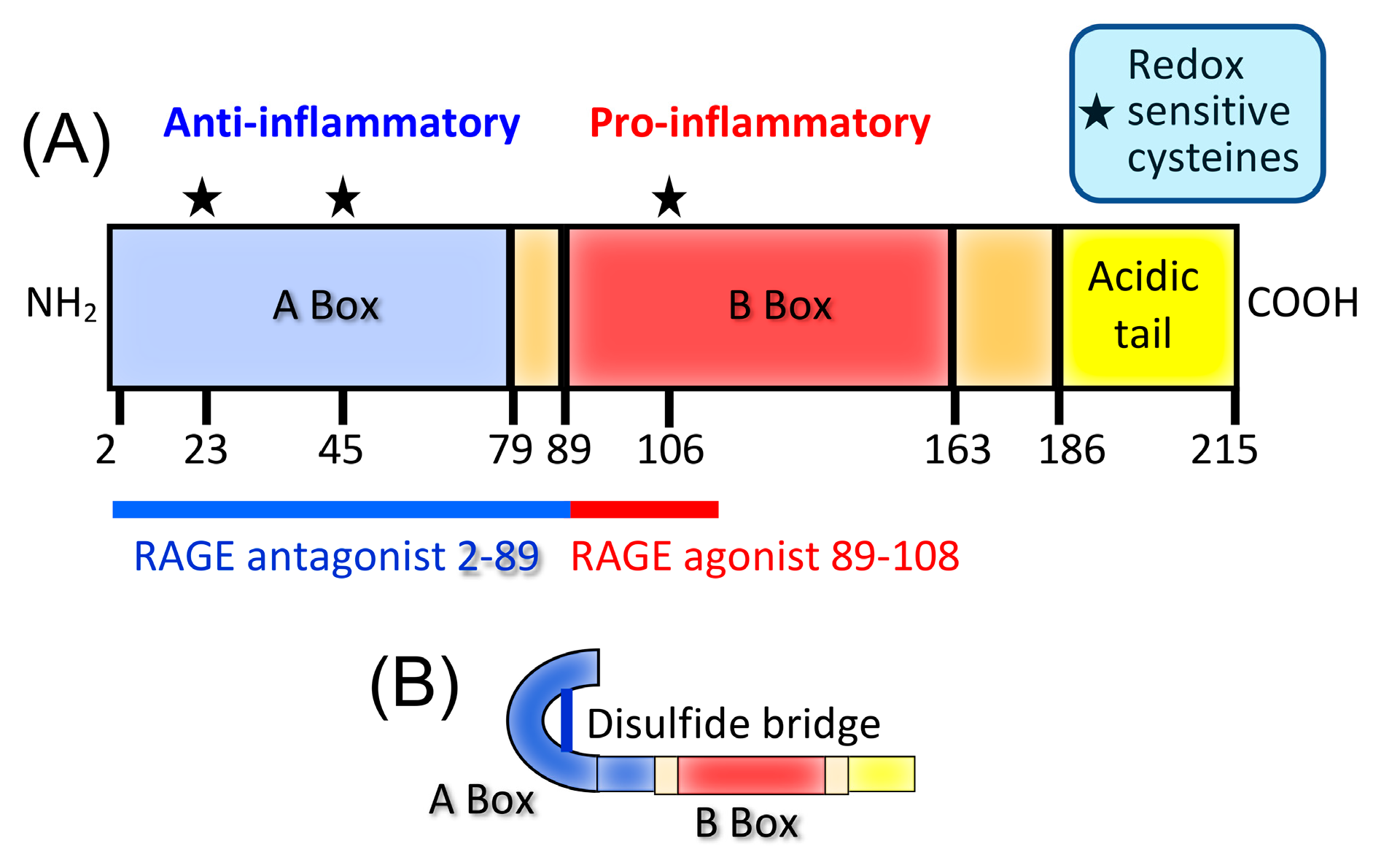
5. HMGB1 Is a Ubiquitously Expressed Protein with Intracellular and Extracellular Functions
6. HMGB1 Predominantly Favors Cancer Progression
7. HMGB1 Drives the Accumulation and Function of MDSCs in Cancer
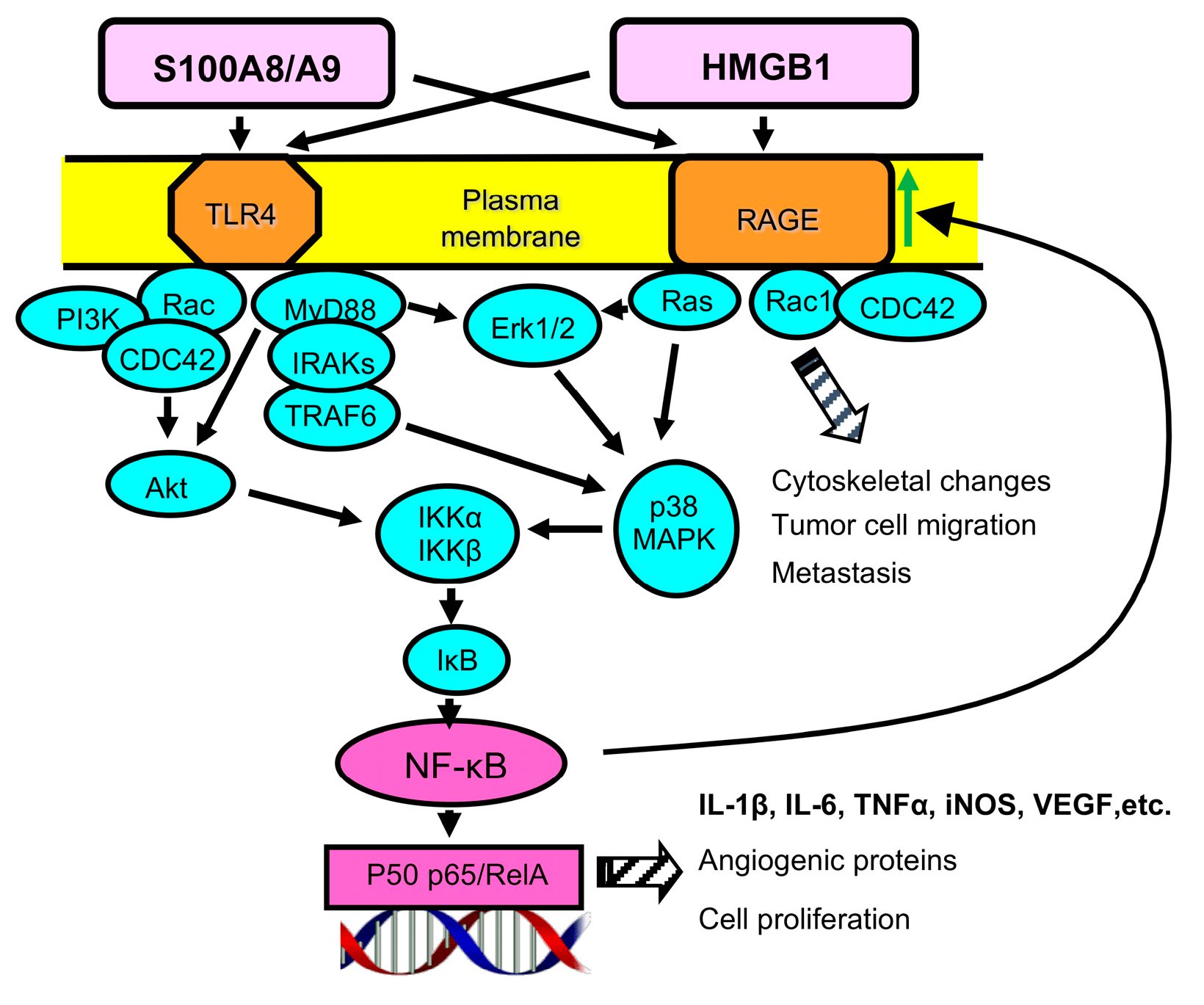
8. Receptor for Advanced Glycation Endproducts (RAGE) Is a Player in Malignancy
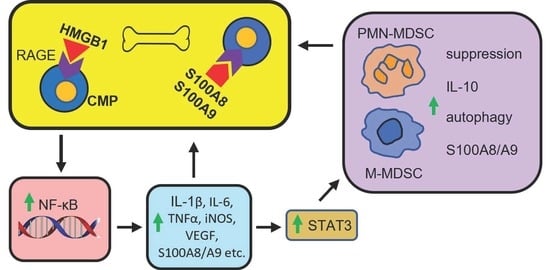
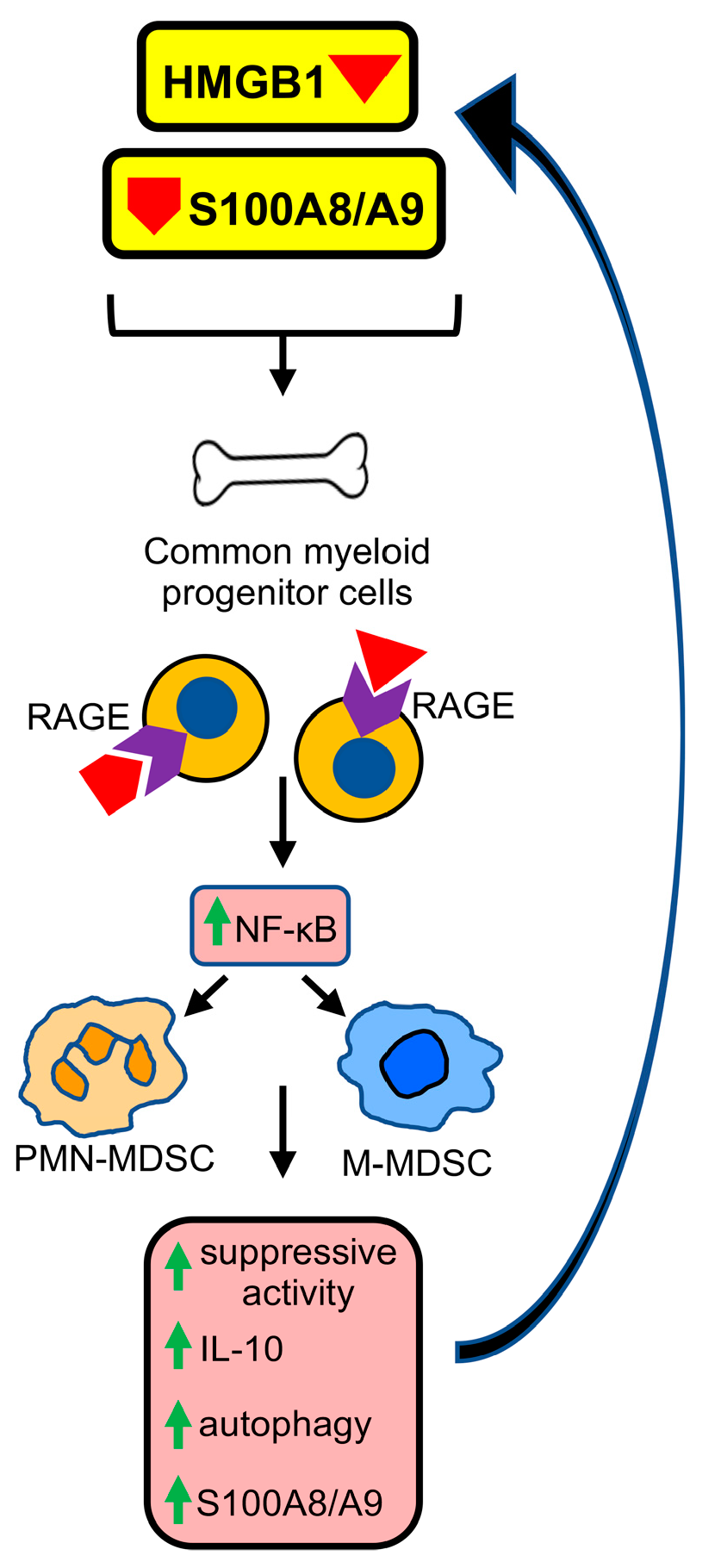
9. RAGE Contributes to the Generation and Pro-Cancer Effects of MDSCs
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, R.; Salehi-Rad, R.; Crosson, W.; Momcilovic, M.; Lim, R.J.; Ong, S.L.; Huang, Z.L.; Zhang, T.; Abascal, J.; Dumitras, C.; et al. Inhibition of Granulocytic Myeloid-Derived Suppressor Cells Overcomes Resistance to Immune Checkpoint Inhibition in LKB1-Deficient Non-Small Cell Lung Cancer. Cancer Res. 2021, 81, 3295–3308. [Google Scholar] [CrossRef]
- Loeuillard, E.; Yang, J.; Buckarma, E.; Wang, J.; Liu, Y.; Conboy, C.; Pavelko, K.D.; Li, Y.; O’Brien, D.; Wang, C.; et al. Targeting tumor-associated macrophages and granulocytic myeloid-derived suppressor cells augments PD-1 blockade in cholangiocarcinoma. J. Clin. Investig. 2020, 130, 5380–5396. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Yu, W.; He, J.; Liu, W.; Yang, J.; Lin, X.; Zhang, Y.; Wang, X.; Jiang, W.; Luo, J.; et al. Reprogramming immunosuppressive myeloid cells facilitates immunotherapy for colorectal cancer. EMBO Mol. Med. 2021, 13, e12798. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhou, C.; Li, Q.; Cheng, X.; Huang, T.; Li, F.; He, L.; Zhang, B.; Tu, S. Targeting depletion of myeloid-derived suppressor cells potentiates PD-L1 blockade efficacy in gastric and colon cancers. Oncoimmunology 2022, 11, 2131084. [Google Scholar] [CrossRef] [PubMed]
- Long, A.H.; Highfill, S.L.; Cui, Y.; Smith, J.P.; Walker, A.J.; Ramakrishna, S.; El-Etriby, R.; Galli, S.; Tsokos, M.G.; Orentas, R.J.; et al. Reduction of MDSCs with All-trans Retinoic Acid Improves CAR Therapy Efficacy for Sarcomas. Cancer Immunol. Res. 2016, 4, 869–880. [Google Scholar] [CrossRef]
- Ostrand-Rosenberg, S.; Fenselau, C. Myeloid-Derived Suppressor Cells: Immune-Suppressive Cells That Impair Antitumor Immunity and Are Sculpted by Their Environment. J. Immunol. 2018, 200, 422–431. [Google Scholar] [CrossRef]
- Sade-Feldman, M.; Kanterman, J.; Klieger, Y.; Ish-Shalom, E.; Olga, M.; Saragovi, A.; Shtainberg, H.; Lotem, M.; Baniyash, M. Clinical Significance of Circulating CD33+CD11b+HLA-DR- Myeloid Cells in Patients with Stage IV Melanoma Treated with Ipilimumab. Clin. Cancer Res. 2016, 22, 5661–5672. [Google Scholar] [CrossRef] [PubMed]
- Santegoets, S.J.; Stam, A.G.; Lougheed, S.M.; Gall, H.; Jooss, K.; Sacks, N.; Hege, K.; Lowy, I.; Scheper, R.J.; Gerritsen, W.R.; et al. Myeloid derived suppressor and dendritic cell subsets are related to clinical outcome in prostate cancer patients treated with prostate GVAX and ipilimumab. J. Immunother. Cancer 2014, 2, 31. [Google Scholar] [CrossRef]
- Meyer, C.; Cagnon, L.; Costa-Nunes, C.M.; Baumgaertner, P.; Montandon, N.; Leyvraz, L.; Michielin, O.; Romano, E.; Speiser, D.E. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol. Immunother. 2014, 63, 247–257. [Google Scholar] [CrossRef]
- Weber, J.; Gibney, G.; Kudchadkar, R.; Yu, B.; Cheng, P.; Martinez, A.J.; Kroeger, J.; Richards, A.; McCormick, L.; Moberg, V.; et al. Phase I/II Study of Metastatic Melanoma Patients Treated with Nivolumab Who Had Progressed after Ipilimumab. Cancer Immunol. Res. 2016, 4, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Weide, B.; Martens, A.; Zelba, H.; Stutz, C.; Derhovanessian, E.; Di Giacomo, A.M.; Maio, M.; Sucker, A.; Schilling, B.; Schadendorf, D.; et al. Myeloid-derived suppressor cells predict survival of patients with advanced melanoma: Comparison with regulatory T cells and NY-ESO-1- or melan-A-specific T cells. Clin. Cancer Res. 2014, 20, 1601–1609. [Google Scholar] [CrossRef]
- Martens, A.; Wistuba-Hamprecht, K.; Geukes Foppen, M.; Yuan, J.; Postow, M.A.; Wong, P.; Romano, E.; Khammari, A.; Dreno, B.; Capone, M.; et al. Baseline Peripheral Blood Biomarkers Associated with Clinical Outcome of Advanced Melanoma Patients Treated with Ipilimumab. Clin. Cancer Res. 2016, 22, 2908–2918. [Google Scholar] [CrossRef]
- Owen, D.H.; Benner, B.; Pilcher, C.; Good, L.; Savardekar, H.; Norman, R.; Ghattas, C.; Shah, M.; Konda, B.; Verschraegen, C.F.; et al. Deep and Durable Response to Nivolumab and Temozolomide in Small-Cell Lung Cancer Associated With an Early Decrease in Myeloid-Derived Suppressor Cells. Clin. Lung Cancer 2021, 22, e487–e497. [Google Scholar] [CrossRef]
- Weber, R.; Fleming, V.; Hu, X.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Front. Immunol. 2018, 9, 1310. [Google Scholar] [CrossRef]
- Mirza, N.; Fishman, M.; Fricke, I.; Dunn, M.; Neuger, A.M.; Frost, T.J.; Lush, R.M.; Antonia, S.; Gabrilovich, D.I. All-trans-retinoic acid improves differentiation of myeloid cells and immune response in cancer patients. Cancer Res. 2006, 66, 9299–9307. [Google Scholar] [CrossRef] [PubMed]
- Tobin, R.P.; Cogswell, D.T.; Cates, V.M.; Davis, D.M.; Borgers, J.S.W.; Van Gulick, R.J.; Katsnelson, E.; Couts, K.L.; Jordan, K.R.; Gao, D.; et al. Targeting MDSC differentiation using ATRA: A phase I/II clinical trial combining pembrolizumab and all-trans retinoic acid for metastatic melanoma. Clin. Cancer Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, Y.; Fishman, M.; Sherman, S.; Wang, X.; Beg, A.A.; Gabrilovich, D.I. Mechanism of all-trans retinoic acid effect on tumor-associated myeloid-derived suppressor cells. Cancer Res. 2007, 67, 11021–11028. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, E.; Kapoor, V.; Jassar, A.S.; Kaiser, L.R.; Albelda, S.M. Gemcitabine selectively eliminates splenic Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clin. Cancer Res. 2005, 11, 6713–6721. [Google Scholar] [CrossRef]
- Vincent, J.; Mignot, G.; Chalmin, F.; Ladoire, S.; Bruchard, M.; Chevriaux, A.; Martin, F.; Apetoh, L.; Rebe, C.; Ghiringhelli, F. 5-Fluorouracil selectively kills tumor-associated myeloid-derived suppressor cells resulting in enhanced T cell-dependent antitumor immunity. Cancer Res. 2010, 70, 3052–3061. [Google Scholar] [CrossRef]
- Grauers Wiktorin, H.; Nilsson, M.S.; Kiffin, R.; Sander, F.E.; Lenox, B.; Rydstrom, A.; Hellstrand, K.; Martner, A. Histamine targets myeloid-derived suppressor cells and improves the anti-tumor efficacy of PD-1/PD-L1 checkpoint blockade. Cancer Immunol. Immunother. 2019, 68, 163–174. [Google Scholar] [CrossRef]
- Mussai, F.; Wheat, R.; Sarrou, E.; Booth, S.; Stavrou, V.; Fultang, L.; Perry, T.; Kearns, P.; Cheng, P.; Keeshan, K.; et al. Targeting the arginine metabolic brake enhances immunotherapy for leukaemia. Int. J. Cancer 2019, 145, 2201–2208. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; Zhou, T.; Schmidt, J.; Jo, M.; et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015, 7, 314ra185. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Lerman, B.; Sakamaki, I.; Wei, G.; Cha, S.C.; Rao, S.S.; Qian, J.; Hailemichael, Y.; Nurieva, R.; Dwyer, K.C.; et al. Generation of a new therapeutic peptide that depletes myeloid-derived suppressor cells in tumor-bearing mice. Nat. Med. 2014, 20, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Krejcik, J.; Casneuf, T.; Nijhof, I.S.; Verbist, B.; Bald, J.; Plesner, T.; Syed, K.; Liu, K.; van de Donk, N.W.; Weiss, B.M.; et al. Daratumumab depletes CD38+ immune regulatory cells, promotes T-cell expansion, and skews T-cell repertoire in multiple myeloma. Blood 2016, 128, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Beury, D.W.; Carter, K.A.; Nelson, C.; Sinha, P.; Hanson, E.; Nyandjo, M.; Fitzgerald, P.J.; Majeed, A.; Wali, N.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cell Survival and Function Are Regulated by the Transcription Factor Nrf2. J. Immunol. 2016, 196, 3470–3478. [Google Scholar] [CrossRef]
- Condamine, T.; Kumar, V.; Ramachandran, I.R.; Youn, J.I.; Celis, E.; Finnberg, N.; El-Deiry, W.S.; Winograd, R.; Vonderheide, R.H.; English, N.R.; et al. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-R-mediated apoptosis. J. Clin. Investig. 2014, 124, 2626–2639. [Google Scholar] [CrossRef]
- Roth, J.; Burwinkel, F.; van den Bos, C.; Goebeler, M.; Vollmer, E.; Sorg, C. MRP8 and MRP14, S-100-like proteins associated with myeloid differentiation, are translocated to plasma membrane and intermediate filaments in a calcium-dependent manner. Blood 1993, 82, 1875–1883. [Google Scholar] [CrossRef]
- Nacken, W.; Roth, J.; Sorg, C.; Kerkhoff, C. S100A9/S100A8: Myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc. Res. Tech. 2003, 60, 569–580. [Google Scholar] [CrossRef]
- Hessian, P.A.; Edgeworth, J.; Hogg, N. MRP-8 and MRP-14, two abundant Ca(2+)-binding proteins of neutrophils and monocytes. J. Leukoc. Biol. 1993, 53, 197–204. [Google Scholar] [CrossRef]
- Gebhardt, C.; Nemeth, J.; Angel, P.; Hess, J. S100A8 and S100A9 in inflammation and cancer. Biochem. Pharmacol. 2006, 72, 1622–1631. [Google Scholar] [CrossRef]
- Wang, S.; Song, R.; Wang, Z.; Jing, Z.; Wang, S.; Ma, J. S100A8/A9 in Inflammation. Front. Immunol. 2018, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Tenbrock, K.; Ludwig, S.; Leukert, N.; Ehrhardt, C.; van Zoelen, M.A.; Nacken, W.; Foell, D.; van der Poll, T.; Sorg, C.; et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 2007, 13, 1042–1049. [Google Scholar] [CrossRef]
- Ichikawa, M.; Williams, R.; Wang, L.; Vogl, T.; Srikrishna, G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol. Cancer Res. 2011, 9, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Turovskaya, O.; Foell, D.; Sinha, P.; Vogl, T.; Newlin, R.; Nayak, J.; Nguyen, M.; Olsson, A.; Nawroth, P.P.; Bierhaus, A.; et al. RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis 2008, 29, 2035–2043. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Segovia, J.A.; Chang, T.H.; Morris, I.R.; Berton, M.T.; Tessier, P.A.; Tardif, M.R.; Cesaro, A.; Bose, S. DAMP molecule S100A9 acts as a molecular pattern to enhance inflammation during influenza A virus infection: Role of DDX21-TRIF-TLR4-MyD88 pathway. PLoS Pathog. 2014, 10, e1003848. [Google Scholar] [CrossRef]
- Prantner, D.; Nallar, S.; Vogel, S.N. The role of RAGE in host pathology and crosstalk between RAGE and TLR4 in innate immune signal transduction pathways. FASEB J. 2020, 34, 15659–15674. [Google Scholar] [CrossRef] [PubMed]
- Hibino, T.; Sakaguchi, M.; Miyamoto, S.; Yamamoto, M.; Motoyama, A.; Hosoi, J.; Shimokata, T.; Ito, T.; Tsuboi, R.; Huh, N.H. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res. 2013, 73, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Srikrishna, G. S100A8 and S100A9: New insights into their roles in malignancy. J. Innate Immun. 2012, 4, 31–40. [Google Scholar] [CrossRef]
- Ryckman, C.; Vandal, K.; Rouleau, P.; Talbot, M.; Tessier, P.A. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 2003, 170, 3233–3242. [Google Scholar] [CrossRef]
- Hermani, A.; Hess, J.; De Servi, B.; Medunjanin, S.; Grobholz, R.; Trojan, L.; Angel, P.; Mayer, D. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin. Cancer Res. 2005, 11, 5146–5152. [Google Scholar] [CrossRef]
- Wang, L.; Chang, E.W.; Wong, S.C.; Ong, S.M.; Chong, D.Q.; Ling, K.L. Increased myeloid-derived suppressor cells in gastric cancer correlate with cancer stage and plasma S100A8/A9 proinflammatory proteins. J. Immunol. 2013, 190, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Gielen, P.R.; Schulte, B.M.; Kers-Rebel, E.D.; Verrijp, K.; Bossman, S.A.; Ter Laan, M.; Wesseling, P.; Adema, G.J. Elevated levels of polymorphonuclear myeloid-derived suppressor cells in patients with glioblastoma highly express S100A8/9 and arginase and suppress T cell function. Neuro-Oncology 2016, 18, 1253–1264. [Google Scholar] [CrossRef]
- Miller, P.; Kidwell, K.M.; Thomas, D.; Sabel, M.; Rae, J.M.; Hayes, D.F.; Hudson, B.I.; El-Ashry, D.; Lippman, M.E. Elevated S100A8 protein expression in breast cancer cells and breast tumor stroma is prognostic of poor disease outcome. Breast Cancer Res. Treat. 2017, 166, 85–94. [Google Scholar] [CrossRef]
- Minami, S.; Sato, Y.; Matsumoto, T.; Kageyama, T.; Kawashima, Y.; Yoshio, K.; Ishii, J.; Matsumoto, K.; Nagashio, R.; Okayasu, I. Proteomic study of sera from patients with bladder cancer: Usefulness of S100A8 and S100A9 proteins. Cancer Genom. Proteom. 2010, 7, 181–189. [Google Scholar]
- Yasar, O.; Akcay, T.; Obek, C.; Turegun, F.A. Significance of S100A8, S100A9 and calprotectin levels in bladder cancer. Scand J Clin. Lab. Investig. 2017, 77, 437–441. [Google Scholar] [CrossRef]
- Mohr, T.; Zwick, A.; Hans, M.C.; Bley, I.A.; Braun, F.L.; Khalmurzaev, O.; Matveev, V.B.; Loertzer, P.; Pryalukhin, A.; Hartmann, A.; et al. The prominent role of the S100A8/S100A9-CD147 axis in the progression of penile cancer. Front. Oncol. 2022, 12, 891511. [Google Scholar] [CrossRef]
- Sinha, P.; Okoro, C.; Foell, D.; Freeze, H.H.; Ostrand-Rosenberg, S.; Srikrishna, G. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol. 2008, 181, 4666–4675. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Corzo, C.A.; Luetteke, N.; Yu, B.; Nagaraj, S.; Bui, M.M.; Ortiz, M.; Nacken, W.; Sorg, C.; Vogl, T.; et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 2008, 205, 2235–2249. [Google Scholar] [CrossRef]
- Doussiere, J.; Bouzidi, F.; Vignais, P.V. The S100A8/A9 protein as a partner for the cytosolic factors of NADPH oxidase activation in neutrophils. Eur. J. Biochem. 2002, 269, 3246–3255. [Google Scholar] [CrossRef]
- Ramachandran, I.R.; Martner, A.; Pisklakova, A.; Condamine, T.; Chase, T.; Vogl, T.; Roth, J.; Gabrilovich, D.; Nefedova, Y. Myeloid-derived suppressor cells regulate growth of multiple myeloma by inhibiting T cells in bone marrow. J. Immunol. 2013, 190, 3815–3823. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Mitsuhashi, A.; Ogino, H.; Kozai, H.; Yoneda, H.; Afroj, T.; Sato, S.; Nokihara, H.; Shinohara, T.; Nishioka, Y. S-1 eliminates MDSCs and enhances the efficacy of PD-1 blockade via regulation of tumor-derived Bv8 and S100A8 in thoracic tumor. Cancer Sci. 2023, 114, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.H.; Lee, K.Y.; Chang, Y.L.; Chan, Y.F.; Kuo, L.W.; Lin, T.Y.; Chung, F.T.; Kuo, C.S.; Yu, C.T.; Lin, S.M.; et al. CD14(+)S100A9(+) monocytic myeloid-derived suppressor cells and their clinical relevance in non-small cell lung cancer. Am. J. Respir. Crit. Care Med. 2012, 186, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zhang, B.; Wang, B.; Zhang, F.; Fan, K.X.; Guo, Y.J. Increased CD14(+)HLA-DR (-/low) myeloid-derived suppressor cells correlate with extrathoracic metastasis and poor response to chemotherapy in non-small cell lung cancer patients. Cancer Immunol. Immunother. 2013, 62, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Drews-Elger, K.; Iorns, E.; Dias, A.; Miller, P.; Ward, T.M.; Dean, S.; Clarke, J.; Campion-Flora, A.; Rodrigues, D.N.; Reis-Filho, J.S.; et al. Infiltrating S100A8+ myeloid cells promote metastatic spread of human breast cancer and predict poor clinical outcome. Breast Cancer Res. Treat. 2014, 148, 41–59. [Google Scholar] [CrossRef]
- Zhao, F.; Hoechst, B.; Duffy, A.; Gamrekelashvili, J.; Fioravanti, S.; Manns, M.P.; Greten, T.F.; Korangy, F. S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology 2012, 136, 176–183. [Google Scholar] [CrossRef]
- Thomas, J.O. HMG1 and 2: Architectural DNA-binding proteins. Biochem. Soc. Trans. 2001, 29, 395–401. [Google Scholar] [CrossRef]
- Thomas, J.O.; Travers, A.A. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001, 26, 167–174. [Google Scholar] [CrossRef]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 1999, 285, 248–251. [Google Scholar] [CrossRef]
- Gardella, S.; Andrei, C.; Ferrera, D.; Lotti, L.V.; Torrisi, M.R.; Bianchi, M.E.; Rubartelli, A. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 2002, 3, 995–1001. [Google Scholar] [CrossRef]
- Ulloa, L.; Ochani, M.; Yang, H.; Tanovic, M.; Halperin, D.; Yang, R.; Czura, C.J.; Fink, M.P.; Tracey, K.J. Ethyl pyruvate prevents lethality in mice with established lethal sepsis and systemic inflammation. Proc. Natl. Acad. Sci. USA 2002, 99, 12351–12356. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Sims, G.P.; Rowe, D.C.; Rietdijk, S.T.; Herbst, R.; Coyle, A.J. HMGB1 and RAGE in inflammation and cancer. Annu. Rev. Immunol. 2010, 28, 367–388. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Tracey, K.J. Targeting HMGB1 in inflammation. Biochim. Biophys. Acta 2010, 1799, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lundback, P.; Ottosson, L.; Erlandsson-Harris, H.; Venereau, E.; Bianchi, M.E.; Al-Abed, Y.; Andersson, U.; Tracey, K.J. Redox modifications of cysteine residues regulate the cytokine activity of HMGB1. Mol. Med. 2021, 27, 58. [Google Scholar] [CrossRef]
- Ye, L.Y.; Chen, W.; Bai, X.L.; Xu, X.Y.; Zhang, Q.; Xia, X.F.; Sun, X.; Li, G.G.; Hu, Q.D.; Fu, Q.H.; et al. Hypoxia-Induced Epithelial-to-Mesenchymal Transition in Hepatocellular Carcinoma Induces an Immunosuppressive Tumor Microenvironment to Promote Metastasis. Cancer Res. 2016, 76, 818–830. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, G.Z.; Wang, Y.; Huang, H.Z.; Li, W.T.; Qu, X.D. Hypoxia-induced HMGB1 expression of HCC promotes tumor invasiveness and metastasis via regulating macrophage-derived IL-6. Exp. Cell Res. 2018, 367, 81–88. [Google Scholar] [CrossRef]
- Ren, Y.; Cao, L.; Wang, L.; Zheng, S.; Zhang, Q.; Guo, X.; Li, X.; Chen, M.; Wu, X.; Furlong, F.; et al. Autophagic secretion of HMGB1 from cancer-associated fibroblasts promotes metastatic potential of non-small cell lung cancer cells via NFkappaB signaling. Cell Death Dis. 2021, 12, 858. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, C.; Li, L.; Jin, X.; Zhang, Z.; Zheng, H.; Pan, J.; Shi, L.; Jiang, Z.; Su, K.; et al. Tumor-derived HMGB1 induces CD62L(dim) neutrophil polarization and promotes lung metastasis in triple-negative breast cancer. Oncogenesis 2020, 9, 82. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, L.; Guo, X.; Li, Z.; Liao, X.; Zhang, X.; Huang, L.; He, J. HMGB1-activated fibroblasts promote breast cancer cells metastasis via RAGE/aerobic glycolysis. Neoplasma 2021, 68, 71–78. [Google Scholar] [CrossRef]
- Wang, J.D.; Wang, Y.Y.; Lin, S.Y.; Chang, C.Y.; Li, J.R.; Huang, S.W.; Chen, W.Y.; Liao, S.L.; Chen, C.J. Exosomal HMGB1 Promoted Cancer Malignancy. Cancers 2021, 13, 877. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, B.L.; Steel, H.C.; Theron, A.J.; Heyman, L.; Smit, T.; Ramdas, Y.; Anderson, R. High Mobility Group Box 1 in Human Cancer. Cells 2020, 9, 1664. [Google Scholar] [CrossRef] [PubMed]
- Amornsupak, K.; Thongchot, S.; Thinyakul, C.; Box, C.; Hedayat, S.; Thuwajit, P.; Eccles, S.A.; Thuwajit, C. HMGB1 mediates invasion and PD-L1 expression through RAGE-PI3K/AKT signaling pathway in MDA-MB-231 breast cancer cells. BMC Cancer 2022, 22, 578. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yue, X.; Liu, H.; Zhu, Y.; Ke, H.; Yang, X.; Yin, S.; Li, Z.; Zhang, Y.; Hu, T.; et al. MAP7D2 reduces CD8(+) cytotoxic T lymphocyte infiltration through MYH9-HMGB1 axis in colorectal cancer. Mol. Ther. 2022, 31, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Ostrand-Rosenberg, S.; Horn, L.A.; Ciavattone, N.G. Radiotherapy Both Promotes and Inhibits Myeloid-Derived Suppressor Cell Function: Novel Strategies for Preventing the Tumor-Protective Effects of Radiotherapy. Front. Oncol. 2019, 9, 215. [Google Scholar] [CrossRef]
- Zhu, L.; Ren, S.; Daniels, M.J.; Qiu, W.; Song, L.; You, T.; Wang, D.; Wang, Z. Exogenous HMGB1 Promotes the Proliferation and Metastasis of Pancreatic Cancer Cells. Front. Med. 2021, 8, 756988. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, M.; Shinde-Jadhav, S.; Mansure, J.J.; Alvarez, F.; Connell, T.; Seuntjens, J.; Piccirillo, C.A.; Kassouf, W. The immune mediated role of extracellular HMGB1 in a heterotopic model of bladder cancer radioresistance. Sci. Rep. 2019, 9, 6348. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Che, X.; Wang, Y.; Li, B.; Li, J.; Dai, W.; Lin, C.P.; Huang, C. High mobility group Box-1 inhibits cancer cell motility and metastasis by suppressing activation of transcription factor CREB and nWASP expression. Oncotarget 2014, 5, 7458–7470. [Google Scholar] [CrossRef]
- Bartling, B.; Hofmann, H.S.; Weigle, B.; Silber, R.E.; Simm, A. Down-regulation of the receptor for advanced glycation end-products (RAGE) supports non-small cell lung carcinoma. Carcinogenesis 2005, 26, 293–301. [Google Scholar] [CrossRef]
- Chung, H.W.; Lim, J.B.; Jang, S.; Lee, K.J.; Park, K.H.; Song, S.Y. Serum high mobility group box-1 is a powerful diagnostic and prognostic biomarker for pancreatic ductal adenocarcinoma. Cancer Sci. 2012, 103, 1714–1721. [Google Scholar] [CrossRef]
- Handke, N.A.; Rupp, A.B.A.; Trimpop, N.; von Pawel, J.; Holdenrieder, S. Soluble High Mobility Group Box 1 (HMGB1) Is a Promising Biomarker for Prediction of Therapy Response and Prognosis in Advanced Lung Cancer Patients. Diagnostics 2021, 11, 356. [Google Scholar] [CrossRef]
- Clasen, K.; Welz, S.; Faltin, H.; Zips, D.; Eckert, F. Dynamics of HMBG1 (High Mobility Group Box 1) during radiochemotherapy correlate with outcome of HNSCC patients. Strahlenther Onkol. 2022, 198, 194–200. [Google Scholar] [CrossRef]
- Kroemer, G.; Kepp, O. Radiochemotherapy-induced elevations of plasma HMGB1 levels predict therapeutic responses in cancer patients. Oncoimmunology 2021, 10, 2005859. [Google Scholar] [CrossRef]
- Ladoire, S.; Enot, D.; Senovilla, L.; Ghiringhelli, F.; Poirier-Colame, V.; Chaba, K.; Semeraro, M.; Chaix, M.; Penault-Llorca, F.; Arnould, L.; et al. The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy 2016, 12, 864–875. [Google Scholar] [CrossRef]
- Parker, K.H.; Sinha, P.; Horn, L.A.; Clements, V.K.; Yang, H.; Li, J.; Tracey, K.J.; Ostrand-Rosenberg, S. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res. 2014, 74, 5723–5733. [Google Scholar] [CrossRef]
- Li, W.; Wu, K.; Zhao, E.; Shi, L.; Li, R.; Zhang, P.; Yin, Y.; Shuai, X.; Wang, G.; Tao, K. HMGB1 recruits myeloid derived suppressor cells to promote peritoneal dissemination of colon cancer after resection. Biochem. Biophys. Res. Commun. 2013, 436, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, J.; Rong, R.; Li, L.; Shang, W.; Song, D.; Feng, G.; Luo, F. HMGB1 promotes myeloid-derived suppressor cells and renal cell carcinoma immune escape. Oncotarget 2017, 8, 63290–63298. [Google Scholar] [CrossRef] [PubMed]
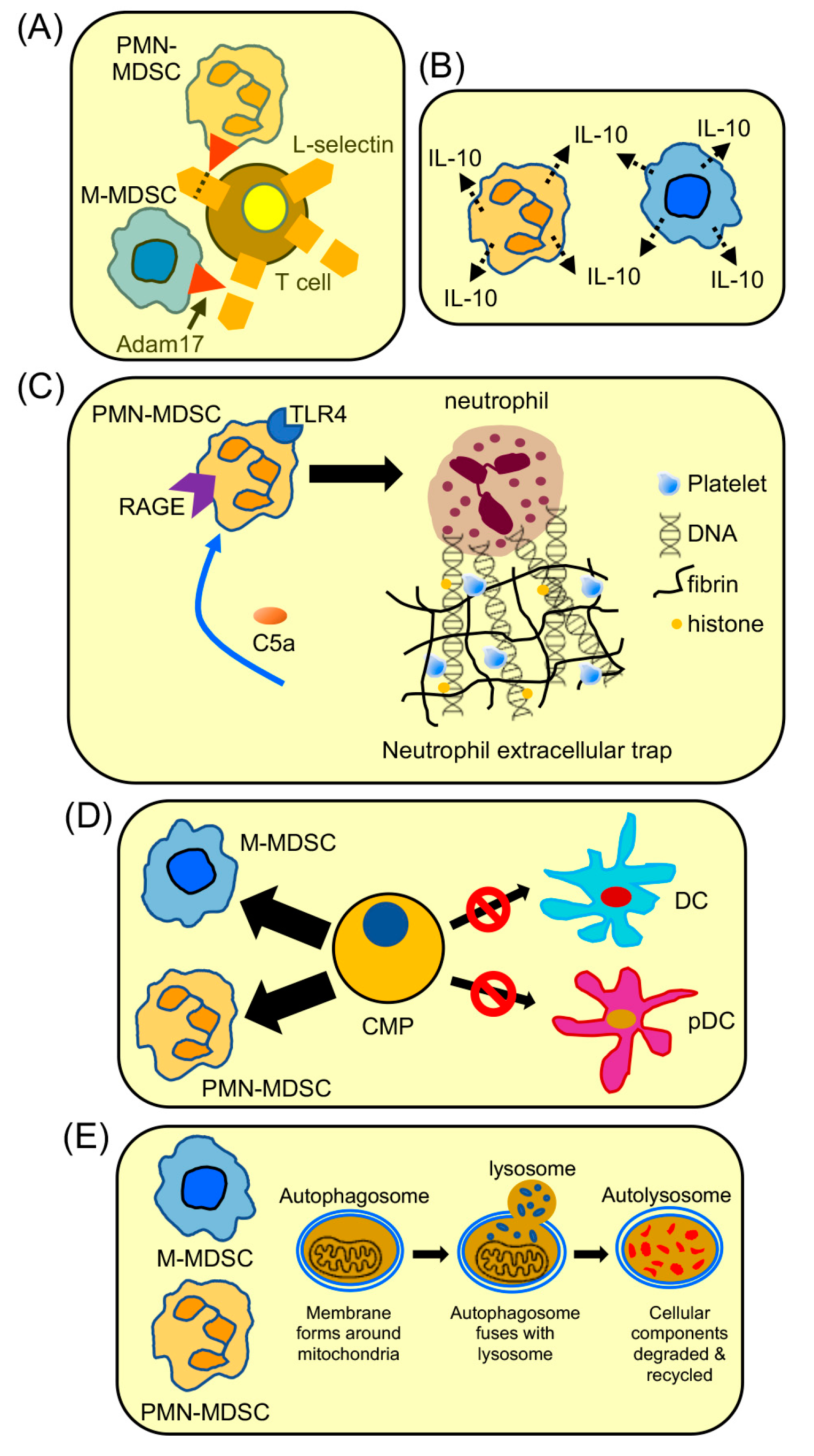
- Hanson, E.M.; Clements, V.K.; Sinha, P.; Ilkovitch, D.; Ostrand-Rosenberg, S. Myeloid-derived suppressor cells down-regulate L-selectin expression on CD4+ and CD8+ T cells. J. Immunol. 2009, 183, 937–944. [Google Scholar] [CrossRef]
- Ku, A.W.; Muhitch, J.B.; Powers, C.A.; Diehl, M.; Kim, M.; Fisher, D.T.; Sharda, A.P.; Clements, V.K.; O’Loughlin, K.; Minderman, H.; et al. Tumor-induced MDSC act via remote control to inhibit L-selectin-dependent adaptive immunity in lymph nodes. eLife 2016, 5, e17375. [Google Scholar] [CrossRef]
- Hubert, P.; Roncarati, P.; Demoulin, S.; Pilard, C.; Ancion, M.; Reynders, C.; Lerho, T.; Bruyere, D.; Lebeau, A.; Radermecker, C.; et al. Extracellular HMGB1 blockade inhibits tumor growth through profoundly remodeling immune microenvironment and enhances checkpoint inhibitor-based immunotherapy. J. Immunother. Cancer 2021, 9, e001966. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Clements, V.K.; Fulton, A.M.; Ostrand-Rosenberg, S. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 2007, 67, 4507–4513. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Espinosa, S.; Morales, X.; Senent, Y.; Alignani, D.; Tavira, B.; Macaya, I.; Ruiz, B.; Moreno, H.; Remirez, A.; Sainz, C.; et al. Complement C5a induces the formation of neutrophil extracellular traps by myeloid-derived suppressor cells to promote metastasis. Cancer Lett. 2022, 529, 70–84. [Google Scholar] [CrossRef]
- Lum, J.J.; Bauer, D.E.; Kong, M.; Harris, M.H.; Li, C.; Lindsten, T.; Thompson, C.B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005, 120, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.H.; Horn, L.A.; Ostrand-Rosenberg, S. High-mobility group box protein 1 promotes the survival of myeloid-derived suppressor cells by inducing autophagy. J. Leukoc. Biol. 2016, 100, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Schmidt, A.M. Characterization and functional analysis of the promoter of RAGE, the receptor for advanced glycation end products. J. Biol. Chem. 1997, 272, 16498–16506. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A.; Blood, D.C.; del Toro, G.; Canet, A.; Lee, D.C.; Qu, W.; Tanji, N.; Lu, Y.; Lalla, E.; Fu, C.; et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 2000, 405, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, C.; Riehl, A.; Durchdewald, M.; Nemeth, J.; Furstenberger, G.; Muller-Decker, K.; Enk, A.; Arnold, B.; Bierhaus, A.; Nawroth, P.P.; et al. RAGE signaling sustains inflammation and promotes tumor development. J. Exp. Med. 2008, 205, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Kwak, T.; Drews-Elger, K.; Ergonul, A.; Miller, P.C.; Braley, A.; Hwang, G.H.; Zhao, D.; Besser, A.; Yamamoto, Y.; Yamamoto, H.; et al. Targeting of RAGE-ligand signaling impairs breast cancer cell invasion and metastasis. Oncogene 2017, 36, 1559–1572. [Google Scholar] [CrossRef]
- Rojas, A.; Schneider, I.; Lindner, C.; Gonzalez, I.; Morales, M.A. The RAGE/multiligand axis: A new actor in tumor biology. Biosci. Rep. 2022, 42, BSR20220395. [Google Scholar] [CrossRef]
- Vernon, P.J.; Loux, T.J.; Schapiro, N.E.; Kang, R.; Muthuswamy, R.; Kalinski, P.; Tang, D.; Lotze, M.T.; Zeh, H.J., 3rd. The receptor for advanced glycation end products promotes pancreatic carcinogenesis and accumulation of myeloid-derived suppressor cells. J. Immunol. 2013, 190, 1372–1379. [Google Scholar] [CrossRef]
- Wuren, T.; Huecksteadt, T.; Beck, E.; Warren, K.; Hoidal, J.; Ostrand-Rosenberg, S.; Sanders, K. The receptor for advanced glycation endproducts (RAGE) decreases survival of tumor-bearing mice by enhancing the generation of lung metastasis-associated myeloid-derived suppressor cells. Cell Immunol. 2021, 365, 104379. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ostrand-Rosenberg, S.; Huecksteadt, T.; Sanders, K. The Receptor for Advanced Glycation Endproducts (RAGE) and Its Ligands S100A8/A9 and High Mobility Group Box Protein 1 (HMGB1) Are Key Regulators of Myeloid-Derived Suppressor Cells. Cancers 2023, 15, 1026. https://doi.org/10.3390/cancers15041026
Ostrand-Rosenberg S, Huecksteadt T, Sanders K. The Receptor for Advanced Glycation Endproducts (RAGE) and Its Ligands S100A8/A9 and High Mobility Group Box Protein 1 (HMGB1) Are Key Regulators of Myeloid-Derived Suppressor Cells. Cancers. 2023; 15(4):1026. https://doi.org/10.3390/cancers15041026
Chicago/Turabian StyleOstrand-Rosenberg, Suzanne, Tom Huecksteadt, and Karl Sanders. 2023. "The Receptor for Advanced Glycation Endproducts (RAGE) and Its Ligands S100A8/A9 and High Mobility Group Box Protein 1 (HMGB1) Are Key Regulators of Myeloid-Derived Suppressor Cells" Cancers 15, no. 4: 1026. https://doi.org/10.3390/cancers15041026
APA StyleOstrand-Rosenberg, S., Huecksteadt, T., & Sanders, K. (2023). The Receptor for Advanced Glycation Endproducts (RAGE) and Its Ligands S100A8/A9 and High Mobility Group Box Protein 1 (HMGB1) Are Key Regulators of Myeloid-Derived Suppressor Cells. Cancers, 15(4), 1026. https://doi.org/10.3390/cancers15041026