Synthesis and Significance of Arachidonic Acid, a Substrate for Cyclooxygenases, Lipoxygenases, and Cytochrome P450 Pathways in the Tumorigenesis of Glioblastoma Multiforme, Including a Pan-Cancer Comparative Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Methodology
3. Arachidonic Acid Biosynthesis and Glioblastoma Multiforme
3.1. Arachidonic Acid Biosynthesis
3.2. Arachidonic Acid Biosynthesis Pathway in Glioblastoma Multiforme Tumors
4. Phospholipase A2 Superfamily and the Release of Arachidonic Acid from Cell Membrane Phospholipids in Glioblastoma Multiforme
4.1. Phospholipase A2 Superfamily
- cytosolic phospholipase A2 (cPLA2),
- calcium-independent phospholipase A2 (iPLA2), and
- secretory phospholipase A2 (sPLA2).
- platelet-activating factor acetyl hydrolases (PAF-AH),
- lysosomal phospholipase A2, and
- adipose phospholipase A2.
4.2. Cytosolic Phospholipase A2 and Calcium-Independent Phospholipase A2 in Glioblastoma Multiforme
4.3. Secretory Phospholipase A2 in Glioblastoma Multiforme
4.4. Pan-Cancer Analysis of Phospholipase A2 Genes and Comparison of GBM Expression against Other Cancers
4.5. Lysophospholipid Acyltransferases in Glioblastoma Multiforme
4.6. Acyl-CoA Thioesterases and Arachidonic Acid C20:4n-6 in Glioblastoma Multiforme
5. Cyclooxygenase Pathway and Prostanoids in Glioblastoma Multiforme
5.1. Cyclooxygenase Pathway
5.2. Cyclooxygenase Pathway and Glioblastoma Multiforme
5.3. Pan-Cancer Analysis of Genes Related to the COX Pathway and GBM
6. Lipoxygenases and Arachidonic Acid in Glioblastoma Multiforme
6.1. Lipoxygenases Pathway
- epidermal lipoxygenase 3/arachidonate lipoxygenase 3 (eLOX3/ALOXE3),
- 5-lipoxygenase/arachidonate 5-lipoxygenase (5-LOX/ALOX5),
- 12S-lipoxygenase/arachidonate 12-lipoxygenase, 12S type (12S-LOX/ALOX12),
- 12R-lipoxygenase/arachidonate 12-lipoxygenase, 12R type (12R-LOX/ALOX12B),
- 15-lipoxygenase-1/arachidonate 15-lipoxygenase (15-LOX-1/ALOX15), also known as 12/15-LOX, and
- 15-lipoxygenase-2/arachidonate 15-lipoxygenase type B (15-LOX-2/ALOX15B).
6.1.1. Epidermal Lipoxygenase 3
6.1.2. 5-Lipoxygenase
6.1.3. 12S-Lipoxygenase
6.1.4. 12R-Lipoxygenase
6.1.5. 15-Lipoxygenases
6.2. Lipoxygenases in Glioblastoma Multiforme
6.2.1. 5-Lipoxygenase Pathway in Glioblastoma Multiforme
6.2.2. 12-Lipoxygenase Pathway in Glioblastoma Multiforme
6.2.3. 15-Lipoxygenase Pathway in Glioblastoma Multiforme
6.3. Pan-Cancer Analysis of Genes Related to LOX Pathway and GBM
7. Cytochrome P450 Pathway in Glioblastoma Multiforme Tumors
7.1. Cytochrome P450 Pathway
7.2. Cytochrome P450 Pathway in Glioblastoma Multiforme Tumors
7.3. Pan-Cancer Analysis of Cytochrome P450 Genes and Comparison of GBM Expression against Other Cancers
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Johnson, D.R.; Omuro, A.M.P.; Ravelo, A.; Sommer, N.; Guerin, A.; Ionescu-Ittu, R.; Shi, S.; Macalalad, A.; Uhm, J.H. Overall survival in patients with glioblastoma before and after bevacizumab approval. Curr. Med. Res. Opin. 2018, 34, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Grech, N.; Dalli, T.; Mizzi, S.; Meilak, L.; Calleja, N.; Zrinzo, A. Rising Incidence of Glioblastoma Multiforme in a Well-Defined Population. Cureus 2020, 12, e8195. [Google Scholar] [CrossRef] [PubMed]
- Bilgin, E.; Duman, B.B.; Altintas, S.; Cil, T.; Gezercan, Y.; Okten, A.I. Predictors of Survival in Turkish Patients with Primary Glioblastoma. Turk. Neurosurg. 2021, 31, 641–653. [Google Scholar] [CrossRef]
- Gomes, R.N.; Felipe da Costa, S.; Colquhoun, A. Eicosanoids and cancer. Clinics 2018, 73, e530s. [Google Scholar] [CrossRef]
- Serhan, C.N.; Levy, B.D. Resolvins in inflammation: Emergence of the pro-resolving superfamily of mediators. J. Clin. Investig. 2018, 128, 2657–2669. [Google Scholar] [CrossRef]
- Straus, D.S.; Pascual, G.; Li, M.; Welch, J.S.; Ricote, M.; Hsiang, C.H.; Sengchanthalangsy, L.L.; Ghosh, G.; Glass, C.K. 15-deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in the NF-kappa B signaling pathway. Proc. Natl. Acad. Sci. USA 2000, 97, 4844–4849. [Google Scholar] [CrossRef]
- Krey, G.; Braissant, O.; L’Horset, F.; Kalkhoven, E.; Perroud, M.; Parker, M.G.; Wahli, W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 1997, 11, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Seifert, M.; Garbe, M.; Friedrich, B.; Mittelbronn, M.; Klink, B. Comparative transcriptomics reveals similarities and differences between astrocytoma grades. BMC Cancer 2015, 15, 952. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- The GTEx Consortium; Ardlie, K.G.; Deluca, D.S.; Segrè, A.V.; Sullivan, T.J.; Young, T.R.; Gelfand, E.T.; Trowbridge, C.A.; Maller, J.B.; Tukiainen, T.; et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar]
- Madhavan, S.; Zenklusen, J.C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 2009, 7, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef]
- Park, W.J.; Kothapalli, K.S.; Lawrence, P.; Tyburczy, C.; Brenna, J.T. An alternate pathway to long-chain polyunsaturates: The FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 2009, 50, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Dennis, E.A.; Cao, J.; Hsu, Y.H.; Magrioti, V.; Kokotos, G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef]
- Leonard, A.E.; Bobik, E.G.; Dorado, J.; Kroeger, P.E.; Chuang, L.T.; Thurmond, J.M.; Parker-Barnes, J.M.; Das, T.; Huang, Y.S.; Mukerji, P. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 2000, 350 Pt 3, 765–770. [Google Scholar] [CrossRef]
- Leonard, A.E.; Kelder, B.; Bobik, E.G.; Chuang, L.T.; Lewis, C.J.; Kopchick, J.J.; Mukerji, P.; Huang, Y.S. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 2002, 37, 733–740. [Google Scholar] [CrossRef]
- Kitazawa, H.; Miyamoto, Y.; Shimamura, K.; Nagumo, A.; Tokita, S. Development of a high-density assay for long-chain fatty acyl-CoA elongases. Lipids 2009, 44, 765–773. [Google Scholar] [CrossRef]
- Shakya, S.; Gromovsky, A.D.; Hale, J.S.; Knudsen, A.M.; Prager, B.; Wallace, L.C.; Penalva, L.O.F.; Brown, H.A.; Kristensen, B.W.; Rich, J.N.; et al. Altered lipid metabolism marks glioblastoma stem and non-stem cells in separate tumor niches. Acta Neuropathol. Commun. 2021, 9, 101. [Google Scholar] [CrossRef]
- Korbecki, J.; Kojder, K.; Jeżewski, D.; Simińska, D.; Tarnowski, M.; Kopytko, P.; Safranow, K.; Gutowska, I.; Goschorska, M.; Kolasa-Wołosiuk, A.; et al. Expression of SCD and FADS2 Is Lower in the Necrotic Core and Growing Tumor Area than in the Peritumoral Area of Glioblastoma Multiforme. Biomolecules 2020, 10, 727. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Simińska, D.; Jeżewski, D.; Kojder, K.; Tomasiak, P.; Tarnowski, M.; Chlubek, D.; Baranowska-Bosiacka, I. Glioblastoma Multiforme Tumors in Women Have a Lower Expression of Fatty Acid Elongases ELOVL2, ELOVL5, ELOVL6, and ELOVL7 than in Men. Brain Sci. 2022, 12, 1356. [Google Scholar] [CrossRef]
- Jalota, A.; Kumar, M.; Das, B.C.; Yadav, A.K.; Chosdol, K.; Sinha, S. A drug combination targeting hypoxia induced chemoresistance and stemness in glioma cells. Oncotarget 2018, 9, 18351–18366. [Google Scholar] [CrossRef] [PubMed]
- Pickard, R.T.; Strifler, B.A.; Kramer, R.M.; Sharp, J.D. Molecular cloning of two new human paralogs of 85-kDa cytosolic phospholipase A2. J. Biol. Chem. 1999, 274, 8823–8831. [Google Scholar] [CrossRef]
- Sundler, R.; Winstedt, D.; Wijkander, J. Acyl-chain selectivity of the 85 kDa phospholipase A2 and of the release process in intact macrophages. Biochem. J. 1994, 301, 455–458. [Google Scholar] [CrossRef]
- Underwood, K.W.; Song, C.; Kriz, R.W.; Chang, X.J.; Knopf, J.L.; Lin, L.L. A novel calcium-independent phospholipase A2, cPLA2-gamma, that is prenylated and contains homology to cPLA2. J. Biol. Chem. 1998, 273, 21926–21932. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, E.J.; Kempner, E.S.; Dennis, E.A. Ca(2+)-independent cytosolic phospholipase A2 from macrophage-like P388D1 cells. Isolation and characterization. J. Biol. Chem. 1994, 269, 9227–9233. [Google Scholar] [CrossRef]
- Jenkins, C.M.; Wolf, M.J.; Mancuso, D.J.; Gross, R.W. Identification of the calmodulin-binding domain of recombinant calcium-independent phospholipase A2beta. implications for structure and function. J. Biol. Chem. 2001, 276, 7129–7135. [Google Scholar] [CrossRef]
- Portilla, D.; Dai, G. Purification of a novel calcium-independent phospholipase A2 from rabbit kidney. J. Biol. Chem. 1996, 271, 15451–15457. [Google Scholar] [CrossRef]
- Hariprasad, G.; Kumar, M.; Srinivasan, A.; Kaur, P.; Singh, T.P.; Jithesh, O. Structural analysis of a group III Glu62-phospholipase A2 from the scorpion, Mesobuthus tamulus: Targeting and reversible inhibition by native peptides. Int. J. Biol. Macromol. 2011, 48, 423–431. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; Sudarshan, S. Inhibition of secretary PLA₂—VRV-PL-VIIIa of Russell’s viper venom by standard aqueous stem bark extract of Mangifera indica L. Trop Biomed. 2015, 32, 24–35. [Google Scholar]
- Krayem, N.; Gargouri, Y. Scorpion venom phospholipases A2: A minireview. Toxicon 2020, 184, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Stadel, J.M.; Jones, C.; Livi, G.P.; Hoyle, K.; Kurdyla, J.; Roshak, A.; McLaughlin, M.M.; Pfarr, D.A.; Comer, S.; Strickler, J.; et al. Recombinant human secretory phospholipase A2: Purification and characterization of the enzyme for active site studies. J. Mol. Recognit. 1992, 5, 145–153. [Google Scholar] [CrossRef]
- Sukocheva, O.; Menschikowski, M.; Hagelgans, A.; Yarla, N.S.; Siegert, G.; Reddanna, P.; Bishayee, A. Current insights into functions of phospholipase A2 receptor in normal and cancer cells: More questions than answers. Semin. Cancer Biol. 2019, 56, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Tokumura, A.; Majima, E.; Kariya, Y.; Tominaga, K.; Kogure, K.; Yasuda, K.; Fukuzawa, K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 2002, 277, 39436–39442. [Google Scholar] [CrossRef] [PubMed]
- Umezu-Goto, M.; Kishi, Y.; Taira, A.; Hama, K.; Dohmae, N.; Takio, K.; Yamori, T.; Mills, G.B.; Inoue, K.; Aoki, J.; et al. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J. Cell Biol. 2002, 158, 227–233. [Google Scholar] [CrossRef]
- Snider, A.J.; Zhang, Z.; Xie, Y.; Meier, K.E. Epidermal growth factor increases lysophosphatidic acid production in human ovarian cancer cells: Roles for phospholipase D2 and receptor transactivation. Am. J. Physiol. Cell Physiol. 2010, 298, C163–C170. [Google Scholar] [CrossRef]
- Fukushima, N.; Ishii, S.; Tsujiuchi, T.; Kagawa, N.; Katoh, K. Comparative analyses of lysophosphatidic acid receptor-mediated signaling. Cell. Mol. Life Sci. 2015, 72, 2377–2394. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, H. Expression of Cytosolic Phospholipase A2 Alpha in Glioblastoma Is Associated with Resistance to Chemotherapy. Am. J. Med. Sci. 2018, 356, 391–398. [Google Scholar] [CrossRef]
- Hernández, M.; Burillo, S.L.; Crespo, M.S.; Nieto, M.L. Secretory phospholipase A2 activates the cascade of mitogen-activated protein kinases and cytosolic phospholipase A2 in the human astrocytoma cell line 1321N1. J. Biol. Chem. 1998, 273, 606–612. [Google Scholar] [CrossRef]
- Hernández, M.; Barrero, M.J.; Alvarez, J.; Montero, M.; Sánchez Crespo, M.; Nieto, M.L. Secretory phospholipase A2 induces phospholipase Cgamma-1 activation and Ca2+ mobilization in the human astrocytoma cell line 1321N1 by a mechanism independent of its catalytic activity. Biochem. Biophys. Res. Commun. 1999, 260, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, S.; Ohno, Y.; Nakamura, S.; Yamada, T.; Noda, Y.; Saio, M.; Iwama, T.; Shimazawa, M.; Hara, H. Temozolomide has anti-tumor effects through the phosphorylation of cPLA2 on glioblastoma cells. Brain Res. 2019, 1723, 146396. [Google Scholar] [CrossRef] [PubMed]
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef]
- Gao, Y.; Han, D.; Sun, L.; Huang, Q.; Gai, G.; Wu, Z.; Meng, W.; Chen, X. PPARα Regulates the Proliferation of Human Glioma Cells through miR-214 and E2F2. Biomed. Res. Int. 2018, 2018, 3842753. [Google Scholar] [CrossRef]
- Leaver, H.A.; Williams, J.R.; Smith, C.; Whittle, I.R. Intracellular oxidation by human glioma cell populations: Effect of arachidonic acid. Prostaglandins Leukot. Essent. Fat. Acids 2004, 70, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Giurdanella, G.; Motta, C.; Muriana, S.; Arena, V.; Anfuso, C.D.; Lupo, G.; Alberghina, M. Cytosolic and calcium-independent phospholipase A2 mediate glioma-enhanced proangiogenic activity of brain endothelial cells. Microvasc. Res. 2011, 81, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Anfuso, C.D.; Motta, C.; Giurdanella, G.; Arena, V.; Alberghina, M.; Lupo, G. Endothelial PKCα-MAPK/ERK-phospholipase A2 pathway activation as a response of glioma in a triple culture model. A new role for pericytes? Biochimie 2014, 99, 77–87. [Google Scholar] [CrossRef]
- Schleicher, S.M.; Thotala, D.K.; Linkous, A.G.; Hu, R.; Leahy, K.M.; Yazlovitskaya, E.M.; Hallahan, D.E. Autotaxin and LPA receptors represent potential molecular targets for the radiosensitization of murine glioma through effects on tumor vasculature. PLoS ONE 2011, 6, e22182. [Google Scholar] [CrossRef]
- Linkous, A.G.; Yazlovitskaya, E.M.; Hallahan, D.E. Cytosolic phospholipase A2 and lysophospholipids in tumor angiogenesis. J. Natl. Cancer Inst. 2010, 102, 1398–1412. [Google Scholar] [CrossRef]
- Mao, P.; Smith, L.; Xie, W.; Wang, M. Dying endothelial cells stimulate proliferation of malignant glioma cells via a caspase 3-mediated pathway. Oncol. Lett. 2013, 5, 1615–1620. [Google Scholar] [CrossRef]
- Lauber, K.; Bohn, E.; Kröber, S.M.; Xiao, Y.J.; Blumenthal, S.G.; Lindemann, R.K.; Marini, P.; Wiedig, C.; Zobywalski, A.; Baksh, S.; et al. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell 2003, 113, 717–730. [Google Scholar] [CrossRef]
- Wu, C.; Su, J.; Wang, X.; Wang, J.; Xiao, K.; Li, Y.; Xiao, Q.; Ling, M.; Xiao, Y.; Qin, C.; et al. Overexpression of the phospholipase A2 group V gene in glioma tumors is associated with poor patient prognosis. Cancer Manag. Res. 2019, 11, 3139–3152. [Google Scholar] [CrossRef] [PubMed]
- Kishi, Y.; Okudaira, S.; Tanaka, M.; Hama, K.; Shida, D.; Kitayama, J.; Yamori, T.; Aoki, J.; Fujimaki, T.; Arai, H. Autotaxin is overexpressed in glioblastoma multiforme and contributes to cell motility of glioblastoma by converting lysophosphatidylcholine to lysophosphatidic acid. J. Biol. Chem. 2006, 281, 17492–17500. [Google Scholar] [CrossRef] [PubMed]
- Hoelzinger, D.B.; Nakada, M.; Demuth, T.; Rosensteel, T.; Reavie, L.B.; Berens, M.E. Autotaxin: A secreted autocrine/paracrine factor that promotes glioma invasion. J. Neurooncol. 2008, 86, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Amaral, R.F.; Geraldo, L.H.M.; Einicker-Lamas, M.; Spohr, T.C.L.d.S.e.; Mendes, F.; Lima, F.R.S. Microglial lysophosphatidic acid promotes glioblastoma proliferation and migration via LPA1 receptor. J. Neurochem. 2021, 156, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Loskutov, Y.V.; Griffin, C.L.; Marinak, K.M.; Bobko, A.; Margaryan, N.V.; Geldenhuys, W.J.; Sarkaria, J.N.; Pugacheva, E.N. LPA signaling is regulated through the primary cilium: A novel target in glioblastoma. Oncogene 2018, 37, 1457–1471. [Google Scholar] [CrossRef] [PubMed]
- Malchinkhuu, E.; Sato, K.; Maehama, T.; Ishiuchi, S.; Yoshimoto, Y.; Mogi, C.; Kimura, T.; Kurose, H.; Tomura, H.; Okajima, F. Role of Rap1B and tumor suppressor PTEN in the negative regulation of lysophosphatidic acid--induced migration by isoproterenol in glioma cells. Mol. Biol. Cell 2009, 20, 5156–5165. [Google Scholar] [CrossRef] [PubMed]
- Bhave, S.R.; Dadey, D.Y.; Karvas, R.M.; Ferraro, D.J.; Kotipatruni, R.P.; Jaboin, J.J.; Hallahan, A.N.; Dewees, T.A.; Linkous, A.G.; Hallahan, D.E.; et al. Autotaxin Inhibition with PF-8380 Enhances the Radiosensitivity of Human and Murine Glioblastoma Cell Lines. Front. Oncol. 2013, 3, 236. [Google Scholar] [CrossRef]
- Valdés-Rives, S.A.; de la Fuente-Granada, M.; Velasco-Velázquez, M.A.; González-Flores, O.; González-Arenas, A. LPA1 receptor activation induces PKCα nuclear translocation in glioblastoma cells. Int. J. Biochem. Cell Biol. 2019, 110, 91–102. [Google Scholar] [CrossRef]
- Valdés-Rives, S.A.; Arcos-Montoya, D.; de la Fuente-Granada, M.; Zamora-Sánchez, C.J.; Arias-Romero, L.E.; Villamar-Cruz, O.; Camacho-Arroyo, I.; Pérez-Tapia, S.M.; González-Arenas, A. LPA1 Receptor Promotes Progesterone Receptor Phosphorylation through PKCα in Human Glioblastoma Cells. Cells 2021, 10, 807. [Google Scholar] [CrossRef]
- Rai, V.; Touré, F.; Chitayat, S.; Pei, R.; Song, F.; Li, Q.; Zhang, J.; Rosario, R.; Ramasamy, R.; Chazin, W.J.; et al. Lysophosphatidic acid targets vascular and oncogenic pathways via RAGE signaling. J. Exp. Med. 2012, 209, 2339–2350. [Google Scholar] [CrossRef]
- Cechin, S.R.; Dunkley, P.R.; Rodnight, R. Signal transduction mechanisms involved in the proliferation of C6 glioma cells induced by lysophosphatidic acid. Neurochem. Res. 2005, 30, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Hernández, M.; Ibeas, E.; Fuentes, L.; Salicio, V.; Arnés, M.; Nieto, M.L. Secreted phospholipase A2-IIA modulates key regulators of proliferation on astrocytoma cells. J. Neurochem. 2009, 111, 988–999. [Google Scholar] [CrossRef]
- Hernández, M.; Martín, R.; García-Cubillas, M.D.; Maeso-Hernández, P.; Nieto, M.L. Secreted PLA2 induces proliferation in astrocytoma through the EGF receptor: Another inflammation-cancer link. Neuro Oncol. 2010, 12, 1014–1023. [Google Scholar] [CrossRef]
- Martín, R.; Cordova, C.; Gutiérrez, B.; Hernández, M.; Nieto, M.L. A dangerous liaison: Leptin and sPLA2-IIA join forces to induce proliferation and migration of astrocytoma cells. PLoS ONE 2017, 12, e0170675. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.J.; Lin, V.T.; Zheng, Y.; Benveniste, E.N.; Lin, F.T. The adaptor protein TRIP6 antagonizes Fas-induced apoptosis but promotes its effect on cell migration. Mol. Cell. Biol. 2010, 30, 5582–5596. [Google Scholar] [CrossRef] [PubMed]
- Annabi, B.; Lachambre, M.P.; Plouffe, K.; Sartelet, H.; Béliveau, R. Modulation of invasive properties of CD133+ glioblastoma stem cells: A role for MT1-MMP in bioactive lysophospholipid signaling. Mol. Carcinog. 2009, 48, 910–919. [Google Scholar] [CrossRef]
- Kita, Y.; Shindou, H.; Shimizu, T. Cytosolic phospholipase A2 and lysophospholipid acyltransferases. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2019, 1864, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.C.; Alexson, S.E. The role Acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog. Lipid Res. 2002, 41, 99–130. [Google Scholar] [CrossRef]
- Hunt, M.C.; Yamada, J.; Maltais, L.J.; Wright, M.W.; Podesta, E.J.; Alexson, S.E. A revised nomenclature for mammalian acyl-CoA thioesterases/hydrolases. J. Lipid Res. 2005, 46, 2029–2032. [Google Scholar] [CrossRef]
- Miyazawa, S.; Furuta, S.; Hashimoto, T. Induction of a novel long-chain acyl-CoA hydrolase in rat liver by administration of peroxisome proliferators. Eur. J. Biochem. 1981, 117, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Poupon, V.; Bègue, B.; Gagnon, J.; Dautry-Varsat, A.; Cerf-Bensussan, N.; Benmerah, A. Molecular cloning and characterization of MT-ACT48, a novel mitochondrial acyl-CoA thioesterase. J. Biol. Chem. 1999, 274, 19188–19194. [Google Scholar] [CrossRef] [PubMed]
- Levin, G.; Duffin, K.L.; Obukowicz, M.G.; Hummert, S.L.; Fujiwara, H.; Needleman, P.; Raz, A. Differential metabolism of dihomo-gamma-linolenic acid and arachidonic acid by cyclo-oxygenase-1 and cyclo-oxygenase-2: Implications for cellular synthesis of prostaglandin E1 and prostaglandin E2. Biochem. J. 2002, 365, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Nakatsugi, S.; Sugimoto, N.; Furukawa, M. Effects of non-steroidal anti-inflammatory drugs on prostaglandin E2 production by cyclooxygenase-2 from endogenous and exogenous arachidonic acid in rat peritoneal macrophages stimulated with lipopolysaccharide. Prostaglandins Leukot. Essent. Fat. Acids 1996, 55, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Hawcroft, G.; Loadman, P.M.; Belluzzi, A.; Hull, M.A. Effect of eicosapentaenoic acid on E-type prostaglandin synthesis and EP4 receptor signaling in human colorectal cancer cells. Neoplasia 2010, 12, 618–627. [Google Scholar] [CrossRef]
- Nugteren, D.H.; Hazelhof, E. Isolation and properties of intermediates in prostaglandin biosynthesis. Biochim. Biophys. Acta 1973, 326, 448–461. [Google Scholar] [CrossRef]
- Hamberg, M.; Svensson, J.; Samuelsson, B. Prostaglandin endoperoxides. A new concept concerning the mode of action and release of prostaglandins. Proc. Natl. Acad. Sci. USA 1974, 71, 3824–3828. [Google Scholar] [CrossRef]
- Yu, R.; Xiao, L.; Zhao, G.; Christman, J.W.; van Breemen, R.B. Competitive enzymatic interactions determine the relative amounts of prostaglandins E2 and D2. J. Pharmacol. Exp. Ther. 2011, 339, 716–725. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, Y.; Law, B.; Qian, S.Y. The first characterization of free radicals formed from cellular COX-catalyzed peroxidation. Free Radic. Biol. Med. 2013, 57, 49–60. [Google Scholar] [CrossRef]
- Engels, F.; Willems, H.; Nijkamp, F.P. Cyclooxygenase-catalyzed formation of 9-hydroxylinoleic acid by guinea pig alveolar macrophages under non-stimulated conditions. FEBS Lett. 1986, 209, 249–253. [Google Scholar] [CrossRef]
- Vangaveti, V.N.; Shashidhar, V.M.; Rush, C.; Malabu, U.H.; Rasalam, R.R.; Collier, F.; Baune, B.T.; Kennedy, R.L. Hydroxyoctadecadienoic acids regulate apoptosis in human THP-1 cells in a PPARγ-dependent manner. Lipids 2014, 49, 1181–1192. [Google Scholar] [CrossRef] [PubMed]
- Alsalem, M.; Wong, A.; Millns, P.; Arya, P.H.; Chan, M.S.; Bennett, A.; Barrett, D.A.; Chapman, V.; Kendall, D.A. The contribution of the endogenous TRPV1 ligands 9-HODE and 13-HODE to nociceptive processing and their role in peripheral inflammatory pain mechanisms. Br. J. Pharmacol. 2013, 168, 1961–1974. [Google Scholar] [CrossRef] [PubMed]
- Obinata, H.; Hattori, T.; Nakane, S.; Tatei, K.; Izumi, T. Identification of 9-hydroxyoctadecadienoic acid and other oxidized free fatty acids as ligands of the G protein-coupled receptor G2A. J. Biol. Chem. 2005, 280, 40676–40683. [Google Scholar] [CrossRef]
- Wang, L.H.; Hajibeigi, A.; Xu, X.M.; Loose-Mitchell, D.; Wu, K.K. Characterization of the promoter of human prostaglandin H synthase-1 gene. Biochem. Biophys. Res. Commun. 1993, 190, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, T.; Miyata, A.; Ihara, H.; Hara, S.; Sugimoto, T.; Takeda, O.; Takahashi, E.; Tanabe, T. Characterization of the human gene (PTGS2) encoding prostaglandin-endoperoxide synthase 2. Eur. J. Biochem. 1994, 221, 889–897. [Google Scholar] [CrossRef]
- Mbonye, U.R.; Wada, M.; Rieke, C.J.; Tang, H.Y.; Dewitt, D.L.; Smith, W.L. The 19-amino acid cassette of cyclooxygenase-2 mediates entry of the protein into the endoplasmic reticulum-associated degradation system. J. Biol. Chem. 2006, 281, 35770–35778. [Google Scholar] [CrossRef]
- Chandrasekharan, N.V.; Dai, H.; Roos, K.L.; Evanson, N.K.; Tomsik, J.; Elton, T.S.; Simmons, D.L. COX-3, a cyclooxygenase-1 variant inhibited by acetaminophen and other analgesic/antipyretic drugs: Cloning, structure, and expression. Proc. Natl. Acad. Sci. USA 2002, 99, 13926–13931. [Google Scholar] [CrossRef]
- Qin, N.; Zhang, S.P.; Reitz, T.L.; Mei, J.M.; Flores, C.M. Cloning, expression, and functional characterization of human cyclooxygenase-1 splicing variants: Evidence for intron 1 retention. J. Pharmacol. Exp. Ther. 2005, 315, 1298–1305. [Google Scholar] [CrossRef]
- Ouellet, M.; Falgueyret, J.P.; Ear, P.H.; Pen, A.; Mancini, J.A.; Riendeau, D.; Percival, M.D. Purification and characterization of recombinant microsomal prostaglandin E synthase-1. Protein Expr. Purif. 2002, 26, 489–495. [Google Scholar] [CrossRef]
- Thorén, S.; Weinander, R.; Saha, S.; Jegerschöld, C.; Pettersson, P.L.; Samuelsson, B.; Hebert, H.; Hamberg, M.; Morgenstern, R.; Jakobsson, P.J. Human microsomal prostaglandin E synthase-1: Purification, functional characterization, and projection structure determination. J. Biol. Chem. 2003, 278, 22199–22209. [Google Scholar] [CrossRef]
- Huang, X.; Yan, W.; Gao, D.; Tong, M.; Tai, H.H.; Zhan, C.G. Structural and functional characterization of human microsomal prostaglandin E synthase-1 by computational modeling and site-directed mutagenesis. Bioorg. Med. Chem. 2006, 14, 3553–3562. [Google Scholar] [CrossRef]
- Murakami, M.; Nakashima, K.; Kamei, D.; Masuda, S.; Ishikawa, Y.; Ishii, T.; Ohmiya, Y.; Watanabe, K.; Kudo, I. Cellular prostaglandin E2 production by membrane-bound prostaglandin E synthase-2 via both cyclooxygenases-1 and -2. J. Biol. Chem. 2003, 278, 37937–37947. [Google Scholar] [CrossRef] [PubMed]
- Tanioka, T.; Nakatani, Y.; Semmyo, N.; Murakami, M.; Kudo, I. Molecular identification of cytosolic prostaglandin E2 synthase that is functionally coupled with cyclooxygenase-1 in immediate prostaglandin E2 biosynthesis. J. Biol. Chem. 2000, 275, 32775–32782. [Google Scholar] [CrossRef] [PubMed]
- Tanioka, T.; Nakatani, Y.; Kobayashi, T.; Tsujimoto, M.; Oh-ishi, S.; Murakami, M.; Kudo, I. Regulation of cytosolic prostaglandin E2 synthase by 90-kDa heat shock protein. Biochem. Biophys. Res. Commun. 2003, 303, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Zhang, Y.; Zhang, M.; Lu, W.J.; Rao, R.; Fan, X.; Redha, R.; Davis, L.; Breyer, R.M.; Harris, R.; et al. Membrane-associated PGE synthase-1 (mPGES-1) is coexpressed with both COX-1 and COX-2 in the kidney. Kidney Int. 2004, 65, 1205–1213. [Google Scholar] [CrossRef]
- Giannico, G.; Mendez, M.; LaPointe, M.C. Regulation of the membrane-localized prostaglandin E synthases mPGES-1 and mPGES-2 in cardiac myocytes and fibroblasts. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H165–H174. [Google Scholar] [CrossRef]
- Ueno, N.; Murakami, M.; Tanioka, T.; Fujimori, K.; Tanabe, T.; Urade, Y.; Kudo, I. Coupling between cyclooxygenase, terminal prostanoid synthase, and phospholipase A2. J. Biol. Chem. 2001, 276, 34918–34927. [Google Scholar] [CrossRef]
- Polet, H.; Levine, L. Metabolism of prostaglandins E, A, and C in serum. J. Biol. Chem. 1975, 250, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.L. Preparation of prostaglandins C: Chemical fixation of prostaglandin A isomerase to a gel support and partition chromatography of prostaglandins A, B and C. Prostaglandins 1974, 5, 283–290. [Google Scholar] [CrossRef]
- Lee, B.R.; Paing, M.H.; Sharma-Walia, N. Cyclopentenone Prostaglandins: Biologically Active Lipid Mediators Targeting Inflammation. Front. Physiol. 2021, 12, 640374. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, F.A.; Wynalda, M.A. Albumin-catalyzed metabolism of prostaglandin D2. Identification of products formed in vitro. J. Biol. Chem. 1983, 258, 11713–11718. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Kondo, M.; Osawa, T.; Shibata, N.; Kobayashi, M.; Uchida, K. 15-deoxy-delta 12,14-prostaglandin J2. A prostaglandin D2 metabolite generated during inflammatory processes. J. Biol. Chem. 2002, 277, 10459–10466. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, I.; Smith, D.L.; Whyte, M.A.; Nelson, J.T.; Cho, D.; Tokes, L.G.; Alvarez, R.; Willis, A.L. On the identification and biological properties of prostaglandin J2. Prostaglandins Leukot. Med. 1984, 16, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Aldini, G.; Ito, S.; Morishita, N.; Shibata, T.; Vistoli, G.; Carini, M.; Uchida, K. Delta12-prostaglandin J2 as a product and ligand of human serum albumin: Formation of an unusual covalent adduct at His146. J. Am. Chem. Soc. 2010, 132, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Straus, D.S.; Glass, C.K. Cyclopentenone prostaglandins: New insights on biological activities and cellular targets. Med. Res. Rev. 2001, 21, 185–210. [Google Scholar] [CrossRef]
- Hsu, P.Y.; Tsai, A.L.; Kulmacz, R.J.; Wang, L.H. Expression, purification, and spectroscopic characterization of human thromboxane synthase. J. Biol. Chem. 1999, 274, 762–769. [Google Scholar] [CrossRef] [PubMed]
- Watkins, G.; Douglas-Jones, A.; Mansel, R.E.; Jiang, W.G. Expression of thromboxane synthase, TBXAS1 and the thromboxane A2 receptor, TBXA2R, in human breast cancer. Int. Semin. Surg. Oncol. 2005, 2, 23. [Google Scholar] [CrossRef]
- Hamberg, M.; Svensson, J.; Samuelsson, B. Thromboxanes: A new group of biologically active compounds derived from prostaglandin endoperoxides. Proc. Natl. Acad. Sci. USA 1975, 72, 2994–2998. [Google Scholar] [CrossRef]
- Matsunobu, T.; Okuno, T.; Yokoyama, C.; Yokomizo, T. Thromboxane A synthase-independent production of 12-hydroxyheptadecatrienoic acid, a BLT2 ligand. J. Lipid Res. 2013, 54, 2979–2987. [Google Scholar] [CrossRef]
- Okuno, T.; Iizuka, Y.; Okazaki, H.; Yokomizo, T.; Taguchi, R.; Shimizu, T. 12(S)-Hydroxyheptadeca-5Z, 8E, 10E-trienoic acid is a natural ligand for leukotriene B4 receptor 2. J. Exp. Med. 2008, 205, 759–766. [Google Scholar] [CrossRef]
- Liu, M.; Saeki, K.; Matsunobu, T.; Okuno, T.; Koga, T.; Sugimoto, Y.; Yokoyama, C.; Nakamizo, S.; Kabashima, K.; Narumiya, S.; et al. 12-Hydroxyheptadecatrienoic acid promotes epidermal wound healing by accelerating keratinocyte migration via the BLT2 receptor. J. Exp. Med. 2014, 211, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Giusti, F.; Casiraghi, M.; Point, E.; Damian, M.; Rieger, J.; Bon, C.L.; Pozza, A.; Moncoq, K.; Banères, J.L.; Catoire, L.J. Structure of the agonist 12-HHT in its BLT2 receptor-bound state. Sci. Rep. 2020, 10, 2630. [Google Scholar] [CrossRef]
- Cathcart, M.C.; Gray, S.G.; Baird, A.M.; Boyle, E.; Gately, K.; Kay, E.; Cummins, R.; Pidgeon, G.P.; O’Byrne, K.J. Prostacyclin synthase expression and epigenetic regulation in nonsmall cell lung cancer. Cancer 2011, 117, 5121–5132. [Google Scholar] [CrossRef] [PubMed]
- Dozier, B.L.; Watanabe, K.; Duffy, D.M. Two pathways for prostaglandin F2 alpha synthesis by the primate periovulatory follicle. Reproduction 2008, 136, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Bresson, E.; Boucher-Kovalik, S.; Chapdelaine, P.; Madore, E.; Harvey, N.; Laberge, P.Y.; Leboeuf, M.; Fortier, M.A. The human aldose reductase AKR1B1 qualifies as the primary prostaglandin F synthase in the endometrium. J. Clin. Endocrinol. Metab. 2011, 96, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Wielinga, P.; Zelcer, N.; van der Heijden, I.; Kuil, A.; de Haas, M.; Wijnholds, J.; Borst, P. The human multidrug resistance protein MRP4 functions as a prostaglandin efflux transporter and is inhibited by nonsteroidal antiinflammatory drugs. Proc. Natl. Acad. Sci. USA 2003, 100, 9244–9249. [Google Scholar] [CrossRef]
- Lu, R.; Kanai, N.; Bao, Y.; Schuster, V.L. Cloning, in vitro expression, and tissue distribution of a human prostaglandin transporter cDNA(hPGT). J. Clin. Investig. 1996, 98, 1142–1149. [Google Scholar] [CrossRef]
- Nomura, T.; Lu, R.; Pucci, M.L.; Schuster, V.L. The two-step model of prostaglandin signal termination: In vitro reconstitution with the prostaglandin transporter and prostaglandin 15 dehydrogenase. Mol. Pharmacol. 2004, 65, 973–978. [Google Scholar] [CrossRef]
- Cattori, V.; van Montfoort, J.E.; Stieger, B.; Landmann, L.; Meijer, D.K.; Winterhalter, K.H.; Meier, P.J.; Hagenbuch, B. Localization of organic anion transporting polypeptide 4 (Oatp4) in rat liver and comparison of its substrate specificity with Oatp1, Oatp2 and Oatp3. Pflugers Arch. 2001, 443, 188–195. [Google Scholar] [CrossRef]
- Chou, W.L.; Chuang, L.M.; Chou, C.C.; Wang, A.H.; Lawson, J.A.; FitzGerald, G.A.; Chang, Z.F. Identification of a novel prostaglandin reductase reveals the involvement of prostaglandin E2 catabolism in regulation of peroxisome proliferator-activated receptor gamma activation. J. Biol. Chem. 2007, 282, 18162–18172. [Google Scholar] [CrossRef]
- Panagopoulos, A.T.; Gomes, R.N.; Almeida, F.G.; da Costa Souza, F.; Veiga, J.C.E.; Nicolaou, A.; Colquhoun, A. The prostanoid pathway contains potential prognostic markers for glioblastoma. Prostaglandins Other Lipid Mediat. 2018, 137, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Granström, E.; Hamberg, M.; Hansson, G.; Kindahl, H. Chemical instability of 15-keto-13,14-dihydro-PGE2: The reason for low assay reliability. Prostaglandins 1980, 19, 933–957. [Google Scholar] [CrossRef] [PubMed]
- Bremme, K.; Eneroth, P.; Gottlieb, C.; Kindahl, H.; Svanborg, K.; Nilsson, B.; Olsson, M.; Bygdeman, M. 15-Keto-13,14-dihydroprostaglandin E2- and F2 alpha-metabolite levels in blood from men and women given prostaglandin E2 orally. Prostaglandins Leukot. Essent. Fat. Acids 1989, 37, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Okumura, T.; Nakayama, R.; Sago, T.; Saito, K. Identification of prostaglandin E metabolites from primary cultures of rat hepatocytes. Biochim. Biophys. Acta 1985, 837, 197–207. [Google Scholar]
- Schepers, L.; Casteels, M.; Vamecq, J.; Parmentier, G.; Van Veldhoven, P.P.; Mannaerts, G.P. Beta-oxidation of the carboxyl side chain of prostaglandin E2 in rat liver peroxisomes and mitochondria. J. Biol. Chem. 1988, 263, 2724–2731. [Google Scholar] [CrossRef]
- Powell, W.S.; Solomon, S. Formation of 20-hydroxyprostaglandins by lungs of pregnant rabbits. J. Biol. Chem. 1978, 253, 4609–4616. [Google Scholar] [CrossRef]
- Hamberg, M.; Samuelsson, B. The structure of the major urinary metabolite of prostaglandin E2 in man. J. Am. Chem. Soc. 1969, 91, 2177–2178. [Google Scholar] [CrossRef]
- Hamberg, M.; Samuelsson, B. On the metabolism of prostaglandins E 1 and E 2 in man. J. Biol. Chem. 1971, 246, 6713–6721. [Google Scholar] [CrossRef]
- Mancini, J.A.; O’Neill, G.P.; Bayly, C.; Vickers, P.J. Mutation of serine-516 in human prostaglandin G/H synthase-2 to methionine or aspirin acetylation of this residue stimulates 15-R-HETE synthesis. FEBS Lett. 1994, 342, 33–37. [Google Scholar] [CrossRef]
- Mulugeta, S.; Suzuki, T.; Hernandez, N.T.; Griesser, M.; Boeglin, W.E.; Schneider, C. Identification and absolute configuration of dihydroxy-arachidonic acids formed by oxygenation of 5S-HETE by native and aspirin-acetylated COX-2. J. Lipid Res. 2010, 51, 575–585. [Google Scholar] [CrossRef]
- Tejera, N.; Boeglin, W.E.; Suzuki, T.; Schneider, C. COX-2-dependent and -independent biosynthesis of dihydroxy-arachidonic acids in activated human leukocytes. J. Lipid Res. 2012, 53, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Lucido, M.J.; Orlando, B.J.; Vecchio, A.J.; Malkowski, M.G. Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with Reversed Stereochemistry. Biochemistry 2016, 55, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Loll, P.J.; Picot, D.; Garavito, R.M. The structural basis of aspirin activity inferred from the crystal structure of inactivated prostaglandin H2 synthase. Nat. Struct. Biol. 1995, 2, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Distrutti, E.; de Lima, O.M.; Romano, M.; Mencarelli, A.; Barbanti, M.; Palazzini, E.; Morelli, A.; Wallace, J.L. Relative contribution of acetylated cyclo-oxygenase (COX)-2 and 5-lipooxygenase (LOX) in regulating gastric mucosal integrity and adaptation to aspirin. FASEB J. 2003, 17, 1171–1173. [Google Scholar] [CrossRef]
- Wada, M.; DeLong, C.J.; Hong, Y.H.; Rieke, C.J.; Song, I.; Sidhu, R.S.; Yuan, C.; Warnock, M.; Schmaier, A.H.; Yokoyama, C.; et al. Enzymes and receptors of prostaglandin pathways with arachidonic acid-derived versus eicosapentaenoic acid-derived substrates and products. J. Biol. Chem. 2007, 282, 22254–22266. [Google Scholar] [CrossRef]
- Mengeaud, V.; Nano, J.L.; Fournel, S.; Rampal, P. Effects of eicosapentaenoic acid, gamma-linolenic acid and prostaglandin E1 on three human colon carcinoma cell lines. Prostaglandins Leukot. Essent. Fat. Acids 1992, 47, 313–319. [Google Scholar] [CrossRef]
- Tabolacci, C.; Lentini, A.; Provenzano, B.; Gismondi, A.; Rossi, S.; Beninati, S. Similar antineoplastic effects of nimesulide, a selective COX-2 inhibitor, and prostaglandin E1 on B16-F10 murine melanoma cells. Melanoma Res. 2010, 20, 273–279. [Google Scholar] [CrossRef]
- Xu, Y.; Qi, J.; Yang, X.; Wu, E.; Qian, S.Y. Free radical derivatives formed from cyclooxygenase-catalyzed dihomo-γ-linolenic acid peroxidation can attenuate colon cancer cell growth and enhance 5-fluorouracil’s cytotoxicity. Redox Biol. 2014, 2, 610–618. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, X.; Zhao, P.; Yang, Z.; Yan, C.; Guo, B.; Qian, S.Y. Knockdown of delta-5-desaturase promotes the anti-cancer activity of dihomo-γ-linolenic acid and enhances the efficacy of chemotherapy in colon cancer cells expressing COX-2. Free Radic. Biol. Med. 2016, 96, 67–77. [Google Scholar] [CrossRef]
- Ishizaki, Y.; Morita, I.; Murota, S. Arachidonic acid metabolism in cultured astrocytes from rat embryo and in C6 glioma cells. Brain Res. 1989, 494, 138–142. [Google Scholar] [CrossRef]
- Deininger, M.H.; Weller, M.; Streffer, J.; Mittelbronn, M.; Meyermann, R. Patterns of cyclooxygenase-1 and -2 expression in human gliomas in vivo. Acta Neuropathol. 1999, 98, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Joki, T.; Heese, O.; Nikas, D.C.; Bello, L.; Zhang, J.; Kraeft, S.K.; Seyfried, N.T.; Abe, T.; Chen, L.B.; Carroll, R.S.; et al. Expression of cyclooxygenase 2 (COX-2) in human glioma and in vitro inhibition by a specific COX-2 inhibitor, NS-398. Cancer Res. 2000, 60, 4926–4931. [Google Scholar] [PubMed]
- Mattila, S.; Tuominen, H.; Koivukangas, J.; Stenbäck, F. The terminal prostaglandin synthases mPGES-1, mPGES-2, and cPGES are all overexpressed in human gliomas. Neuropathology 2009, 29, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Giese, A.; Hagel, C.; Kim, E.L.; Zapf, S.; Djawaheri, J.; Berens, M.E.; Westphal, M. Thromboxane synthase regulates the migratory phenotype of human glioma cells. Neuro Oncol. 1999, 1, 3–13. [Google Scholar] [CrossRef]
- Castelli, M.G.; Butti, G.; Chiabrando, C.; Cozzi, E.; Fanelli, R.; Gaetani, P.; Silvani, V.; Paoletti, P. Arachidonic acid metabolic profiles in human meningiomas and gliomas. J. Neurooncol. 1987, 5, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Jiang, X.; Xue, D.; Chen, D. Glioma prostaglandin levels correlate with brain edema. J. Tongji Med. Univ. 1998, 18, 115–118. [Google Scholar] [PubMed]
- Lo, H.W.; Cao, X.; Zhu, H.; Ali-Osman, F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol. Cancer Res. 2010, 8, 232–245. [Google Scholar] [CrossRef]
- Xu, K.; Shu, H.K. Transcription factor interactions mediate EGF-dependent COX-2 expression. Mol. Cancer Res. 2013, 11, 875–886. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, Y.; Zhang, H.; Liu, X.; Du, W.; Li, Y.; Zhang, J.; Chen, L.; Jiang, C. HGF/MET signaling promotes glioma growth via up-regulation of Cox-2 expression and PGE2 production. Int. J. Clin. Exp. Pathol. 2015, 8, 3719–3726. [Google Scholar]
- Nakano, Y.; Kuroda, E.; Kito, T.; Yokota, A.; Yamashita, U. Induction of macrophagic prostaglandin E2 synthesis by glioma cells. J. Neurosurg. 2006, 104, 574–582. [Google Scholar] [CrossRef]
- Huang, N.; Chen, S.; Deng, J.; Huang, Q.; Liao, P.; Wang, F.; Cheng, Y. Overexpression of S100A9 in human glioma and in-vitro inhibition by aspirin. Eur. J. Cancer Prev. 2013, 22, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Fiebich, B.L.; Hüll, M.; Lieb, K.; Gyufko, K.; Berger, M.; Bauer, J. Prostaglandin E2 induces interleukin-6 synthesis in human astrocytoma cells. J. Neurochem. 1997, 68, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Venza, I.; Visalli, M.; Fortunato, C.; Ruggeri, M.; Ratone, S.; Caffo, M.; Caruso, G.; Alafaci, C.; Tomasello, F.; Teti, D.; et al. PGE2 induces interleukin-8 derepression in human astrocytoma through coordinated DNA demethylation and histone hyperacetylation. Epigenetics 2012, 7, 1315–1330. [Google Scholar] [CrossRef] [PubMed]
- Kardosh, A.; Blumenthal, M.; Wang, W.J.; Chen, T.C.; Schönthal, A.H. Differential effects of selective COX-2 inhibitors on cell cycle regulation and proliferation of glioblastoma cell lines. Cancer Biol. Ther. 2004, 3, 55–62. [Google Scholar] [CrossRef]
- Payner, T.; Leaver, H.A.; Knapp, B.; Whittle, I.R.; Trifan, O.C.; Miller, S.; Rizzo, M.T. Microsomal prostaglandin E synthase-1 regulates human glioma cell growth via prostaglandin E2–dependent activation of type II protein kinase A. Mol. Cancer Ther. 2006, 5, 1817–1826. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.T.; Miyake, J.A.; Gomes, R.N.; Feitoza, F.; Stevannato, P.B.; da Cunha, A.S.; Serachi, F.O.; Panagopoulos, A.T.; Colquhoun, A. Cyclooxygenase Inhibition Alters Proliferative, Migratory, and Invasive Properties of Human Glioblastoma Cells In Vitro. Int. J. Mol. Sci. 2021, 22, 4297. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, T.; Yu, S.; Liu, C.; He, M.; Hu, C. Prostaglandin E2 increases migration and proliferation of human glioblastoma cells by activating transient receptor potential melastatin 7 channels. J. Cell Mol. Med. 2018, 22, 6327–6337. [Google Scholar] [CrossRef]
- Annabi, B.; Laflamme, C.; Sina, A.; Lachambre, M.P.; Béliveau, R. A MT1-MMP/NF-kappaB signaling axis as a checkpoint controller of COX-2 expression in CD133(+) U87 glioblastoma cells. J. Neuroinflamm. 2009, 6, 8. [Google Scholar] [CrossRef]
- Wu, M.; Guan, J.; Li, C.; Gunter, S.; Nusrat, L.; Ng, S.; Dhand, K.; Morshead, C.; Kim, A.; Das, S. Aberrantly activated Cox-2 and Wnt signaling interact to maintain cancer stem cells in glioblastoma. Oncotarget 2017, 8, 82217–82230. [Google Scholar] [CrossRef]
- El-Sayed, M.; Taha, M.M. Immunohistochemical expression of cycloxygenase-2 in astrocytoma: Correlation with angiogenesis, tumor progression and survival. Turk. Neurosurg. 2011, 21, 27–35. [Google Scholar] [CrossRef]
- Rong, X.; Huang, B.; Qiu, S.; Li, X.; He, L.; Peng, Y. Tumor-associated macrophages induce vasculogenic mimicry of glioblastoma multiforme through cyclooxygenase-2 activation. Oncotarget 2016, 7, 83976–83986. [Google Scholar] [CrossRef] [PubMed]
- Addison, C.L.; Daniel, T.O.; Burdick, M.D.; Liu, H.; Ehlert, J.E.; Xue, Y.Y.; Buechi, L.; Walz, A.; Richmond, A.; Strieter, R.M. The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J. Immunol. 2000, 165, 5269–5277. [Google Scholar] [CrossRef] [PubMed]
- Ochs, K.; Ott, M.; Rauschenbach, K.J.; Deumelandt, K.; Sahm, F.; Opitz, C.A.; von Deimling, A.; Wick, W.; Platten, M. Tryptophan-2,3-dioxygenase is regulated by prostaglandin E2 in malignant glioma via a positive signaling loop involving prostaglandin E receptor-4. J. Neurochem. 2016, 136, 1142–1154. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Kohanbash, G.; Fellows-Mayle, W.; Hamilton, R.L.; Komohara, Y.; Decker, S.A.; Ohlfest, J.R.; Okada, H. COX-2 blockade suppresses gliomagenesis by inhibiting myeloid-derived suppressor cells. Cancer Res. 2011, 71, 2664–2674. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Holt, D.; Kundu, N.; Reader, J.; Goloubeva, O.; Take, Y.; Fulton, A.M. A prostaglandin E (PGE) receptor EP4 antagonist protects natural killer cells from PGE2-mediated immunosuppression and inhibits breast cancer metastasis. Oncoimmunology 2013, 2, e22647. [Google Scholar] [CrossRef]
- Park, A.; Lee, Y.; Kim, M.S.; Kang, Y.J.; Park, Y.J.; Jung, H.; Kim, T.D.; Lee, H.G.; Choi, I.; Yoon, S.R. Prostaglandin E2 Secreted by Thyroid Cancer Cells Contributes to Immune Escape Through the Suppression of Natural Killer (NK) Cell Cytotoxicity and NK Cell Differentiation. Front. Immunol. 2018, 9, 1859. [Google Scholar] [CrossRef]
- Sharma, S.; Stolina, M.; Yang, S.C.; Baratelli, F.; Lin, J.F.; Atianzar, K.; Luo, J.; Zhu, L.; Lin, Y.; Huang, M.; et al. Tumor cyclooxygenase 2-dependent suppression of dendritic cell function. Clin. Cancer Res. 2003, 9, 961–968. [Google Scholar]
- Yuan, X.L.; Chen, L.; Li, M.X.; Dong, P.; Xue, J.; Wang, J.; Zhang, T.T.; Wang, X.A.; Zhang, F.M.; Ge, H.L.; et al. Elevated expression of Foxp3 in tumor-infiltrating Treg cells suppresses T-cell proliferation and contributes to gastric cancer progression in a COX-2-dependent manner. Clin. Immunol. 2010, 134, 277–288. [Google Scholar] [CrossRef]
- Liu, L.; Ge, D.; Ma, L.; Mei, J.; Liu, S.; Zhang, Q.; Ren, F.; Liao, H.; Pu, Q.; Wang, T.; et al. Interleukin-17 and prostaglandin E2 are involved in formation of an M2 macrophage-dominant microenvironment in lung cancer. J. Thorac. Oncol. 2012, 7, 1091–1100. [Google Scholar] [CrossRef]
- Brocard, E.; Oizel, K.; Lalier, L.; Pecqueur, C.; Paris, F.; Vallette, F.M.; Oliver, L. Radiation-induced PGE2 sustains human glioma cells growth and survival through EGF signaling. Oncotarget 2015, 6, 6840–6849. [Google Scholar] [CrossRef]
- Cook, P.J.; Thomas, R.; Kingsley, P.J.; Shimizu, F.; Montrose, D.C.; Marnett, L.J.; Tabar, V.S.; Dannenberg, A.J.; Benezra, R. Cox-2-derived PGE2 induces Id1-dependent radiation resistance and self-renewal in experimental glioblastoma. Neuro Oncol. 2016, 18, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.T.; Lo, W.L.; Chen, P.Y.; Ko, C.Y.; Chuang, J.Y.; Kao, T.J.; Yang, W.B.; Chang, K.Y.; Hung, C.Y.; Kikkawa, U.; et al. Reprogramming of arachidonate metabolism confers temozolomide resistance to glioblastoma through enhancing mitochondrial activity in fatty acid oxidation. J. Biomed. Sci. 2022, 29, 21. [Google Scholar] [CrossRef]
- Lombardi, F.; Augello, F.R.; Artone, S.; Ayroldi, E.; Giusti, I.; Dolo, V.; Cifone, M.G.; Cinque, B.; Palumbo, P. Cyclooxygenase-2 Upregulated by Temozolomide in Glioblastoma Cells Is Shuttled In Extracellular Vesicles Modifying Recipient Cell Phenotype. Front. Oncol. 2022, 12, 933746. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Yu, Y.; He, S.; Cheng, J.; Gong, Y.; Zhang, Z.; Yang, X.; Xu, B.; Liu, X.; Li, C.Y.; et al. Dying glioma cells establish a proangiogenic microenvironment through a caspase 3 dependent mechanism. Cancer Lett. 2017, 385, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Desmarais, G.; Charest, G.; Therriault, H.; Shi, M.; Fortin, D.; Bujold, R.; Mathieu, D.; Paquette, B. Infiltration of F98 glioma cells in Fischer rat brain is temporary stimulated by radiation. Int. J. Radiat. Biol. 2016, 92, 444–450. [Google Scholar] [CrossRef]
- Desmarais, G.; Charest, G.; Fortin, D.; Bujold, R.; Mathieu, D.; Paquette, B. Cyclooxygenase-2 inhibitor prevents radiation-enhanced infiltration of F98 glioma cells in brain of Fischer rat. Int. J. Radiat. Biol. 2015, 91, 624–633. [Google Scholar] [CrossRef]
- Ferreira, M.T.; Gomes, R.N.; Panagopoulos, A.T.; de Almeida, F.G.; Veiga, J.C.E.; Colquhoun, A. Opposing roles of PGD2 in GBM. Prostaglandins Other Lipid Mediat. 2018, 134, 66–76. [Google Scholar] [CrossRef]
- Keyaki, A.; Handa, H.; Yamashita, J.; Tokuriki, Y.; Otsuka, S.; Yamasaki, T.; Gi, H. Growth-inhibitory effect of prostaglandin D2 on mouse glioma cells. J. Neurosurg. 1984, 61, 912–917. [Google Scholar] [CrossRef]
- Westphal, M.; Neuss, M.; Herrmann, H.D. Prostaglandins: Antiproliferative effect of PGD 2 on cultured human glioma cells. Acta Neurochir. 1986, 83, 56–61. [Google Scholar] [CrossRef]
- Cho, W.H.; Choi, C.H.; Park, J.Y.; Kang, S.K.; Kim, Y.K. 15-deoxy-(Delta12,14)-prostaglandin J2 (15d-PGJ2) induces cell death through caspase-independent mechanism in A172 human glioma cells. Neurochem. Res. 2006, 31, 1247–1254. [Google Scholar] [CrossRef]
- Nakahata, N.; Abe, M.T.; Nakanishi, H. PGJ2 and delta 12PGJ2 inhibit cell growth accompanied with inhibition of phosphoinositide turnover in human astrocytoma cells. Prostaglandins 1990, 40, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Morosetti, R.; Servidei, T.; Mirabella, M.; Rutella, S.; Mangiola, A.; Maira, G.; Mastrangelo, R.; Koeffler, H.P. The PPARgamma ligands PGJ2 and rosiglitazone show a differential ability to inhibit proliferation and to induce apoptosis and differentiation of human glioblastoma cell lines. Int. J. Oncol. 2004, 25, 493–502. [Google Scholar]
- Obara, Y.; Kurose, H.; Nakahata, N. Thromboxane A2 promotes interleukin-6 biosynthesis mediated by an activation of cyclic AMP-response element-binding protein in 1321N1 human astrocytoma cells. Mol. Pharmacol. 2005, 68, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Yoshizato, K.; Zapf, S.; Westphal, M.; Berens, M.E.; Giese, A. Thromboxane synthase inhibitors induce apoptosis in migration-arrested glioma cells. Neurosurgery 2002, 50, 343–354. [Google Scholar] [PubMed]
- Schmidt, N.O.; Ziu, M.; Cargioli, T.; Westphal, M.; Giese, A.; Black, P.M.; Carroll, R.S. Inhibition of thromboxane synthase activity improves glioblastoma response to alkylation chemotherapy. Transl. Oncol. 2010, 3, 43–49. [Google Scholar] [CrossRef]
- Schauff, A.K.; Kim, E.L.; Leppert, J.; Nadrowitz, R.; Wuestenberg, R.; Brockmann, M.A.; Giese, A. Inhibition of invasion-associated thromboxane synthase sensitizes experimental gliomas to gamma-radiation. J. Neurooncol. 2009, 91, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Shono, T.; Tofilon, P.J.; Bruner, J.M.; Owolabi, O.; Lang, F.F. Cyclooxygenase-2 expression in human gliomas: Prognostic significance and molecular correlations. Cancer Res. 2001, 61, 4375–4381. [Google Scholar] [PubMed]
- Wang, X.; Chen, Y.; Zhang, S.; Zhang, L.; Liu, X.; Zhang, L.; Li, X.; Chen, D. Co-expression of COX-2 and 5-LO in primary glioblastoma is associated with poor prognosis. J. Neurooncol. 2015, 125, 277–285. [Google Scholar] [CrossRef]
- Zohn, I.E.; Klinger, M.; Karp, X.; Kirk, H.; Symons, M.; Chrzanowska-Wodnicka, M.; Der, C.J.; Kay, R.J. G2A is an oncogenic G protein-coupled receptor. Oncogene 2000, 19, 3866–3877. [Google Scholar] [CrossRef]
- Nam, D.H.; Park, K.; Park, C.; Im, Y.H.; Kim, M.H.; Lee, S.; Hong, S.C.; Shin, H.J.; Kim, J.H.; Eoh, W.; et al. Intracranial inhibition of glioma cell growth by cyclooxygenase-2 inhibitor celecoxib. Oncol. Rep. 2004, 11, 263–268. [Google Scholar] [CrossRef]
- Tuettenberg, J.; Grobholz, R.; Korn, T.; Wenz, F.; Erber, R.; Vajkoczy, P. Continuous low-dose chemotherapy plus inhibition of cyclooxygenase-2 as an antiangiogenic therapy of glioblastoma multiforme. J. Cancer Res. Clin. Oncol. 2005, 131, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yang, X.; Liu, P.; Zhou, J.; Luo, J.; Wang, H.; Li, A.; Zhou, Y. Association between nonsteroidal anti-inflammatory drugs use and risk of central nervous system tumors: A dose-response meta analysis. Oncotarget 2017, 8, 102486–102498. [Google Scholar] [CrossRef] [PubMed]
- Amirian, E.S.; Ostrom, Q.T.; Armstrong, G.N.; Lai, R.K.; Gu, X.; Jacobs, D.I.; Jalali, A.; Claus, E.B.; Barnholtz-Sloan, J.S.; Il’yasova, D.; et al. Aspirin, NSAIDs, and Glioma Risk: Original Data from the Glioma International Case-Control Study and a Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2019, 28, 555–562. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, Y.; Wang, J.; Xie, L.; Li, T.; He, Y.; Peng, Q.; Qin, X.; Li, S. Association between nonsteroidal anti-inflammatory drug use and brain tumour risk: A meta-analysis. Br. J. Clin. Pharmacol. 2014, 78, 58–68. [Google Scholar] [CrossRef]
- Funk, C.D.; Funk, L.B.; FitzGerald, G.A.; Samuelsson, B. Characterization of human 12-lipoxygenase genes. Proc. Natl. Acad. Sci. USA 1992, 89, 3962–3966. [Google Scholar] [CrossRef]
- Krieg, P.; Marks, F.; Fürstenberger, G. A gene cluster encoding human epidermis-type lipoxygenases at chromosome 17p13.1: Cloning, physical mapping, and expression. Genomics 2001, 73, 323–330. [Google Scholar] [CrossRef]
- Jisaka, M.; Kim, R.B.; Boeglin, W.E.; Brash, A.R. Identification of amino acid determinants of the positional specificity of mouse 8S-lipoxygenase and human 15S-lipoxygenase-2. J. Biol. Chem. 2000, 275, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Fürstenberger, G.; Marks, F.; Krieg, P. Arachidonate 8(S)-lipoxygenase. Prostaglandins Other Lipid Mediat. 2002, 68–69, 235–243. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Yu, Z.; Schneider, C.; Boeglin, W.E.; Marnett, L.J.; Brash, A.R. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc. Natl. Acad. Sci. USA 2003, 100, 9162–9167. [Google Scholar] [CrossRef]
- Zheng, Y.; Brash, A.R. Dioxygenase activity of epidermal lipoxygenase-3 unveiled: Typical and atypical features of its catalytic activity with natural and synthetic polyunsaturated fatty acids. J. Biol. Chem. 2010, 285, 39866–39875. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Schneider, C.; Boeglin, W.E.; Brash, A.R. Human and mouse eLOX3 have distinct substrate specificities: Implications for their linkage with lipoxygenases in skin. Arch. Biochem. Biophys. 2006, 455, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Gregus, A.M.; Dumlao, D.S.; Wei, S.C.; Norris, P.C.; Catella, L.C.; Meyerstein, F.G.; Buczynski, M.W.; Steinauer, J.J.; Fitzsimmons, B.L.; Yaksh, T.L.; et al. Systematic analysis of rat 12/15-lipoxygenase enzymes reveals critical role for spinal eLOX3 hepoxilin synthase activity in inflammatory hyperalgesia. FASEB J. 2013, 27, 1939–1949. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.B.; Zhang, Y.; Mayer, A.L.; Fujiwara, H.; Stothard, A.I.; Graham, M.J.; Swarts, B.M.; DeBosch, B.J. Hepatocyte ALOXE3 is induced during adaptive fasting and enhances insulin sensitivity by activating hepatic PPARγ. JCI Insight 2018, 3, e120794. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, J.; Wang, C.; Cheng, K.K.; Xu, H.; Li, Q.; Hua, T.; Jiang, X.; Sheng, L.; Mao, J.; et al. miR-18a promotes glioblastoma development by down-regulating ALOXE3-mediated ferroptotic and anti-migration activities. Oncogenesis 2021, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.S.; Rokach, J. Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid. Biochim. Biophys. Acta 2015, 1851, 340–355. [Google Scholar] [CrossRef]
- Pace-Asciak, C.R. Pathophysiology of the hepoxilins. Biochim. Biophys. Acta 2015, 1851, 383–396. [Google Scholar] [CrossRef]
- Yu, Z.; Schneider, C.; Boeglin, W.E.; Brash, A.R. Mutations associated with a congenital form of ichthyosis (NCIE) inactivate the epidermal lipoxygenases 12R-LOX and eLOX3. Biochim. Biophys. Acta 2005, 1686, 238–247. [Google Scholar] [CrossRef]
- Wang, T.; Xu, C.; Zhou, X.; Li, C.; Zhang, H.; Lian, B.Q.; Lee, J.J.; Shen, J.; Liu, Y.; Lian, C.G. Homozygous ALOXE3 Nonsense Variant Identified in a Patient with Non-Bullous Congenital Ichthyosiform Erythroderma Complicated by Superimposed Bullous Majocchi’s Granuloma: The Consequences of Skin Barrier Dysfunction. Int. J. Mol. Sci. 2015, 16, 21791–21801. [Google Scholar] [CrossRef]
- Hotz, A.; Kopp, J.; Bourrat, E.; Oji, V.; Komlosi, K.; Giehl, K.; Bouadjar, B.; Bygum, A.; Tantcheva-Poor, I.; Hellström Pigg, M.; et al. Meta-Analysis of Mutations in ALOX12B or ALOXE3 Identified in a Large Cohort of 224 Patients. Genes 2021, 12, 80. [Google Scholar] [CrossRef]
- Ueda, N.; Kaneko, S.; Yoshimoto, T.; Yamamoto, S. Purification of arachidonate 5-lipoxygenase from porcine leukocytes and its reactivity with hydroperoxyeicosatetraenoic acids. J. Biol. Chem. 1986, 261, 7982–7988. [Google Scholar] [CrossRef] [PubMed]
- Mancini, J.A.; Abramovitz, M.; Cox, M.E.; Wong, E.; Charleson, S.; Perrier, H.; Wang, Z.; Prasit, P.; Vickers, P.J. 5-lipoxygenase-activating protein is an arachidonate binding protein. FEBS Lett. 1993, 318, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Häfner, A.K.; Gerstmeier, J.; Hörnig, M.; George, S.; Ball, A.K.; Schröder, M.; Garscha, U.; Werz, O.; Steinhilber, D. Characterization of the interaction of human 5-lipoxygenase with its activating protein FLAP. Biochim. Biophys. Acta 2015, 1851, 1465–1472. [Google Scholar] [CrossRef] [PubMed]
- Narala, V.R.; Adapala, R.K.; Suresh, M.V.; Brock, T.G.; Peters-Golden, M.; Reddy, R.C. Leukotriene B4 is a physiologically relevant endogenous peroxisome proliferator-activated receptor-alpha agonist. J. Biol. Chem. 2010, 285, 22067–22074. [Google Scholar] [CrossRef]
- Falgueyret, J.; Riendeau, D. LTA4-derived 5-oxo-eicosatetraenoic acid: pH-dependent formation and interaction with the LTB4 receptor of human polymorphonuclear leukocytes. Biochim. Biophys. Acta 2000, 1484, 51–58. [Google Scholar] [CrossRef]
- Chiba, N.; Imai, H.; Narashima, K.; Arai, M.; Oshima, G.; Kunimoto, M.; Nakagawa, Y. Cellular glutathione peroxidase as a predominant scavenger of hydroperoxyeicosatetraenoic acids in rabbit alveolar macrophages. Biol. Pharm. Bull. 1999, 22, 1047–1051. [Google Scholar] [CrossRef]
- Erlemann, K.R.; Cossette, C.; Grant, G.E.; Lee, G.J.; Patel, P.; Rokach, J.; Powell, W.S. Regulation of 5-hydroxyeicosanoid dehydrogenase activity in monocytic cells. Biochem. J. 2007, 403, 157–165. [Google Scholar] [CrossRef]
- Patel, P.; Cossette, C.; Anumolu, J.R.; Erlemann, K.R.; Grant, G.E.; Rokach, J.; Powell, W.S. Substrate selectivity of 5-hydroxyeicosanoid dehydrogenase and its inhibition by 5-hydroxy-Delta6-long-chain fatty acids. J. Pharmacol. Exp. Ther. 2009, 329, 335–341. [Google Scholar] [CrossRef]
- Nagendra Reddy, C.; Ye, Q.; Patel, P.; Sivendran, S.; Chourey, S.; Wang, R.; Anumolu, J.R.; Grant, G.E.; Powell, W.S.; Rokach, J. Design and synthesis of affinity chromatography ligands for the purification of 5-hydroxyeicosanoid dehydrogenase. Bioorg. Med. Chem. 2017, 25, 116–125. [Google Scholar] [CrossRef]
- Hosoi, T.; Koguchi, Y.; Sugikawa, E.; Chikada, A.; Ogawa, K.; Tsuda, N.; Suto, N.; Tsunoda, S.; Taniguchi, T.; Ohnuki, T. Identification of a novel human eicosanoid receptor coupled to Gi/o. J. Biol. Chem. 2002, 277, 31459–31465. [Google Scholar] [CrossRef]
- Jones, C.E.; Holden, S.; Tenaillon, L.; Bhatia, U.; Seuwen, K.; Tranter, P.; Turner, J.; Kettle, R.; Bouhelal, R.; Charlton, S.; et al. Expression and characterization of a 5-oxo-6E,8Z,11Z,14Z-eicosatetraenoic acid receptor highly expressed on human eosinophils and neutrophils. Mol. Pharmacol. 2003, 63, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Kalyvianaki, K.; Drosou, I.; Notas, G.; Castanas, E.; Kampa, M. Enhanced OXER1 expression is indispensable for human cancer cell migration. Biochem. Biophys. Res. Commun. 2021, 584, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N. Lipoxin biosynthesis and its impact in inflammatory and vascular events. Biochim. Biophys. Acta 1994, 1212, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Rådmark, O.; Shimizu, T.; Jörnvall, H.; Samuelsson, B. Leukotriene A4 hydrolase in human leukocytes. Purification and properties. J. Biol. Chem. 1984, 259, 12339–12345. [Google Scholar] [CrossRef]
- Rudberg, P.C.; Tholander, F.; Andberg, M.; Thunnissen, M.M.; Haeggström, J.Z. Leukotriene A4 hydrolase: Identification of a common carboxylate recognition site for the epoxide hydrolase and aminopeptidase substrates. J. Biol. Chem. 2004, 279, 27376–27382. [Google Scholar] [CrossRef] [PubMed]
- Paige, M.; Wang, K.; Burdick, M.; Park, S.; Cha, J.; Jeffery, E.; Sherman, N.; Shim, Y.M. Role of leukotriene A4 hydrolase aminopeptidase in the pathogenesis of emphysema. J. Immunol. 2014, 192, 5059–5068. [Google Scholar] [CrossRef]
- Yokomizo, T.; Izumi, T.; Chang, K.; Takuwa, Y.; Shimizu, T. A G-protein-coupled receptor for leukotriene B4 that mediates chemotaxis. Nature 1997, 387, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Yokomizo, T.; Kato, K.; Terawaki, K.; Izumi, T.; Shimizu, T. A second leukotriene B4 receptor, BLT2. A new therapeutic target in inflammation and immunological disorders. J. Exp. Med. 2000, 192, 421–432. [Google Scholar] [CrossRef]
- Lam, B.K.; Penrose, J.F.; Freeman, G.J.; Austen, K.F. Expression cloning of a cDNA for human leukotriene C4 synthase, an integral membrane protein conjugating reduced glutathione to leukotriene A4. Proc. Natl. Acad. Sci. USA 1994, 91, 7663–7667. [Google Scholar] [CrossRef]
- Ago, H.; Kanaoka, Y.; Irikura, D.; Lam, B.K.; Shimamura, T.; Austen, K.F.; Miyano, M. Crystal structure of a human membrane protein involved in cysteinyl leukotriene biosynthesis. Nature 2007, 448, 609–612. [Google Scholar] [CrossRef]
- Strid, T.; Svartz, J.; Franck, N.; Hallin, E.; Ingelsson, B.; Söderström, M.; Hammarström, S. Distinct parts of leukotriene C4 synthase interact with 5-lipoxygenase and 5-lipoxygenase activating protein. Biochem. Biophys. Res. Commun. 2009, 381, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Wickham, S.; West, M.B.; Cook, P.F.; Hanigan, M.H. Gamma-glutamyl compounds: Substrate specificity of gamma-glutamyl transpeptidase enzymes. Anal. Biochem. 2011, 414, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Kubota, I.; Okamura, N.; Iwata, H.; Tsujimoto, M.; Nakazato, H.; Nishihara, T.; Noguchi, T. Purification and characterization of human microsomal dipeptidase. J. Biochem. 1989, 105, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Habib, G.M.; Shi, Z.Z.; Cuevas, A.A.; Lieberman, M.W. Identification of two additional members of the membrane-bound dipeptidase family. FASEB J. 2003, 17, 1313–1315. [Google Scholar] [CrossRef]
- Reddanna, P.; Prabhu, K.S.; Whelan, J.; Reddy, C.C. Carboxypeptidase A-catalyzed direct conversion of leukotriene C4 to leukotriene F4. Arch. Biochem. Biophys. 2003, 413, 158–163. [Google Scholar] [CrossRef]
- Bernström, K.; Hammarström, S. A novel leukotriene formed by transpeptidation of leukotriene E. Biochem. Biophys. Res. Commun. 1982, 109, 800–804. [Google Scholar] [CrossRef]
- Jedlitschky, G.; Keppler, D. Transport of leukotriene C4 and structurally related conjugates. Vitam. Horm. 2002, 64, 153–184. [Google Scholar]
- Zhang, D.W.; Nunoya, K.; Vasa, M.; Gu, H.M.; Cole, S.P.; Deeley, R.G. Mutational analysis of polar amino acid residues within predicted transmembrane helices 10 and 16 of multidrug resistance protein 1 (ABCC1): Effect on substrate specificity. Drug Metab. Dispos. 2006, 34, 539–546. [Google Scholar] [CrossRef]
- Johnson, Z.L.; Chen, J. Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1. Cell 2017, 168, 1075–1085. [Google Scholar] [CrossRef]
- Cui, Y.; König, J.; Buchholz, J.K.; Spring, H.; Leier, I.; Keppler, D. Drug resistance and ATP-dependent conjugate transport mediated by the apical multidrug resistance protein, MRP2, permanently expressed in human and canine cells. Mol. Pharmacol. 1999, 55, 929–937. [Google Scholar]
- Kamisako, T.; Leier, I.; Cui, Y.; König, J.; Buchholz, U.; Hummel-Eisenbeiss, J.; Keppler, D. Transport of monoglucuronosyl and bisglucuronosyl bilirubin by recombinant human and rat multidrug resistance protein 2. Hepatology 1999, 30, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Cui, Y.; König, J.; Risch, A.; Jäger, B.; Drings, P.; Bartsch, H.; Keppler, D.; Nies, A.T. Identification and functional characterization of the natural variant MRP3-Arg1297His of human multidrug resistance protein 3 (MRP3/ABCC3). Pharmacogenetics 2004, 14, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Rius, M.; Hummel-Eisenbeiss, J.; Keppler, D. ATP-dependent transport of leukotrienes B4 and C4 by the multidrug resistance protein ABCC4 (MRP4). J. Pharmacol. Exp. Ther. 2008, 324, 86–94. [Google Scholar] [CrossRef]
- Belinsky, M.G.; Chen, Z.S.; Shchaveleva, I.; Zeng, H.; Kruh, G.D. Characterization of the drug resistance and transport properties of multidrug resistance protein 6 (MRP6, ABCC6). Cancer Res. 2002, 62, 6172–6177. [Google Scholar] [PubMed]
- Chen, Z.S.; Hopper-Borge, E.; Belinsky, M.G.; Shchaveleva, I.; Kotova, E.; Kruh, G.D. Characterization of the transport properties of human multidrug resistance protein 7 (MRP7, ABCC10). Mol. Pharmacol. 2003, 63, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.S.; Guo, Y.; Belinsky, M.G.; Kotova, E.; Kruh, G.D. Transport of bile acids, sulfated steroids, estradiol 17-beta-D-glucuronide, and leukotriene C4 by human multidrug resistance protein 8 (ABCC11). Mol. Pharmacol. 2005, 67, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lee, T.K.; Meier, P.J.; Ballatori, N. Identification of glutathione as a driving force and leukotriene C4 as a substrate for oatp1, the hepatic sinusoidal organic solute transporter. J. Biol. Chem. 1998, 273, 16184–16191. [Google Scholar] [CrossRef]
- Yokomizo, T.; Kato, K.; Hagiya, H.; Izumi, T.; Shimizu, T. Hydroxyeicosanoids bind to and activate the low affinity leukotriene B4 receptor, BLT2. J. Biol. Chem. 2001, 276, 12454–12459. [Google Scholar] [CrossRef]
- Lynch, K.R.; O’Neill, G.P.; Liu, Q.; Im, D.S.; Sawyer, N.; Metters, K.M.; Coulombe, N.; Abramovitz, M.; Figueroa, D.J.; Zeng, Z.; et al. Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature 1999, 399, 789–793. [Google Scholar] [CrossRef]
- Heise, C.E.; O’Dowd, B.F.; Figueroa, D.J.; Sawyer, N.; Nguyen, T.; Im, D.S.; Stocco, R.; Bellefeuille, J.N.; Abramovitz, M.; Cheng, R.; et al. Characterization of the human cysteinyl leukotriene 2 receptor. J. Biol. Chem. 2000, 275, 30531–30536. [Google Scholar] [CrossRef]
- Nothacker, H.P.; Wang, Z.; Zhu, Y.; Reinscheid, R.K.; Lin, S.H.; Civelli, O. Molecular cloning and characterization of a second human cysteinyl leukotriene receptor: Discovery of a subtype selective agonist. Mol. Pharmacol. 2000, 58, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Kanaoka, Y.; Maekawa, A.; Austen, K.F. Identification of GPR99 protein as a potential third cysteinyl leukotriene receptor with a preference for leukotriene E4 ligand. J. Biol. Chem. 2013, 288, 10967–10972. [Google Scholar] [CrossRef] [PubMed]
- Ciana, P.; Fumagalli, M.; Trincavelli, M.L.; Verderio, C.; Rosa, P.; Lecca, D.; Ferrario, S.; Parravicini, C.; Capra, V.; Gelosa, P.; et al. The orphan receptor GPR17 identified as a new dual uracil nucleotides/cysteinyl-leukotrienes receptor. EMBO J. 2006, 25, 4615–4627. [Google Scholar] [CrossRef] [PubMed]
- Qi, A.D.; Harden, T.K.; Nicholas, R.A. Is GPR17 a P2Y/leukotriene receptor? examination of uracil nucleotides, nucleotide sugars, and cysteinyl leukotrienes as agonists of GPR17. J. Pharmacol. Exp. Ther. 2013, 347, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Simon, K.; Merten, N.; Schröder, R.; Hennen, S.; Preis, P.; Schmitt, N.K.; Peters, L.; Schrage, R.; Vermeiren, C.; Gillard, M.; et al. The Orphan Receptor GPR17 Is Unresponsive to Uracil Nucleotides and Cysteinyl Leukotrienes. Mol. Pharmacol. 2017, 91, 518–532. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, A.; Balestrieri, B.; Austen, K.F.; Kanaoka, Y. GPR17 is a negative regulator of the cysteinyl leukotriene 1 receptor response to leukotriene D4. Proc. Natl. Acad. Sci. USA 2009, 106, 11685–11690. [Google Scholar] [CrossRef]
- Wheelan, P.; Zirrolli, J.A.; Morelli, J.G.; Murphy, R.C. Metabolism of leukotriene B4 by cultured human keratinocytes. Formation of glutathione conjugates and dihydro metabolites. J. Biol. Chem. 1993, 268, 25439–25448. [Google Scholar] [CrossRef]
- Yokomizo, T.; Ogawa, Y.; Uozumi, N.; Kume, K.; Izumi, T.; Shimizu, T. cDNA cloning, expression, and mutagenesis study of leukotriene B4 12-hydroxydehydrogenase. J. Biol. Chem. 1996, 271, 2844–2850. [Google Scholar] [CrossRef]
- Tobin, D.M.; Roca, F.J.; Ray, J.P.; Ko, D.C.; Ramakrishnan, L. An enzyme that inactivates the inflammatory mediator leukotriene B4 restricts mycobacterial infection. PLoS ONE 2013, 8, e67828. [Google Scholar] [CrossRef]
- Wainwright, S.L.; Powell, W.S. Mechanism for the formation of dihydro metabolites of 12-hydroxyeicosanoids. Conversion of leukotriene B4 and 12-hydroxy-5,8,10,14-eicosatetraenoic acid to 12-oxo intermediates. J. Biol. Chem. 1991, 266, 20899–20906. [Google Scholar] [CrossRef]
- Hagmann, W.; Korte, M. Hepatic uptake and metabolic disposition of leukotriene B4 in rats. Biochem. J. 1990, 267, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Berry, K.A.; Borgeat, P.; Gosselin, J.; Flamand, L.; Murphy, R.C. Urinary metabolites of leukotriene B4 in the human subject. J. Biol. Chem. 2003, 278, 24449–24460. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, S.; Falck, J.R.; Yadagiri, P.; Powell, W.S. Metabolism of 12(S)-hydroxy-5,8,10,14-eicosatetraenoic acid and other hydroxylated fatty acids by the reductase pathway in porcine polymorphonuclear leukocytes. Biochemistry 1990, 29, 10126–10135. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Müller, J.; Leier, I.; Jedlitschky, G.; Ball, H.A.; Moore, K.P.; Taylor, G.W.; Williams, R.; Keppler, D. Metabolism of cysteinyl leukotrienes in monkey and man. Eur. J. Biochem. 1990, 194, 309–315. [Google Scholar] [CrossRef]
- Chang, W.C.; Ning, C.C.; Lin, M.T.; Huang, J.D. Epidermal growth factor enhances a microsomal 12-lipoxygenase activity in A431 cells. J. Biol. Chem. 1992, 267, 3657–3666. [Google Scholar] [CrossRef]
- Wecksler, A.T.; Kenyon, V.; Deschamps, J.D.; Holman, T.R. Substrate specificity changes for human reticulocyte and epithelial 15-lipoxygenases reveal allosteric product regulation. Biochemistry 2008, 47, 7364–7375. [Google Scholar] [CrossRef]
- Ikei, K.N.; Yeung, J.; Apopa, P.L.; Ceja, J.; Vesci, J.; Holman, T.R.; Holinstat, M. Investigations of human platelet-type 12-lipoxygenase: Role of lipoxygenase products in platelet activation. J. Lipid Res. 2012, 53, 2546–2559. [Google Scholar] [CrossRef]
- Yeung, J.; Tourdot, B.E.; Adili, R.; Green, A.R.; Freedman, C.J.; Fernandez-Perez, P.; Yu, J.; Holman, T.R.; Holinstat, M. 12(S)-HETrE, a 12-Lipoxygenase Oxylipin of Dihomo-γ-Linolenic Acid, Inhibits Thrombosis via Gαs Signaling in Platelets. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 2068–2077. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, W.; Giroux, C.; Cai, Y.; Ekambaram, P.; Dilly, A.K.; Hsu, A.; Zhou, S.; Maddipati, K.R.; Liu, J.; et al. Identification of the orphan G protein-coupled receptor GPR31 as a receptor for 12-(S)-hydroxyeicosatetraenoic acid. J. Biol. Chem. 2011, 286, 33832–33840. [Google Scholar] [CrossRef]
- Sun, L.; Xu, Y.W.; Han, J.; Liang, H.; Wang, N.; Cheng, Y. 12/15-Lipoxygenase metabolites of arachidonic acid activate PPARγ: A possible neuroprotective effect in ischemic brain. J. Lipid Res. 2015, 56, 502–514. [Google Scholar] [CrossRef]
- Falgueyret, J.P.; Leblanc, Y.; Riendeau, D. Stereoselective carbonyl reductases from rat skin and leukocyte microsomes converting 12-ketoeicosatetraenoic acid to 12(S)-HETE. FEBS Lett. 1990, 262, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Pace-Asciak, C.R. Hemoglobin- and hemin-catalyzed transformation of 12L-hydroperoxy-5,8,10,14-eicosatetraenoic acid. Biochim. Biophys. Acta 1984, 793, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Reynaud, D.; Demin, P.; Pace-Asciak, C.R. Hepoxilin A3 formation in the rat pineal gland selectively utilizes (12S)-hydroperoxyeicosatetraenoic acid (HPETE), but not (12R)-HPETE. J. Biol. Chem. 1994, 269, 23976–23980. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.; Patabhiraman, S.; Ciccoli, R.; Ishdorj, G.; Schwarz, K.; Petrucev, B.; Kühn, H.; Haeggström, J.Z. The rat leukocyte-type 12-lipoxygenase exhibits an intrinsic hepoxilin A3 synthase activity. J. Biol. Chem. 2004, 279, 29023–29030. [Google Scholar] [CrossRef] [PubMed]
- Krieg, P.; Rosenberger, S.; de Juanes, S.; Latzko, S.; Hou, J.; Dick, A.; Kloz, U.; van der Hoeven, F.; Hausser, I.; Esposito, I.; et al. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J. Investig. Dermatol. 2013, 133, 172–180. [Google Scholar] [CrossRef]
- Pace-Asciak, C.R.; Lee, W.S. Purification of hepoxilin epoxide hydrolase from rat liver. J. Biol. Chem. 1989, 264, 9310–9313. [Google Scholar] [CrossRef]
- Laneuville, O.; Chang, M.; Reddy, C.C.; Corey, E.J.; Pace-Asciak, C.R. Isozyme specificity in the conversion of hepoxilin A3 (HxA3) into a glutathionyl hepoxilin (HxA3-C) by the Yb2 subunit of rat liver glutathione S-transferase. J. Biol. Chem. 1990, 265, 21415–21418. [Google Scholar] [CrossRef]
- Pace-Asciak, C.R.; Laneuville, O.; Chang, M.; Reddy, C.C.; Su, W.G.; Corey, E.J. New products in the hepoxilin pathway: Isolation of 11-glutathionyl hepoxilin A3 through reaction of hepoxilin A3 with glutathione S-transferase. Biochem. Biophys. Res. Commun. 1989, 163, 1230–1234. [Google Scholar] [CrossRef]
- Pace-Asciak, C.R.; Laneuville, O.; Su, W.G.; Corey, E.J.; Gurevich, N.; Wu, P.; Carlen, P.L. A glutathione conjugate of hepoxilin A3: Formation and action in the rat central nervous system. Proc. Natl. Acad. Sci. USA 1990, 87, 3037–3041. [Google Scholar] [CrossRef]
- Cronin, A.; Decker, M.; Arand, M. Mammalian soluble epoxide hydrolase is identical to liver hepoxilin hydrolase. J. Lipid Res. 2011, 52, 712–719. [Google Scholar] [CrossRef]
- Gregus, A.M.; Doolen, S.; Dumlao, D.S.; Buczynski, M.W.; Takasusuki, T.; Fitzsimmons, B.L.; Hua, X.Y.; Taylor, B.K.; Dennis, E.A.; Yaksh, T.L. Spinal 12-lipoxygenase-derived hepoxilin A3 contributes to inflammatory hyperalgesia via activation of TRPV1 and TRPA1 receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 6721–6726. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Rao, G.N. Emerging role of 12/15-Lipoxygenase (ALOX15) in human pathologies. Prog. Lipid Res. 2019, 73, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Siangjong, L.; Goldman, D.H.; Kriska, T.; Gauthier, K.M.; Smyth, E.M.; Puli, N.; Kumar, G.; Falck, J.R.; Campbell, W.B. Vascular hepoxilin and trioxilins mediate vasorelaxation through TP receptor inhibition in mouse arteries. Acta Physiol. 2017, 219, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Boeglin, W.E.; Kim, R.B.; Brash, A.R. A 12R-lipoxygenase in human skin: Mechanistic evidence, molecular cloning, and expression. Proc. Natl. Acad. Sci. USA 1998, 95, 6744–6749. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, Y.; Lu, J.; Zhao, P.; Zhang, X.; Tan, L.; Li, J.; Xiao, C.; Zeng, L.; He, X. Identification of the first congenital ichthyosis case caused by a homozygous deletion in the ALOX12B gene due to chromosome 17 mixed uniparental disomy. Front. Genet. 2022, 13, 931833. [Google Scholar] [CrossRef]
- Bryant, R.W.; Bailey, J.M.; Schewe, T.; Rapoport, S.M. Positional specificity of a reticulocyte lipoxygenase. Conversion of arachidonic acid to 15-S-hydroperoxy-eicosatetraenoic acid. J. Biol. Chem. 1982, 257, 6050–6055. [Google Scholar] [CrossRef]
- Sigal, E.; Dicharry, S.; Highland, E.; Finkbeiner, W.E. Cloning of human airway 15-lipoxygenase: Identity to the reticulocyte enzyme and expression in epithelium. Am. J. Physiol. 1992, 262, L392–L398. [Google Scholar] [CrossRef]
- Brash, A.R.; Boeglin, W.E.; Chang, M.S. Discovery of a second 15S-lipoxygenase in humans. Proc. Natl. Acad. Sci. USA 1997, 94, 6148–6152. [Google Scholar] [CrossRef]
- Altmann, R.; Hausmann, M.; Spöttl, T.; Gruber, M.; Bull, A.W.; Menzel, K.; Vogl, D.; Herfarth, H.; Schölmerich, J.; Falk, W.; et al. 13-Oxo-ODE is an endogenous ligand for PPARgamma in human colonic epithelial cells. Biochem. Pharmacol. 2007, 74, 612–622. [Google Scholar] [CrossRef]
- Umeno, A.; Sakashita, M.; Sugino, S.; Murotomi, K.; Okuzawa, T.; Morita, N.; Tomii, K.; Tsuchiya, Y.; Yamasaki, K.; Horie, M.; et al. Comprehensive analysis of PPARγ agonist activities of stereo-, regio-, and enantio-isomers of hydroxyoctadecadienoic acids. Biosci. Rep. 2020, 40, BSR20193767. [Google Scholar] [CrossRef]
- Brunnström, A.; Hamberg, M.; Griffiths, W.J.; Mannervik, B.; Claesson, H.E. Biosynthesis of 14,15-hepoxilins in human l1236 Hodgkin lymphoma cells and eosinophils. Lipids 2011, 46, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Feltenmark, S.; Gautam, N.; Brunnström, A.; Griffiths, W.; Backman, L.; Edenius, C.; Lindbom, L.; Björkholm, M.; Claesson, H.E. Eoxins are proinflammatory arachidonic acid metabolites produced via the 15-lipoxygenase-1 pathway in human eosinophils and mast cells. Proc. Natl. Acad. Sci. USA 2008, 105, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Green, A.R.; Freedman, C.; Tena, J.; Tourdot, B.E.; Liu, B.; Holinstat, M.; Holman, T.R. 5S,15S-Dihydroperoxyeicosatetraenoic Acid (5,15-diHpETE) as a Lipoxin Intermediate: Reactivity and Kinetics with Human Leukocyte 5-Lipoxygenase, Platelet 12-Lipoxygenase, and Reticulocyte 15-Lipoxygenase-1. Biochemistry 2018, 57, 6726–6734. [Google Scholar] [CrossRef] [PubMed]
- Steinhilber, D.; Roth, H.J. New series of lipoxins isolated from human eosinophils. FEBS Lett. 1989, 255, 143–148. [Google Scholar] [CrossRef]
- Tornhamre, S.; Elmqvist, A.; Lindgren, J.A. 15-Lipoxygenation of leukotriene A4. Studies Of 12- and 15-lipoxygenase efficiency to catalyze lipoxin formation. Biochim. Biophys. Acta 2000, 1484, 298–306. [Google Scholar] [CrossRef]
- Fiore, S.; Maddox, J.F.; Perez, H.D.; Serhan, C.N. Identification of a human cDNA encoding a functional high affinity lipoxin A4 receptor. J. Exp. Med. 1994, 180, 253–260. [Google Scholar] [CrossRef]
- Romano, M.; Recchia, I.; Recchiuti, A. Lipoxin receptors. Sci. World J. 2007, 7, 1393–1412. [Google Scholar] [CrossRef]
- Schaldach, C.M.; Riby, J.; Bjeldanes, L.F. Lipoxin A4: A new class of ligand for the Ah receptor. Biochemistry 1999, 38, 7594–7600. [Google Scholar] [CrossRef]
- Russell, R.; Gori, I.; Pellegrini, C.; Kumar, R.; Achtari, C.; Canny, G.O. Lipoxin A4 is a novel estrogen receptor modulator. FASEB J. 2011, 25, 4326–4337. [Google Scholar] [CrossRef]
- Boado, R.J.; Pardridge, W.M.; Vinters, H.V.; Black, K.L. Differential expression of arachidonate 5-lipoxygenase transcripts in human brain tumors: Evidence for the expression of a multitranscript family. Proc. Natl. Acad. Sci. USA 1992, 89, 9044–9048. [Google Scholar] [CrossRef]
- Golubic, M.; Prayson, R.A.; Vargo, L.; Bondar, J.; Barnett, G.H. Increased expression of 5-lipoxygenase in glioblastoma multiforme. Adv. Exp. Med. Biol. 2003, 525, 205–208. [Google Scholar] [PubMed]
- Nathoo, N.; Prayson, R.A.; Bondar, J.; Vargo, L.; Arrigain, S.; Mascha, E.J.; Suh, J.H.; Barnett, G.H.; Golubic, M. Increased expression of 5-lipoxygenase in high-grade astrocytomas. Neurosurgery 2006, 58, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yu, S.C.; Jiang, J.Y.; Porter, G.W.; Zhao, L.T.; Wang, Z.; Tan, H.; Cui, Y.H.; Qian, C.; Ping, Y.F.; et al. An inhibitor of arachidonate 5-lipoxygenase, Nordy, induces differentiation and inhibits self-renewal of glioma stem-like cells. Stem. Cell Rev. Rep. 2011, 7, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Simmet, T.; Luck, W.; Winking, M.; Delank, W.K.; Peskar, B.A. Identification and characterization of cysteinyl-leukotriene formation in tissue slices from human intracranial tumors: Evidence for their biosynthesis under in vivo conditions. J. Neurochem. 1990, 54, 2091–2099. [Google Scholar] [CrossRef] [PubMed]
- Black, K.L.; Hoff, J.T.; McGillicuddy, J.E.; Gebarski, S.S. Increased leukotriene C4 and vasogenic edema surrounding brain tumors in humans. Ann. Neurol. 1986, 19, 592–595. [Google Scholar] [CrossRef]
- Jin, T.B.; Li, X.L.; Yang, H.; Jiri, M.; Shi, X.G.; Yuan, D.Y.; Kang, L.L.; Li, S.Q. Association of polymorphisms in FLT3, EGFR, ALOX5, and NEIL3 with glioblastoma in the Han Chinese population. Med. Oncol. 2013, 30, 718. [Google Scholar] [CrossRef]
- Kim, J.A.; Chung, Y.J.; Lee, Y.S. Intracellular Ca2+ mediates lipoxygenase-induced proliferation of U-373 MG human astrocytoma cells. Arch. Pharm. Res. 1998, 21, 664–670. [Google Scholar] [CrossRef]
- Lim, J.Y.; Oh, J.H.; Jung, J.R.; Kim, S.M.; Ryu, C.H.; Kim, H.T.; Jeun, S.S. MK886-induced apoptosis depends on the 5-LO expression level in human malignant glioma cells. J. Neurooncol. 2010, 97, 339–346. [Google Scholar] [CrossRef]
- Souza, F.D.C.; Ferreira, M.T.; Colquhoun, A. Influence of Lipoxygenase Inhibition on Glioblastoma Cell Biology. Int. J. Mol. Sci. 2020, 21, 8395. [Google Scholar] [CrossRef]
- Xu, J.P.; Liu, H.; Bian, X.W.; Chen, J.H.; Zhou, X.D.; Wu, Y.Z. Effect of nordy on biological behaviors of malignant glioma cell line U87MG and the analysis of differential expression proteome. Zhonghua Bing Li Xue Za Zhi 2007, 36, 609–613. [Google Scholar]
- Ishii, K.; Zaitsu, M.; Yonemitsu, N.; Kan, Y.; Hamasaki, Y.; Matsuo, M. 5-lipoxygenase pathway promotes cell proliferation in human glioma cell lines. Clin. Neuropathol. 2009, 28, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Gáti, I.; Bergström, M.; Csóka, K.; Muhr, C.; Carlsson, J. Effects of the 5-lipoxygenase inhibitors AA-863 and U-60,257 on human glioma cell lines. Prostaglandins Leukot. Essent. Fat. Acids 1990, 40, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Piromkraipak, P.; Parakaw, T.; Phuagkhaopong, S.; Srihirun, S.; Chongthammakun, S.; Chaithirayanon, K.; Vivithanaporn, P. Cysteinyl leukotriene receptor antagonists induce apoptosis and inhibit proliferation of human glioblastoma cells by downregulating B-cell lymphoma 2 and inducing cell cycle arrest. Can. J. Physiol. Pharmacol. 2018, 96, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J. Blood-brain barrier structure and function and the challenges for CNS drug delivery. J. Inherit. Metab. Dis. 2013, 36, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Chio, C.C.; Baba, T.; Black, K.L. Selective blood-tumor barrier disruption by leukotrienes. J. Neurosurg. 1992, 77, 407–410. [Google Scholar] [CrossRef]
- Black, K.L.; King, W.A.; Ikezaki, K. Selective opening of the blood-tumor barrier by intracarotid infusion of leukotriene C4. J. Neurosurg. 1990, 72, 912–916. [Google Scholar] [CrossRef]
- Black, P.; Hand, C.M.; Vender, J.R.; Finkelstein, S.D. Chemotherapy in experimental brain tumor, part 2: Pretreatment with leukotriene C4 prolongs survival. J. Neurooncol. 1998, 36, 7–19. [Google Scholar] [CrossRef]
- Fellner, S.; Bauer, B.; Miller, D.S.; Schaffrik, M.; Fankhänel, M.; Spruss, T.; Bernhardt, G.; Graeff, C.; Färber, L.; Gschaidmeier, H.; et al. Transport of paclitaxel (Taxol) across the blood-brain barrier in vitro and in vivo. J. Clin. Investig. 2002, 110, 1309–1318. [Google Scholar] [CrossRef]
- Doan, P.; Nguyen, P.; Murugesan, A.; Subramanian, K.; Konda Mani, S.; Kalimuthu, V.; Abraham, B.G.; Stringer, B.W.; Balamuthu, K.; Yli-Harja, O.; et al. Targeting Orphan G Protein-Coupled Receptor 17 with T0 Ligand Impairs Glioblastoma Growth. Cancers 2021, 13, 3773. [Google Scholar] [CrossRef]
- Nguyen, P.; Doan, P.; Rimpilainen, T.; Konda Mani, S.; Murugesan, A.; Yli-Harja, O.; Candeias, N.R.; Kandhavelu, M. Synthesis and Preclinical Validation of Novel Indole Derivatives as a GPR17 Agonist for Glioblastoma Treatment. J. Med. Chem. 2021, 64, 10908–10918. [Google Scholar] [CrossRef]
- Hu, Y.; Luo, H.; Zhu, X.; Guo, H. CRNDE/ETS1/GPR17 Facilitates the Proliferation, Migration, and Invasion of Glioma. Comput. Math. Methods Med. 2021, 2021, 7566365. [Google Scholar] [CrossRef] [PubMed]
- Terpinskaya, T.I.; Osipov, A.V.; Kryukova, E.V.; Kudryavtsev, D.S.; Kopylova, N.V.; Yanchanka, T.L.; Palukoshka, A.F.; Gondarenko, E.A.; Zhmak, M.N.; Tsetlin, V.I.; et al. α-Conotoxins and α-Cobratoxin Promote, while Lipoxygenase and Cyclooxygenase Inhibitors Suppress the Proliferation of Glioma C6 Cells. Mar. Drugs 2021, 19, 118. [Google Scholar] [CrossRef]
- Ding, X.Z.; Tong, W.G.; Adrian, T.E. 12-lipoxygenase metabolite 12(S)-HETE stimulates human pancreatic cancer cell proliferation via protein tyrosine phosphorylation and ERK activation. Int. J. Cancer 2001, 94, 630–636. [Google Scholar] [CrossRef]
- Guo, A.M.; Liu, X.; Al-Wahab, Z.; Maddippati, K.R.; Ali-Fehmi, R.; Scicli, A.G.; Munkarah, A.R. Role of 12-lipoxygenase in regulation of ovarian cancer cell proliferation and survival. Cancer Chemother. Pharmacol. 2011, 68, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y.; Yoshimoto, T. Arachidonic acid converts the glutathione depletion-induced apoptosis to necrosis by promoting lipid peroxidation and reducing caspase-3 activity in rat glioma cells. Arch. Biochem. Biophys. 2002, 400, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Strakova, N.; Ehrmann, J.; Dzubak, P.; Bouchal, J.; Kolar, Z. The synthetic ligand of peroxisome proliferator-activated receptor-gamma ciglitazone affects human glioblastoma cell lines. J. Pharmacol. Exp. Ther. 2004, 309, 1239–1247. [Google Scholar] [CrossRef]
- Grommes, C.; Landreth, G.E.; Sastre, M.; Beck, M.; Feinstein, D.L.; Jacobs, A.H.; Schlegel, U.; Heneka, M.T. Inhibition of in vivo glioma growth and invasion by peroxisome proliferator-activated receptor gamma agonist treatment. Mol. Pharmacol. 2006, 70, 1524–1533. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, L.; Liu, Y.; Li, P.; Jiang, G.; Gao, X.; Zhang, Y.; Jiang, C.; Zhu, W.; Han, H.; et al. Activation of PPARγ mediates icaritin-induced cell cycle arrest and apoptosis in glioblastoma multiforme. Biomed. Pharmacother. 2018, 100, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Viita, H.; Pacholska, A.; Ahmad, F.; Tietäväinen, J.; Naarala, J.; Hyvärinen, A.; Wirth, T.; Ylä-Herttuala, S. 15-Lipoxygenase-1 induces lipid peroxidation and apoptosis, and improves survival in rat malignant glioma. In Vivo 2012, 26, 1–8. [Google Scholar]
- Yuan, H.; Li, M.Y.; Ma, L.T.; Hsin, M.K.; Mok, T.S.; Underwood, M.J.; Chen, G.G. 15-Lipoxygenases and its metabolites 15(S)-HETE and 13(S)-HODE in the development of non-small cell lung cancer. Thorax 2010, 65, 321–326. [Google Scholar] [CrossRef]
- Liu, L.; Li, X.; Shi, J.; Li, L.; Wang, J.; Luo, Z.Z. Effects of FPR2 gene silencing on the proliferation, migration and invasion of human glioma U87 cells. Zhonghua Zhong Liu Za Zhi 2018, 40, 659–666. [Google Scholar] [PubMed]
- He, H.Q.; Ye, R.D. The Formyl Peptide Receptors: Diversity of Ligands and Mechanism for Recognition. Molecules 2017, 22, 455. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L.; Edson, K.Z.; Totah, R.A.; Rettie, A.E. Cytochrome P450 ω-Hydroxylases in Inflammation and Cancer. Adv. Pharmacol. 2015, 74, 223–262. [Google Scholar]
- Rifkind, A.B.; Lee, C.; Chang, T.K.; Waxman, D.J. Arachidonic acid metabolism by human cytochrome P450s 2C8, 2C9, 2E1, and 1A2: Regioselective oxygenation and evidence for a role for CYP2C enzymes in arachidonic acid epoxygenation in human liver microsomes. Arch. Biochem. Biophys. 1995, 320, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.; Jansson, I.; Stoilov, I.; Sarfarazi, M.; Schenkman, J.B. Metabolism of retinoids and arachidonic acid by human and mouse cytochrome P450 1b1. Drug Metab. Dispos. 2004, 32, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.S.; Helvig, C.; Taimi, M.; Ramshaw, H.A.; Collop, A.H.; Amad, M.; White, J.A.; Petkovich, M.; Jones, G.; Korczak, B. CYP2U1, a novel human thymus- and brain-specific cytochrome P450, catalyzes omega- and (omega-1)-hydroxylation of fatty acids. J. Biol. Chem. 2004, 279, 6305–6314. [Google Scholar] [CrossRef]
- Lasker, J.M.; Chen, W.B.; Wolf, I.; Bloswick, B.P.; Wilson, P.D.; Powell, P.K. Formation of 20-hydroxyeicosatetraenoic acid, a vasoactive and natriuretic eicosanoid, in human kidney. Role of Cyp4F2 and Cyp4A11. J. Biol. Chem. 2000, 275, 4118–4126. [Google Scholar] [CrossRef]
- Gainer, J.V.; Bellamine, A.; Dawson, E.P.; Womble, K.E.; Grant, S.W.; Wang, Y.; Cupples, L.A.; Guo, C.Y.; Demissie, S.; O’Donnell, C.J.; et al. Functional variant of CYP4A11 20-hydroxyeicosatetraenoic acid synthase is associated with essential hypertension. Circulation 2005, 111, 63–69. [Google Scholar] [CrossRef]
- Fer, M.; Corcos, L.; Dréano, Y.; Plée-Gautier, E.; Salaün, J.P.; Berthou, F.; Amet, Y. Cytochromes P450 from family 4 are the main omega hydroxylating enzymes in humans: CYP4F3B is the prominent player in PUFA metabolism. J. Lipid Res. 2008, 49, 2379–2389. [Google Scholar] [CrossRef]
- Bylund, J.; Ericsson, J.; Oliw, E.H. Analysis of cytochrome P450 metabolites of arachidonic and linoleic acids by liquid chromatography-mass spectrometry with ion trap MS. Anal. Biochem. 1998, 265, 55–68. [Google Scholar] [CrossRef]
- Brash, A.R.; Boeglin, W.E.; Capdevila, J.H.; Yeola, S.; Blair, I.A. 7-HETE, 10-HETE, and 13-HETE are major products of NADPH-dependent arachidonic acid metabolism in rat liver microsomes: Analysis of their stereochemistry, and the stereochemistry of their acid-catalyzed rearrangement. Arch. Biochem. Biophys. 1995, 321, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.A.; Balazy, M.; Margiotta, P.; Huang, D.D.; Falck, J.R.; McGiff, J.C. Cytochrome P-450-dependent HETEs: Profile of biological activity and stimulation by vasoactive peptides. Am. J. Physiol. 1996, 271, R863–R869. [Google Scholar] [CrossRef] [PubMed]
- Bylund, J.; Kunz, T.; Valmsen, K.; Oliw, E.H. Cytochromes P450 with bisallylic hydroxylation activity on arachidonic and linoleic acids studied with human recombinant enzymes and with human and rat liver microsomes. J. Pharmacol. Exp. Ther. 1998, 284, 51–60. [Google Scholar]
- Bednar, M.M.; Gross, C.E.; Russell, S.R.; Fuller, S.P.; Ahern, T.P.; Howard, D.B.; Falck, J.R.; Reddy, K.M.; Balazy, M. 16(R)-hydroxyeicosatetraenoic acid, a novel cytochrome P450 product of arachidonic acid, suppresses activation of human polymorphonuclear leukocyte and reduces intracranial pressure in a rabbit model of thromboembolic stroke. Neurosurgery 2000, 47, 1410–1419. [Google Scholar] [CrossRef]
- Garcia, V.; Gilani, A.; Shkolnik, B.; Pandey, V.; Zhang, F.F.; Dakarapu, R.; Gandham, S.K.; Reddy, N.R.; Graves, J.P.; Gruzdev, A.; et al. 20-HETE Signals Through G-Protein-Coupled Receptor GPR75 (Gq) to Affect Vascular Function and Trigger Hypertension. Circ. Res. 2017, 120, 1776–1788. [Google Scholar] [CrossRef]
- Wen, H.; Östman, J.; Bubb, K.J.; Panayiotou, C.; Priestley, J.V.; Baker, M.D.; Ahluwalia, A. 20-Hydroxyeicosatetraenoic acid (20-HETE) is a novel activator of transient receptor potential vanilloid 1 (TRPV1) channel. J. Biol. Chem. 2012, 287, 13868–13876. [Google Scholar] [CrossRef]
- Tunaru, S.; Bonnavion, R.; Brandenburger, I.; Preussner, J.; Thomas, D.; Scholich, K.; Offermanns, S. 20-HETE promotes glucose-stimulated insulin secretion in an autocrine manner through FFAR1. Nat. Commun. 2018, 9, 177. [Google Scholar] [CrossRef]
- Ng, V.Y.; Huang, Y.; Reddy, L.M.; Falck, J.R.; Lin, E.T.; Kroetz, D.L. Cytochrome P450 eicosanoids are activators of peroxisome proliferator-activated receptor alpha. Drug Metab. Dispos. 2007, 35, 1126–1134. [Google Scholar] [CrossRef]
- Dai, D.; Zeldin, D.C.; Blaisdell, J.A.; Chanas, B.; Coulter, S.J.; Ghanayem, B.I.; Goldstein, J.A. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics 2001, 11, 597–607. [Google Scholar] [CrossRef]
- Kato, Y.; Mukai, Y.; Rane, A.; Inotsume, N.; Toda, T. The Inhibitory Effect of Telmisartan on the Metabolism of Arachidonic Acid by CYP2C9 and CYP2C8: An in Vitro Study. Biol. Pharm. Bull. 2017, 40, 1409–1415. [Google Scholar] [CrossRef]
- Wu, S.; Moomaw, C.R.; Tomer, K.B.; Falck, J.R.; Zeldin, D.C. Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J. Biol. Chem. 1996, 271, 3460–3468. [Google Scholar] [CrossRef] [PubMed]
- Stark, K.; Dostalek, M.; Guengerich, F.P. Expression and purification of orphan cytochrome P450 4X1 and oxidation of anandamide. FEBS J. 2008, 275, 3706–3717. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Herrnreiter, A.; Pfister, S.L.; Gauthier, K.M.; Falck, B.A.; Falck, J.R.; Campbell, W.B. GPR40 is a low-affinity epoxyeicosatrienoic acid receptor in vascular cells. J. Biol. Chem. 2018, 293, 10675–10691. [Google Scholar] [CrossRef]
- Yang, C.; Kwan, Y.W.; Au, A.L.; Poon, C.C.; Zhang, Q.; Chan, S.W.; Lee, S.M.; Leung, G.P. 14,15-Epoxyeicosatrienoic acid induces vasorelaxation through the prostaglandin EP2 receptors in rat mesenteric artery. Prostaglandins Other Lipid Mediat. 2010, 93, 44–51. [Google Scholar] [CrossRef]
- Liu, X.; Qian, Z.Y.; Xie, F.; Fan, W.; Nelson, J.W.; Xiao, X.; Kaul, S.; Barnes, A.P.; Alkayed, N.J. Functional screening for G protein-coupled receptor targets of 14,15-epoxyeicosatrienoic acid. Prostaglandins Other Lipid Mediat. 2017, 132, 31–40. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Schmelzer, K.; Lee, T.S.; Fang, X.; Zhu, Y.; Spector, A.A.; Gill, S.; Morisseau, C.; Hammock, B.D.; et al. The antiinflammatory effect of laminar flow: The role of PPARgamma, epoxyeicosatrienoic acids, and soluble epoxide hydrolase. Proc. Natl. Acad. Sci. USA 2005, 102, 16747–16752. [Google Scholar] [CrossRef]
- Spector, A.A.; Norris, A.W. Action of epoxyeicosatrienoic acids on cellular function. Am. J. Physiol. Cell Physiol. 2007, 292, C996–C1012. [Google Scholar] [CrossRef] [PubMed]
- Karara, A.; Dishman, E.; Falck, J.R.; Capdevila, J.H. Endogenous epoxyeicosatrienoyl-phospholipids. A novel class of cellular glycerolipids containing epoxidized arachidonate moieties. J. Biol. Chem. 1991, 266, 7561–7569. [Google Scholar] [CrossRef]
- Somani, S.T.; Zeigler, M.; Fay, E.E.; Leahy, M.; Bermudez, B.; Totah, R.A.; Hebert, M.F. Changes in erythrocyte membrane epoxyeicosatrienoic, dihydroxyeicosatrienoic, and hydroxyeicosatetraenoic acids during pregnancy. Life Sci. 2021, 264, 118590. [Google Scholar] [CrossRef]
- Edin, M.L.; Hamedani, B.G.; Gruzdev, A.; Graves, J.P.; Lih, F.B.; Arbes, S.J., 3rd; Singh, R.; Orjuela Leon, A.C.; Bradbury, J.A.; DeGraff, L.M.; et al. Epoxide hydrolase 1 (EPHX1) hydrolyzes epoxyeicosanoids and impairs cardiac recovery after ischemia. J. Biol. Chem. 2018, 293, 3281–3292. [Google Scholar] [CrossRef]
- Fang, X.; Hu, S.; Xu, B.; Snyder, G.D.; Harmon, S.; Yao, J.; Liu, Y.; Sangras, B.; Falck, J.R.; Weintraub, N.L.; et al. 14,15-Dihydroxyeicosatrienoic acid activates peroxisome proliferator-activated receptor-alpha. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H55–H63. [Google Scholar] [CrossRef]
- Le Quéré, V.; Plée-Gautier, E.; Potin, P.; Madec, S.; Salaün, J.P. Human CYP4F3s are the main catalysts in the oxidation of fatty acid epoxides. J. Lipid Res. 2004, 45, 1446–1458. [Google Scholar] [CrossRef] [PubMed]
- Moreland, K.T.; Procknow, J.D.; Sprague, R.S.; Iverson, J.L.; Lonigro, A.J.; Stephenson, A.H. Cyclooxygenase (COX)-1 and COX-2 participate in 5,6-epoxyeicosatrienoic acid-induced contraction of rabbit intralobar pulmonary arteries. J. Pharmacol. Exp. Ther. 2007, 321, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Rand, A.A.; Barnych, B.; Morisseau, C.; Cajka, T.; Lee, K.S.S.; Panigrahy, D.; Hammock, B.D. Cyclooxygenase-derived proangiogenic metabolites of epoxyeicosatrienoic acids. Proc. Natl. Acad. Sci. USA 2017, 114, 4370–4375. [Google Scholar] [CrossRef] [PubMed]
- Barnych, B.; Rand, A.A.; Cajka, T.; Lee, K.S.S.; Hammock, B.D. Synthesis of cyclooxygenase metabolites of 8,9-epoxyeicosatrienoic acid (EET): 11- and 15-hydroxy 8,9-EETs. Org. Biomol. Chem. 2017, 15, 4308–4313. [Google Scholar] [CrossRef] [PubMed]
- Rand, A.A.; Rajamani, A.; Kodani, S.D.; Harris, T.R.; Schlatt, L.; Barnych, B.; Passerini, A.G.; Hammock, B.D. Epoxyeicosatrienoic acid (EET)-stimulated angiogenesis is mediated by epoxy hydroxyeicosatrienoic acids (EHETs) formed from COX-2. J. Lipid Res. 2019, 60, 1996–2005. [Google Scholar] [CrossRef]
- Hildreth, K.; Kodani, S.D.; Hammock, B.D.; Zhao, L. Cytochrome P450-derived linoleic acid metabolites EpOMEs and DiHOMEs: A review of recent studies. J. Nutr. Biochem. 2020, 86, 108484. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Chen, P.; Guo, M.; Wygle, D.; Edwards, P.A.; Falck, J.R.; Roman, R.J.; Scicli, A.G. Inhibitors of cytochrome P450 4A suppress angiogenic responses. Am. J. Pathol. 2005, 166, 615–624. [Google Scholar] [CrossRef]
- Guo, A.M.; Sheng, J.; Scicli, G.M.; Arbab, A.S.; Lehman, N.L.; Edwards, P.A.; Falck, J.R.; Roman, R.J.; Scicli, A.G. Expression of CYP4A1 in U251 human glioma cell induces hyperproliferative phenotype in vitro and rapidly growing tumors in vivo. J. Pharmacol. Exp. Ther. 2008, 327, 10–19. [Google Scholar] [CrossRef]
- Angara, K.; Rashid, M.H.; Shankar, A.; Ara, R.; Iskander, A.; Borin, T.F.; Jain, M.; Achyut, B.R.; Arbab, A.S. Vascular mimicry in glioblastoma following anti-angiogenic and anti-20-HETE therapies. Histol. Histopathol. 2017, 32, 917–928. [Google Scholar] [PubMed]
- Guo, M.; Roman, R.J.; Falck, J.R.; Edwards, P.A.; Scicli, A.G. Human U251 glioma cell proliferation is suppressed by HET0016 [N-hydroxy-N′-(4-butyl-2-methylphenyl)formamidine], a selective inhibitor of CYP4A. J. Pharmacol. Exp. Ther. 2005, 315, 526–533. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Chen, H.; Zhang, J.; Zhang, J.; Qin, T.; Duan, C.; Chen, X.; Liu, Y.; Zhou, X.; et al. Inhibition of CYP4A by a novel flavonoid FLA-16 prolongs survival and normalizes tumor vasculature in glioma. Cancer Lett. 2017, 402, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wu, J.; Hu, M.; Wu, J.; Zhu, Q.; Yang, Z.; Xie, X.; Feng, Y.Q.; Yue, J. Glutamate affects the CYP1B1- and CYP2U1-mediated hydroxylation of arachidonic acid metabolism via astrocytic mGlu5 receptor. Int. J. Biochem. Cell Biol. 2019, 110, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Zagorac, D.; Jakovcevic, D.; Gebremedhin, D.; Harder, D.R. Antiangiogenic effect of inhibitors of cytochrome P450 on rats with glioblastoma multiforme. J. Cereb. Blood Flow Metab. 2008, 28, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Lin, J.H.; Arcuino, G.; Gao, Q.; Yang, J.; Nedergaard, M. Glutamate release promotes growth of malignant gliomas. Nat. Med. 2001, 7, 1010–1015. [Google Scholar] [CrossRef]
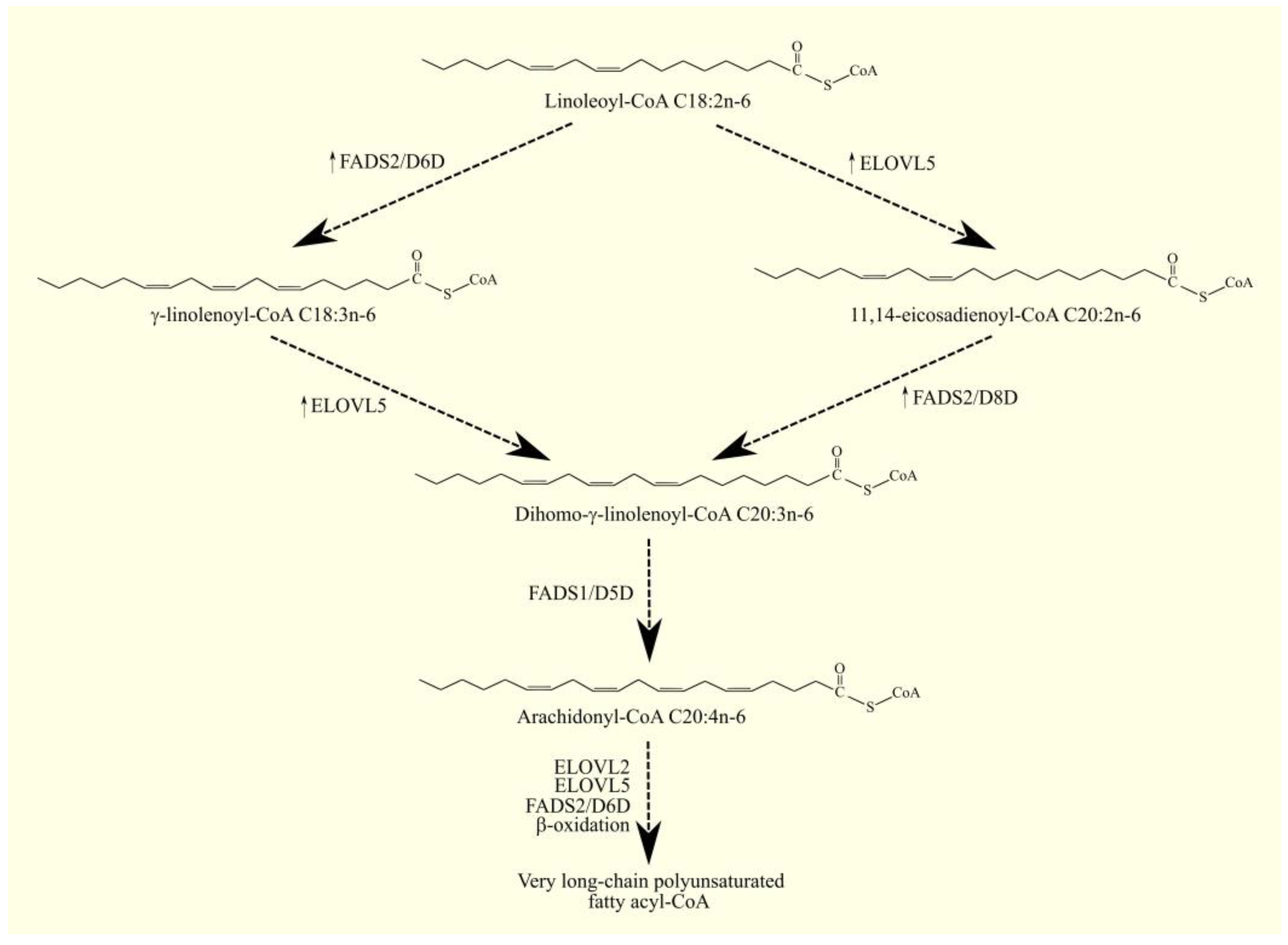
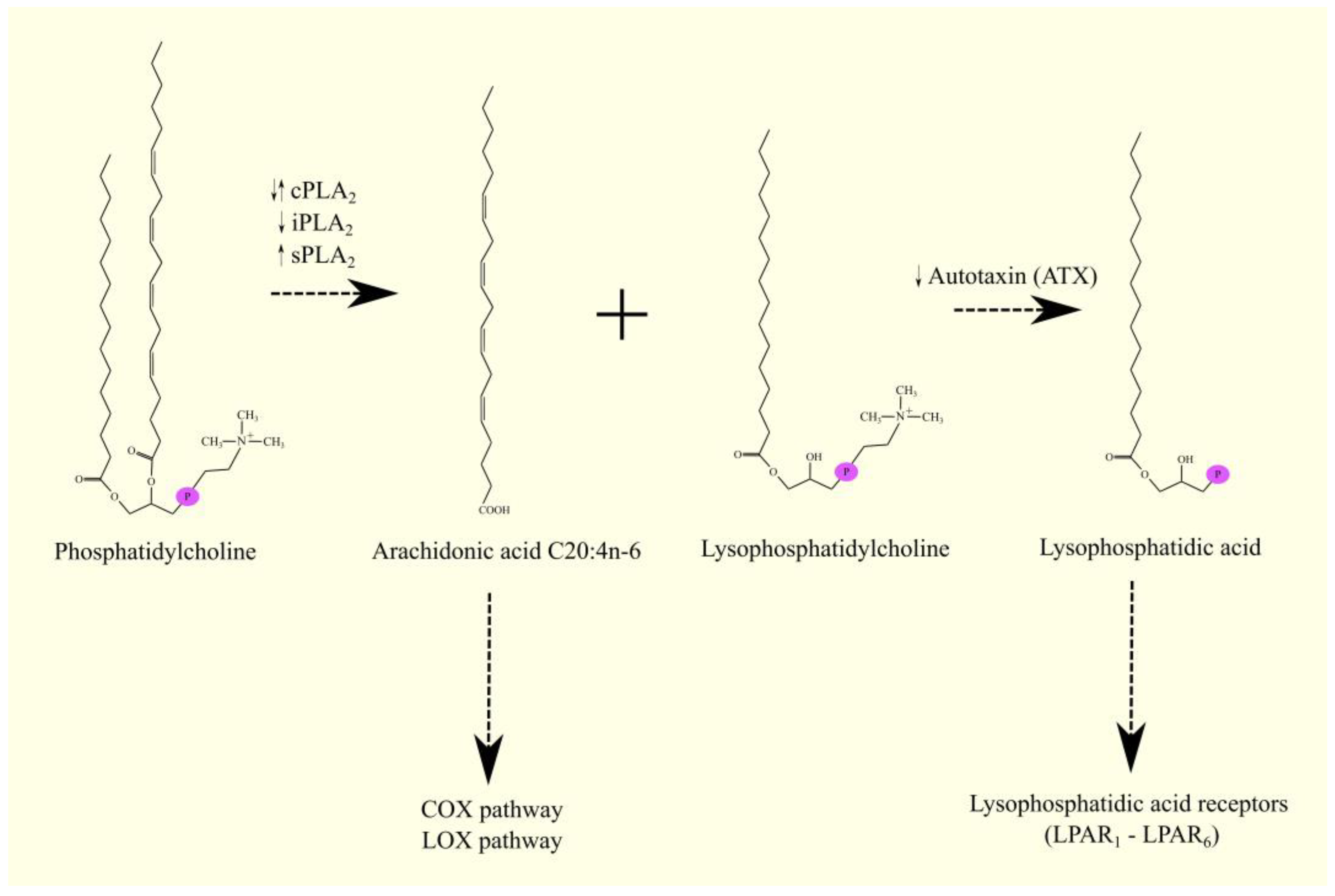
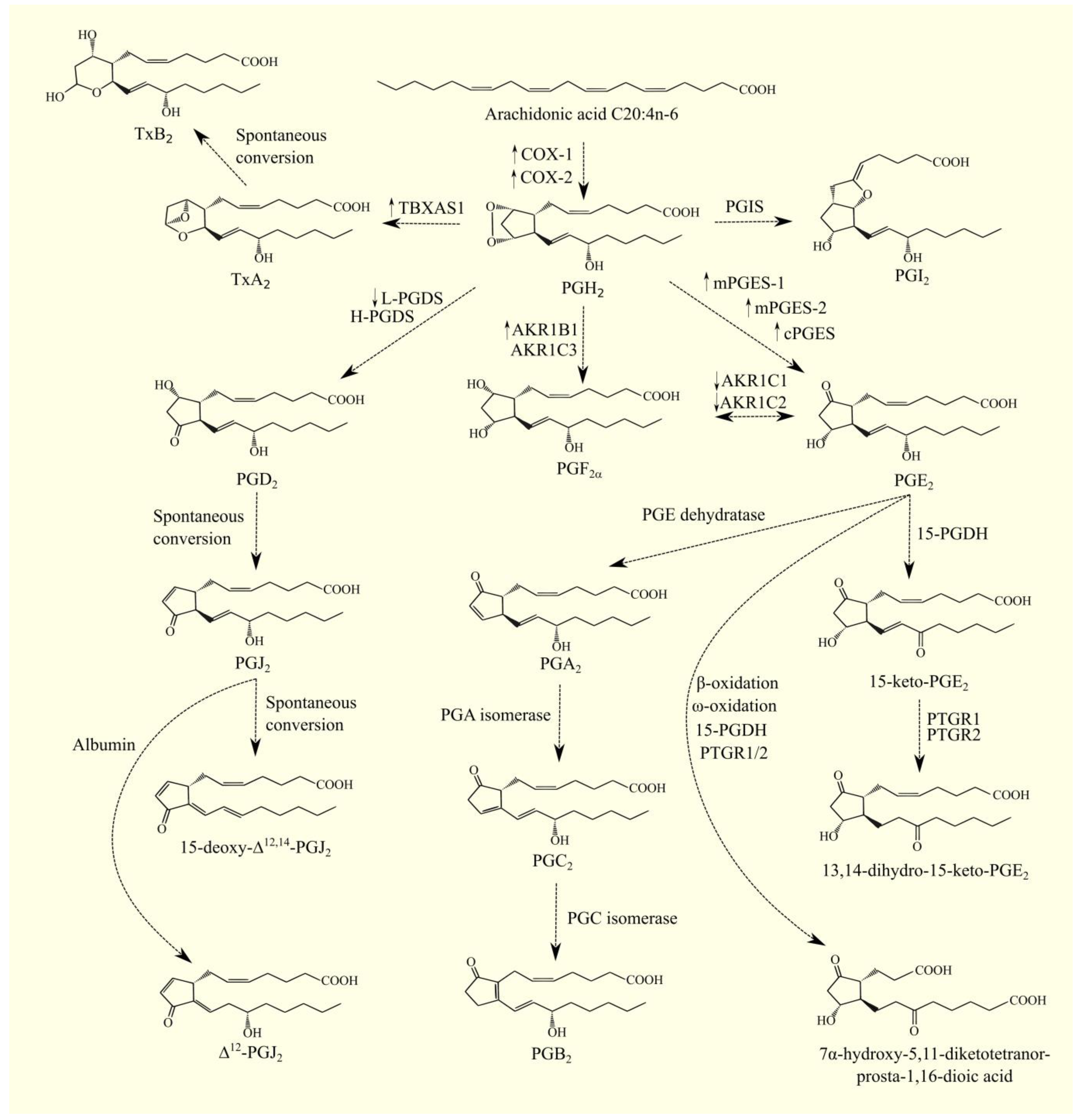


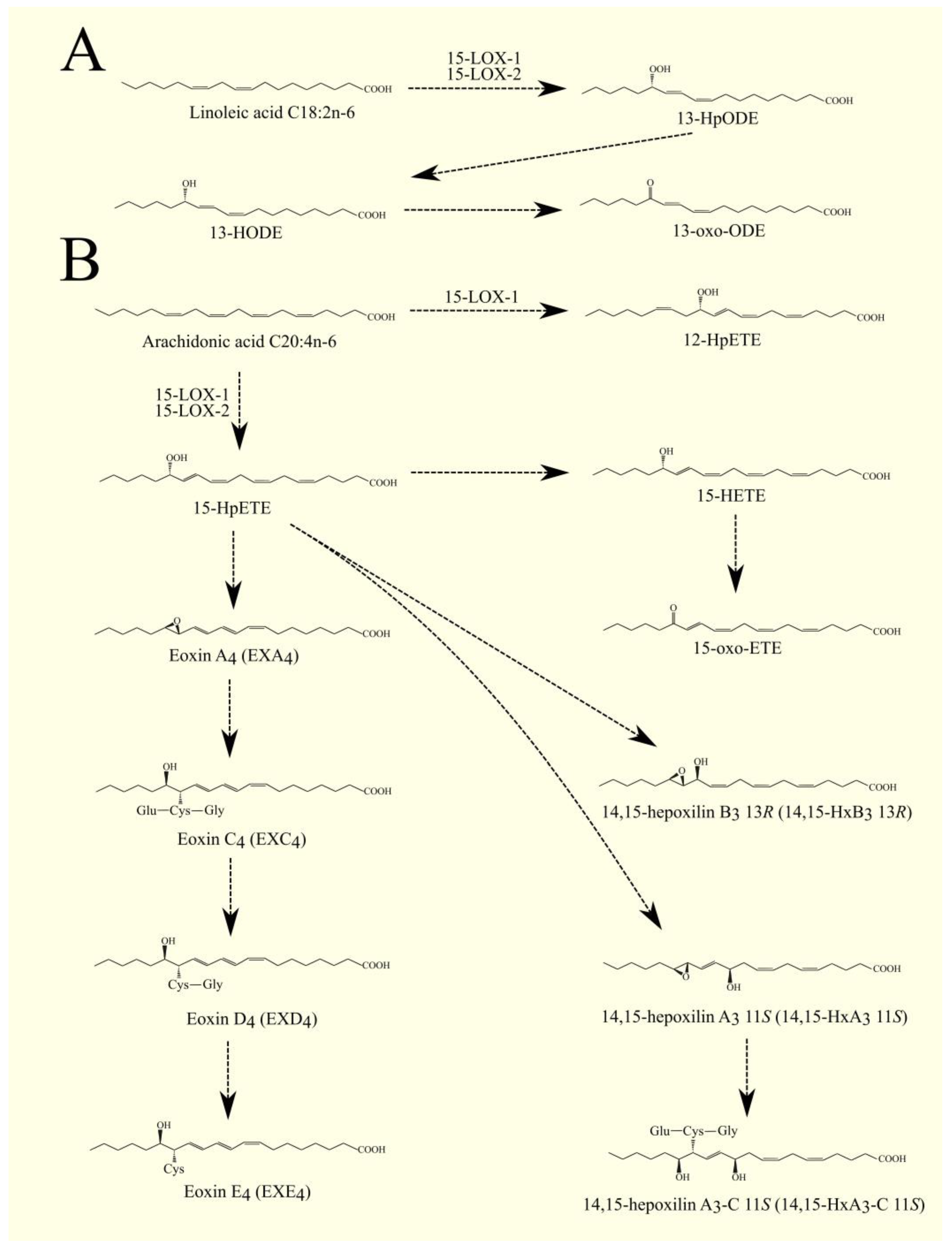

| Name | Expression Level in GBM Tumor Relative to Healthy Tissue | Impact on Prognosis with Higher Expression in GBM Tumors | |
|---|---|---|---|
| Source | GEPIA [9] | Seifert et al. [8] | GEPIA [9] |
| cPLA2 | |||
| cPLA2α/PLA2G4A | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis |
| cPLA2β/PLA2G4B | Lower expression in the tumor | Expression does not change | No significant impact on prognosis |
| cPLA2γ/PLA2G4C | Expression does not change | Lower expression in the tumor | No significant impact on prognosis |
| cPLA2δ/PLA2G4D | Expression does not change | Expression does not change | No significant impact on prognosis |
| cPLA2ε/PLA2G4E | Expression does not change | Expression does not change | No significant impact on prognosis |
| cPLA2ζ/PLA2G4F | Expression does not change | Expression does not change | No significant impact on prognosis |
| iPLA2 | |||
| iPLA2β/PLA2G6 | Expression does not change | Lower expression in the tumor | No significant impact on prognosis |
| iPLA2γ/PNPLA8 | Expression does not change | Expression does not change | No significant impact on prognosis |
| iPLA2δ/PNPLA6 | Expression does not change | Lower expression in the tumor | No significant impact on prognosis |
| iPLA2ε/PNPLA3 | Expression does not change | Expression does not change | No significant impact on prognosis |
| iPLA2ζ/PNPLA2 | Expression does not change | Expression does not change | Worse prognosis p = 0.087 |
| iPLA2η/PNPLA4 | Expression does not change | Expression does not change | Worse prognosis |
| Name | Expression Level in GBM Tumor Relative to Healthy Tissue | Impact on Prognosis with Higher Expression in GBM Tumors | ||
|---|---|---|---|---|
| Source | GEPIA [9] | Seifert et al. [8] | GEPIA [9] | Wu et al. [52] |
| PLA2G1B | Expression does not change | Expression does not change | Worse prognosis p = 0.078 | Worse prognosis |
| PLA2G2A | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis | No significant impact on prognosis |
| PLA2G2D | Expression does not change | Expression does not change | No significant impact on prognosis | No significant impact on prognosis |
| PLA2G2E | Expression does not change | Expression does not change | N/A | Worse prognosis |
| PLA2G2F | Expression does not change | Expression does not change | N/A | No significant impact on prognosis |
| PLA2G3 | Expression does not change | Expression does not change | No significant impact on prognosis | Worse prognosis |
| PLA2G5 | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis | Worse prognosis |
| PLA2G7 | Expression does not change | Expression does not change | No significant impact on prognosis | No significant impact on prognosis |
| PLA2G10 | Expression does not change | Expression does not change | N/A | No significant impact on prognosis |
| PLA2G12A | Higher expression in the tumor | Expression does not change | No significant impact on prognosis | No significant impact on prognosis |
| PLA2G12B | Expression does not change | Expression does not change | N/A | No significant impact on prognosis |
| PLA2G15 | Higher expression in the tumor | Expression does not change | Worse prognosis | No significant impact on prognosis |
| PLA2G16 | Expression does not change | Expression does not change | No significant impact on prognosis | No significant impact on prognosis |
| PLA2R1/PLA2R1 | Expression does not change | Expression does not change | Worse prognosis | |
| Name of Cancer | cPLA2α/PLA2G4A | cPLA2β/PLA2G4B | cPLA2γ/PLA2G4C | cPLA2δ/PLA2G4D | cPLA2ε/PLA2G4E | cPLA2ζ/PLA2G4F | iPLA2β/PLA2G6 | iPLA2γ/PNPLA8 | iPLA2δ/PNPLA6 | iPLA2ε/PNPLA3 | iPLA2ζ/PNPLA2 | iPLA2η/PNPLA4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adrenocortical carcinoma (ACC) | ↓ | ↓ | = | = | = | = | ↓ | = | = | = | = | = |
| Bladder urothelial carcinoma (BLCA) | ↓ | = | ↓ | = | = | = | = | = | = | = | = | = |
| Breast invasive carcinoma (BRCA) | ↓ | ↓ | = | = | = | = | ↓ | = | = | = | ↓ | = |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) | = | ↓ | ↓ | = | = | ↑ | ↓ | = | = | = | = | = |
| Cholangiocarcinoma (CHOL) | = | = | ↑ | = | = | ↑ | ↑ | = | ↑ | ↓ | ↑ | = |
| Colon adenocarcinoma (COAD) | ↓ | ↓ | ↓ | = | = | ↑ | ↓ | = | = | = | = | ↑ |
| Lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) | = | = | ↑ | = | = | = | = | = | = | = | ↓ | ↑ |
| Esophageal carcinoma (ESCA) | = | ↓ | = | = | = | ↓ | = | = | = | = | = | = |
| Glioblastoma multiforme (GBM) | ↑ | ↓ | = | = | = | = | = | = | = | = | = | = |
| Head and neck squamous cell carcinoma (HNSC) | = | ↓ | = | = | = | = | = | = | = | = | = | = |
| Kidney chromophobe (KICH) | = | ↓ | = | = | = | ↑ | = | = | = | = | = | = |
| Kidney renal clear cell carcinoma (KIRC) | ↓ | = | = | = | = | ↓ | = | = | = | = | = | = |
| Kidney renal papillary cell carcinoma (KIRP) | = | = | = | = | = | ↓ | = | = | = | = | = | = |
| Acute myeloid leukemia (LAML) | ↑ | ↑ | ↑ | = | = | = | ↑ | = | ↑ | = | ↑ | ↓ |
| Brain lower grade glioma (LGG) | = | ↓ | = | = | = | = | = | ↑ | = | = | = | = |
| Liver hepatocellular carcinoma (LIHC) | = | ↓ | ↑ | = | = | = | = | = | = | = | = | = |
| Lung adenocarcinoma (LUAD) | ↑ | ↓ | ↓ | = | = | ↓ | ↓ | = | ↓ | = | ↓ | = |
| Lung squamous cell carcinoma (LUSC) | = | ↓ | ↓ | = | = | ↓ | ↓ | = | ↓ | = | ↓ | ↑ |
| Ovarian serous cystadenocarcinoma (OV) | = | ↓ | = | = | = | = | ↓ | = | = | = | = | = |
| Pancreatic adenocarcinoma (PAAD) | ↑ | = | ↑ | = | = | = | = | ↑ | ↑ | = | = | = |
| Pheochromocytoma and paraganglioma (PCPG) | ↓ | = | = | = | = | = | = | = | = | = | ↓ | = |
| Prostate adenocarcinoma (PRAD) | = | ↓ | = | = | = | = | ↓ | = | = | = | = | = |
| Rectum adenocarcinoma (READ) | ↓ | ↓ | ↓ | = | = | ↑ | ↓ | = | = | = | = | ↑ |
| Sarcoma (SARC) | = | = | = | = | = | ↓ | = | = | = | = | = | = |
| Skin cutaneous melanoma (SKCM) | = | ↓ | ↑ | ↓ | ↓ | ↓ | ↓ | = | = | ↓ | ↓ | = |
| Stomach adenocarcinoma (STAD) | ↑ | ↓ | = | = | = | = | ↓ | = | = | = | = | = |
| Testicular germ cell tumors (TGCT) | = | ↓ | ↓ | = | = | ↑ | ↓ | = | ↓ | = | ↓ | ↑ |
| Thyroid carcinoma (THCA) | = | ↓ | = | = | = | ↓ | ↓ | = | = | = | ↓ | = |
| Thymoma (THYM) | = | = | ↑ | = | = | ↑ | ↑ | ↑ | = | = | = | ↑ |
| Uterine corpus endometrial carcinoma (UCEC) | = | ↓ | ↓ | = | = | = | ↓ | = | = | = | = | = |
| Uterine carcinosarcoma (UCS) | ↓ | ↓ | ↓ | = | = | = | ↓ | = | = | = | ↓ | = |
| Name of Cancer | PLA2G1B | PLA2G2A | PLA2G2D | PLA2G2E | PLA2G2F | PLA2G3 | PLA2G5 | PLA2G7 | PLA2G10 | PLA2G12A | PLA2G12B | PLA2G15 | PLA2G16 | PLA2R1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adrenocortical carcinoma (ACC) | ↑ | ↓ | = | = | = | = | = | = | = | = | = | = | = | = |
| Bladder urothelial carcinoma (BLCA) | = | ↓ | = | = | ↑ | = | ↓ | ↑ | = | = | = | = | = | = |
| Breast invasive carcinoma (BRCA) | = | ↓ | = | = | = | = | ↓ | = | = | = | = | = | ↓ | ↓ |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) | ↓ | ↓ | = | = | = | = | = | ↑ | = | = | = | = | = | ↓ |
| Cholangiocarcinoma (CHOL) | = | ↓ | = | = | = | = | = | = | = | = | ↓ | = | = | = |
| Colon adenocarcinoma (COAD) | = | = | = | = | = | = | ↓ | ↑ | ↑ | = | ↑ | = | = | = |
| Lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) | = | = | ↑ | = | = | = | = | ↑ | = | ↑ | = | ↑ | ↑ | = |
| Esophageal carcinoma (ESCA) | ↓ | ↓ | = | = | = | ↓ | ↓ | ↑ | ↑ | = | = | = | ↑ | = |
| Glioblastoma multiforme (GBM) | = | ↑ | = | = | = | = | ↑ | = | = | ↑ | = | ↑ | = | = |
| Head and neck squamous cell carcinoma (HNSC) | = | ↓ | = | = | = | = | = | ↑ | = | = | = | = | ↓ | = |
| Kidney chromophobe (KICH) | = | = | = | = | = | = | = | = | = | = | = | = | = | ↓ |
| Kidney renal clear cell carcinoma (KIRC) | = | = | = | = | = | = | = | ↑ | = | = | = | = | ↑ | ↓ |
| Kidney renal papillary cell carcinoma (KIRP) | = | = | ↑ | = | = | = | = | ↑ | = | = | ↓ | = | ↑ | ↓ |
| Acute myeloid leukemia (LAML) | = | = | = | = | = | ↓ | = | = | = | ↓ | = | = | = | = |
| Brain lower grade glioma (LGG) | = | = | = | = | = | = | = | = | = | = | = | = | = | = |
| Liver hepatocellular carcinoma (LIHC) | = | ↓ | = | = | = | = | = | = | = | = | = | = | = | = |
| Lung adenocarcinoma (LUAD) | ↓ | ↓ | ↑ | = | = | ↓ | ↓ | = | = | = | = | = | = | = |
| Lung squamous cell carcinoma (LUSC) | ↓ | ↓ | ↑ | = | = | = | ↓ | = | ↓ | = | = | = | ↓ | = |
| Ovarian serous cystadenocarcinoma (OV) | ↓ | ↓ | = | = | = | = | ↓ | = | = | = | = | = | ↓ | ↓ |
| Pancreatic adenocarcinoma (PAAD) | ↓ | ↓ | = | = | = | = | ↑ | ↑ | ↑ | = | = | ↑ | ↑ | ↑ |
| Pheochromocytoma and paraganglioma (PCPG) | ↓ | ↓ | = | = | = | = | = | = | = | = | = | = | = | = |
| Prostate adenocarcinoma (PRAD) | = | ↑ | = | = | = | = | = | ↑ | = | ↑ | = | = | = | = |
| Rectum adenocarcinoma (READ) | = | = | = | = | = | = | ↓ | ↑ | ↑ | = | ↑ | = | = | = |
| Sarcoma (SARC) | = | = | = | = | = | = | = | = | = | = | ↓ | = | = | ↓ |
| Skin cutaneous melanoma (SKCM) | = | ↓ | ↑ | = | ↓ | ↓ | = | ↑ | = | = | = | = | ↑ | ↓ |
| Stomach adenocarcinoma (STAD) | ↓ | ↑ | = | = | = | = | = | ↑ | ↑ | = | = | = | = | = |
| Testicular germ cell tumors (TGCT) | ↓ | ↓ | ↑ | = | = | = | ↓ | ↑ | ↓ | = | = | = | ↓ | = |
| Thyroid carcinoma (THCA) | = | ↓ | = | = | = | = | = | = | = | = | ↑ | = | ↑ | ↓ |
| Thymoma (THYM) | = | = | = | = | = | = | = | ↑ | = | ↑ | = | ↑ | ↑ | ↑ |
| Uterine corpus endometrial carcinoma (UCEC) | ↓ | ↓ | = | = | = | = | ↓ | ↑ | ↑ | ↑ | = | = | = | ↓ |
| Uterine carcinosarcoma (UCS) | ↓ | ↓ | = | = | = | = | ↓ | ↑ | = | = | = | = | = | ↓ |
| Name | Biochemical Significance | Expression Level in GBM Tumor Relative to Healthy Tissue | Impact on Prognosis with Higher Expression in GBM Tumors | |||
|---|---|---|---|---|---|---|
| Source | GEPIA [9] | Seifert et al. [8] | Other Data Source | GEPIA [9] | Other Data Source | |
| COX-1 | PGH2 synthesis from ARA | Higher expression in the tumor | Higher expression in the tumor | Higher expression in the tumor [141] | No significant impact on prognosis | No significant impact on prognosis [121] |
| COX-2 | PGH2 synthesis from ARA | Expression does not change | Expression does not change | Higher expression in the tumor [141,142] | No significant impact on prognosis | Worse prognosis [160,187,188] |
| mPGES-1 | PGE2 synthesis from PGH2 | Expression does not change | Expression does not change | Higher expression in the tumor [143] | Worse prognosis | Worse prognosis [121] |
| mPGES-2 | PGE2 synthesis from PGH2 | Expression does not change | Expression does not change | Higher expression in the tumor [143] | No significant impact on prognosis | No significant impact on prognosis [121] |
| cPGES | PGE2 synthesis from PGH2 | Higher expression in the tumor | Expression does not change | Higher expression in the tumor [143] | No significant impact on prognosis | |
| H-PGDS | Synthesis of PGD2 from PGH2 | Higher expression in the tumor | Lower expression in the tumor | No significant impact on prognosis | ||
| L-PGDS | Synthesis of PGD2 from PGH2 | Lower expression in the tumor | Lower expression in the tumor | No significant impact on prognosis | ||
| TBXAS1 | TxA2 synthesis from PGH2 | Higher expression in the tumor | Higher expression in the tumor | Higher expression in the tumor [144] | No significant impact on prognosis | |
| AKR1B1 | PGF2α synthesis from PGH2 | Higher expression in the tumor | Higher expression in the tumor | Worse prognosis | ||
| AKR1C1 | PGF2α synthesis from PGE2 | Lower expression in the tumor | Lower expression in the tumor | No significant impact on prognosis | ||
| AKR1C2 | PGF2α synthesis from PGE2 | Lower expression in the tumor | Expression does not change | No significant impact on prognosis | ||
| AKR1C3 | PGF2α synthesis from PGH2 | Expression does not change | Expression does not change | No significant impact on prognosis | ||
| PGIS | PGIF2 synthesis from PGH2 | Expression does not change | Expression does not change | No significant impact on prognosis | ||
| MRP4 | Secretion of prostaglandins from the cell | Higher expression in the tumor | Expression does not change | No significant impact on prognosis | ||
| PGT/SLCO2A1 | Uptake of prostaglandins into the cell | Expression does not change | Expression does not change | No significant impact on prognosis | ||
| 15-PGDH | First degradation reaction/formation of PPARγ ligand from PGE2 | Expression does not change | Expression does not change | No significant impact on prognosis | Better prognosis [121] | |
| PTGR1 | Second degradation/inactivation reaction of PPARγ ligand made from PGE2 | Expression does not change | Expression does not change | No significant impact on prognosis | Worse prognosis [121] | |
| PTGR2 | Second degradation/inactivation reaction of PPARγ ligand made from PGE2 | Expression does not change | Expression does not change | No significant impact on prognosis | No significant impact on prognosis | |
| Name of Cancer | COX-1/PTGS1 | COX-2/PTGS2 | mPGES-1/PTGES | mPGES-2/PTGES2 | cPGES/PTGES3 | H-PGDS/HPGDS | L-PGDS/PTGDS | TBXAS1 | AKR1B1 | AKR1C1 | AKR1C2 | AKR1C3 | PGIS/PTGIS | MRP4/ABCC4 | PGT/SLCO2A1 | 15-PGDH/HPGD | PTGR1 | PTGR2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adrenocortical carcinoma (ACC) | = | = | = | = | ↑ | = | ↓ | ↓ | ↓ | = | ↓ | = | ↓ | = | ↓ | ↑ | ↓ | = |
| Bladder urothelial carcinoma (BLCA) | ↓ | ↓ | = | = | = | ↓ | ↓ | = | = | ↓ | = | = | ↓ | ↓ | ↓ | ↓ | = | = |
| Breast invasive carcinoma (BRCA) | = | ↓ | = | = | = | = | ↓ | = | = | ↓ | ↓ | ↓ | ↓ | = | = | ↓ | = | = |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) | = | = | = | = | = | = | ↓ | = | = | = | = | = | ↓ | = | ↓ | ↓ | = | = |
| Cholangiocarcinoma (CHOL) | ↑ | = | ↑ | ↑ | ↑ | = | = | = | ↑ | = | = | ↑ | = | ↑ | ↑ | ↓ | ↓ | = |
| Colon adenocarcinoma (COAD) | ↓ | = | = | = | ↑ | ↓ | ↓ | ↑ | ↓ | ↓ | ↓ | = | ↓ | = | ↓ | ↓ | = | = |
| Lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) | = | ↓ | ↑ | ↑ | ↑ | = | ↑ | ↓ | ↑ | = | = | = | = | = | ↑ | ↓ | ↑ | ↑ |
| Esophageal carcinoma (ESCA) | ↓ | ↑ | = | = | = | = | ↓ | = | = | ↓ | ↓ | ↑ | ↓ | ↑ | = | ↓ | ↓ | = |
| Glioblastoma multiforme (GBM) | ↑ | = | = | = | ↑ | ↑ | ↓ | ↑ | ↑ | ↓ | ↓ | = | = | ↑ | = | = | = | = |
| Head and neck squamous cell carcinoma (HNSC) | = | = | = | = | = | = | ↓ | = | ↑ | = | = | = | = | = | = | ↓ | = | = |
| Kidney chromophobe (KICH) | = | = | = | = | = | = | ↓ | = | = | = | ↓ | ↓ | ↓ | = | ↓ | = | ↓ | = |
| Kidney renal clear cell carcinoma (KIRC) | ↑ | = | ↓ | = | = | = | ↓ | ↑ | = | = | = | = | = | = | = | ↓ | ↓ | = |
| Kidney renal papillary cell carcinoma (KIRP) | = | ↓ | = | = | = | = | ↓ | = | ↑ | ↑ | ↑ | = | ↓ | = | ↓ | ↓ | = | = |
| Acute myeloid leukemia (LAML) | = | ↑ | = | ↓ | ↓ | ↑ | ↓ | ↑ | ↑ | ↓ | ↓ | ↑ | = | ↓ | = | = | ↑ | ↓ |
| Brain lower grade glioma (LGG) | ↑ | = | = | = | = | ↑ | = | ↑ | = | = | = | ↑ | = | ↑ | = | = | = | ↑ |
| Liver hepatocellular carcinoma (LIHC) | = | = | = | = | ↑ | = | = | = | = | ↑ | ↑ | ↑ | ↓ | = | = | ↓ | = | = |
| Lung adenocarcinoma (LUAD) | = | = | ↑ | = | = | = | ↓ | = | = | = | = | = | ↓ | = | ↓ | ↓ | = | = |
| Lung squamous cell carcinoma (LUSC) | = | = | ↑ | = | = | ↓ | ↓ | ↓ | = | ↑ | ↑ | ↑ | ↓ | = | ↓ | ↓ | = | = |
| Ovarian serous cystadenocarcinoma (OV) | ↑ | = | = | = | = | = | ↑ | = | = | ↓ | ↓ | ↓ | ↓ | = | = | ↑ | = | = |
| Pancreatic adenocarcinoma (PAAD) | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | = |
| Pheochromocytoma and paraganglioma (PCPG) | = | = | ↑ | = | = | = | ↓ | = | ↓ | = | = | = | = | = | = | = | ↓ | = |
| Prostate adenocarcinoma (PRAD) | = | ↓ | = | = | = | = | ↓ | = | ↓ | ↓ | ↓ | = | ↓ | ↑ | ↓ | = | = | = |
| Rectum adenocarcinoma (READ) | ↓ | ↓ | ↑ | = | ↑ | = | ↓ | ↑ | ↓ | ↓ | ↓ | ↑ | ↓ | = | ↓ | ↓ | = | = |
| Sarcoma (SARC) | ↓ | = | = | = | = | = | = | = | = | = | = | = | = | = | = | ↓ | = | = |
| Skin cutaneous melanoma (SKCM) | ↓ | = | ↓ | = | = | ↓ | = | ↑ | ↑ | ↓ | ↓ | ↓ | = | = | ↓ | ↓ | = | = |
| Stomach adenocarcinoma (STAD) | = | = | = | = | ↑ | = | = | = | = | ↓ | ↓ | = | = | = | = | = | = | = |
| Testicular germ cell tumors (TGCT) | = | = | ↓ | = | ↑ | = | ↓ | ↑ | = | ↓ | ↓ | ↓ | = | = | = | = | ↑ | ↓ |
| Thyroid carcinoma (THCA) | = | = | = | = | = | = | ↓ | = | = | ↓ | ↓ | ↓ | ↓ | = | ↓ | = | = | = |
| Thymoma (THYM) | ↓ | ↓ | = | ↑ | ↑ | ↑ | ↑ | ↓ | ↑ | = | ↑ | ↑ | = | = | ↑ | = | ↑ | ↑ |
| Uterine corpus endometrial carcinoma (UCEC) | ↑ | = | = | = | = | ↓ | ↓ | = | = | ↓ | = | = | ↓ | = | ↓ | ↓ | = | = |
| Uterine carcinosarcoma (UCS) | = | = | ↑ | = | = | = | ↓ | = | = | = | = | = | ↓ | = | ↓ | ↓ | = | = |
| Name | Biochemical Significance | Expression Level In GBM Tumors Relative To Healthy Tissue | Impact on Prognosis with Higher Expression in GBM Tumors | |
|---|---|---|---|---|
| Source | GEPIA [9] | Seifert et al. [8] | GEPIA [9] | |
| eLOX3/ALOXE3 | Production of hepoxilins/hydroxy-epoxyeicosatrienoic acid and oxo-ETE from HpETE | Lower expression in the tumor | Expression does not change | No significant impact on prognosis |
| 5-LOX/ALOX5 | 5-HpETE production from ARA; the first enzyme in leukotrienes and the 5-oxo-ETE synthesis pathway; synthesis of lipoxins from 15-HpETE and 15-HETE | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis |
| FLAP/ALOX5AP | Substrate carrier for 5-LOX | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis |
| 12S-LOX/ALOX12 | 12S-HpETE production from ARA; the first enzyme in the hepoxilin production pathway; production of lipoxins from LTA4 | Expression does not change | Expression does not change | No significant impact on prognosis |
| 12R-LOX/ALOX12B | 12R-HpETE production from ARA | Expression does not change | Expression does not change | No significant impact on prognosis |
| 15-LOX-1/ALOX15 | 15-HpETE production from ARA; 12-HpETE production from ARA; production of lipoxins, eoxins, 15-oxo-ETE and 15-HETE; production of 13-HpODE from linoleic acid C18:2n-6 | Expression does not change | Expression does not change | No significant impact on prognosis |
| 15-LOX-2/ALOX15B | 15-HpETE production from ARA; production of 15-HpETE, lipoxins, eoxins, 15-oxo-ETE and 15-HETE | Expression does not change | Expression does not change | No significant impact on prognosis |
| LTA4H | LTB4 production from LTA4 | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis |
| LTC4S | LTC4 production from LTA4 | Higher expression in the tumor | Expression does not change | No significant impact on prognosis |
| GGT1 | LTD4 production from LTC4 | Expression does not change | Expression does not change | Worse prognosis |
| GGT5 | LTD4 production from LTC4 | Higher expression in the tumor | Higher expression in the tumor | Worse prognosis (p = 0.055) |
| DPEP1 | LTE4 production from LTD4 | Higher expression in the tumor | Expression does not change | No significant impact on prognosis |
| DPEP2 | LTE4 production from LTD4 | Expression does not change | Expression does not change | No significant impact on prognosis |
| EPHX2 | Conversion of hepoxilins into trioxilin | Expression does not change | Expression does not change | Worse prognosis (p = 0.072) |
| Receptors | ||||
| LTB4R1 | LTB4 receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| LTB4R2 | LTB4 receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| CYSLTR1 | Cysteinyl-leukotrienes receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| CYSLTR2 | Cysteinyl-leukotrienes receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| OXER1 | 5-oxo-ETE receptor | Expression does not change | Expression does not change | Worse prognosis |
| ALX/FPR2 | LXA4 receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| GPR17 | Cysteinyl-leukotrienes receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| GPR31 | 12S-HETE receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| OXGR1/GPR99 | LTE4 receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| G2A/GPR132 | 5-HETE, 12-HETE, 15-HETE, 9-HODE receptor | Expression does not change | Expression does not change | Worse prognosis (p = 0.052) |
| Name of Cancer | eLOX3/ALOXE3 | 5-LOX/ALOX5 | FLAP/ALOX5AP | 12S-LOX/ALOX12 | 12R-LOX/ALOX12B | 15-LOX-1/ALOX15 | 15-LOX-2/ALOX15B | LTA4H/LTA4H | LTC4S/LTC4S | GGT1 | GGT5 | DPEP1 | DPEP2 | EPHX2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adrenocortical carcinoma (ACC) | = | ↓ | ↓ | = | = | = | ↓ | = | = | = | ↓ | = | = | ↓ |
| Bladder urothelial carcinoma (BLCA) | = | = | ↓ | = | = | = | = | = | ↓ | = | ↓ | = | = | ↓ |
| Breast invasive carcinoma (BRCA) | = | = | = | = | = | = | ↓ | = | = | = | = | = | = | = |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) | = | = | = | = | = | = | = | = | ↓ | = | ↓ | = | = | ↓ |
| Cholangiocarcinoma (CHOL) | = | ↑ | ↑ | = | = | = | = | ↑ | ↑ | = | = | = | = | ↓ |
| Colon adenocarcinoma (COAD) | = | = | = | = | = | = | = | = | ↓ | ↑ | = | ↑ | ↓ | = |
| Lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) | = | ↓ | ↓ | ↓ | = | = | ↑ | = | = | = | ↑ | = | ↓ | = |
| Esophageal carcinoma (ESCA) | = | = | = | ↓ | = | = | ↓ | = | = | = | = | = | = | ↓ |
| Glioblastoma multiforme (GBM) | ↓ | ↑ | ↑ | = | = | = | = | ↑ | ↑ | = | ↑ | ↑ | = | = |
| Head and neck squamous cell carcinoma (HNSC) | ↑ | = | = | ↓ | = | = | = | = | = | = | ↑ | = | = | ↓ |
| Kidney chromophobe (KICH) | = | = | = | = | = | = | = | = | ↓ | ↓ | ↓ | ↓ | = | = |
| Kidney renal clear cell carcinoma (KIRC) | = | ↑ | ↑ | = | = | = | = | = | = | ↑ | = | ↓ | ↑ | ↓ |
| Kidney renal papillary cell carcinoma (KIRP) | = | ↑ | ↑ | = | = | = | ↑ | = | = | ↑ | ↓ | ↓ | ↑ | = |
| Acute myeloid leukemia (LAML) | = | ↑ | ↑ | = | = | = | = | ↑ | ↑ | = | ↑ | = | ↑ | ↓ |
| Brain lower grade glioma (LGG) | = | ↑ | ↑ | = | = | = | = | = | ↑ | = | ↑ | = | = | = |
| Liver hepatocellular carcinoma (LIHC) | = | = | = | = | = | = | = | = | = | = | ↓ | = | = | ↓ |
| Lung adenocarcinoma (LUAD) | = | ↓ | ↓ | = | = | = | ↓ | = | ↓ | = | = | = | ↓ | = |
| Lung squamous cell carcinoma (LUSC) | = | ↓ | ↓ | = | = | = | ↓ | ↓ | ↓ | ↓ | = | = | ↓ | = |
| Ovarian serous cystadenocarcinoma (OV) | = | ↑ | ↑ | = | = | = | = | = | = | = | ↓ | = | = | ↓ |
| Pancreatic adenocarcinoma (PAAD) | = | ↑ | ↑ | = | = | = | ↑ | ↑ | ↑ | ↑ | ↑ | ↓ | ↑ | ↓ |
| Pheochromocytoma and paraganglioma (PCPG) | = | = | ↓ | = | = | = | = | = | = | = | ↓ | = | = | ↓ |
| Prostate adenocarcinoma (PRAD) | = | = | = | = | = | = | = | = | = | ↑ | = | = | = | = |
| Rectum adenocarcinoma (READ) | = | = | = | = | = | = | = | = | ↓ | ↑ | = | ↑ | = | = |
| Sarcoma (SARC) | = | = | = | = | = | = | = | = | = | ↓ | = | ↓ | = | ↓ |
| Skin cutaneous melanoma (SKCM) | ↓ | ↓ | = | ↓ | ↓ | = | ↓ | = | ↓ | = | ↓ | = | = | ↓ |
| Stomach adenocarcinoma (STAD) | = | = | ↑ | = | = | = | = | = | = | = | = | = | = | = |
| Testicular germ cell tumors (TGCT) | ↓ | = | ↑ | = | = | ↓ | = | = | ↓ | = | = | ↓ | = | ↓ |
| Thyroid carcinoma (THCA) | = | ↑ | ↑ | = | = | = | ↑ | = | = | = | = | = | = | = |
| Thymoma (THYM) | = | ↓ | ↓ | = | = | = | = | = | ↑ | = | ↑ | ↑ | ↓ | ↑ |
| Uterine corpus endometrial carcinoma (UCEC) | = | ↓ | = | = | = | = | = | = | ↓ | ↑ | ↓ | = | = | ↓ |
| Uterine carcinosarcoma (UCS) | = | ↓ | ↓ | = | = | = | = | = | ↓ | = | ↓ | = | = | ↓ |
| Name | Expression Level in GBM Tumor Relative to Healthy Tissue | Impact on Prognosis with Higher Expression in GBM Tumors | |
|---|---|---|---|
| Source | GEPIA [9] | Seifert et al. [8] | GEPIA [9] |
| CYP1A2 | Expression does not change | Expression does not change | N/A |
| CYP1B1 | Expression does not change | Expression does not change | No significant impact on prognosis |
| CYP2C8 | Lower expression in the tumor | Expression does not change | No significant impact on prognosis |
| CYP2C9 | Expression does not change | Expression does not change | N/A |
| CYP2C19 | Expression does not change | Expression does not change | N/A |
| CYP2J2 | Expression does not change | Expression does not change | No significant impact on prognosis |
| CYP2U1 | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis |
| CYP3A4 | Expression does not change | Expression does not change | Worse prognosis p = 0.07 |
| CYP4A11 | Expression does not change | Expression does not change | No significant impact on prognosis |
| CYP4F2 | Expression does not change | Expression does not change | No significant impact on prognosis |
| CYP4F3 | Expression does not change | N/A | No significant impact on prognosis |
| CYP4X1 | Lower expression in the tumor | Lower expression in the tumor | No significant impact on prognosis |
| GPR75 | Expression does not change | Expression does not change | No significant impact on prognosis |
| Name of Cancer | CYP1A2 | CYP1B1 | CYP2C8 | CYP2C9 | CYP2C19 | CYP2J2 | CYP2U1 | CYP3A4 | CYP4A11 | CYP4F2 | CYP4F3 | CYP4X1 | GPR75 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adrenocortical carcinoma (ACC) | = | ↓ | ↓ | = | = | = | = | = | = | = | = | = | = |
| Bladder urothelial carcinoma (BLCA) | = | ↓ | = | = | = | = | ↓ | = | = | = | = | = | = |
| Breast invasive carcinoma (BRCA) | = | = | = | = | = | = | = | = | = | = | = | = | = |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) | = | ↓ | = | = | = | ↑ | ↓ | = | = | = | ↑ | = | = |
| Cholangiocarcinoma (CHOL) | ↓ | ↑ | ↓ | ↓ | ↓ | ↓ | = | ↓ | ↓ | ↓ | ↓ | = | = |
| Colon adenocarcinoma (COAD) | = | ↓ | = | = | = | ↑ | = | = | = | ↑ | ↑ | = | = |
| Lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) | = | = | = | = | = | = | ↑ | = | = | = | ↓ | = | = |
| Esophageal carcinoma (ESCA) | = | = | = | ↓ | = | ↓ | = | = | = | = | ↓ | ↓ | = |
| Glioblastoma multiforme (GBM) | = | = | ↓ | = | = | = | ↑ | = | = | = | = | ↓ | = |
| Head and neck squamous cell carcinoma (HNSC) | = | = | = | = | = | ↓ | = | = | = | = | = | ↓ | = |
| Kidney chromophobe (KICH) | = | ↓ | = | = | = | = | = | = | ↓ | ↓ | ↓ | ↓ | = |
| Kidney renal clear cell carcinoma (KIRC) | = | ↓ | = | = | = | ↑ | = | = | ↓ | ↓ | ↓ | = | = |
| Kidney renal papillary cell carcinoma (KIRP) | = | ↓ | = | = | = | = | = | = | ↓ | ↓ | ↓ | ↓ | = |
| Acute myeloid leukemia (LAML) | = | ↑ | = | = | = | = | ↑ | = | = | = | = | = | ↑ |
| Brain lower grade glioma (LGG) | = | = | ↓ | = | = | = | ↑ | = | = | = | = | ↓ | ↑ |
| Liver hepatocellular carcinoma (LIHC) | ↓ | = | ↓ | ↓ | ↓ | = | = | ↓ | ↓ | = | = | = | = |
| Lung adenocarcinoma (LUAD) | = | = | = | = | = | ↑ | = | = | = | = | = | = | = |
| Lung squamous cell carcinoma (LUSC) | = | = | = | = | = | = | = | = | = | = | = | = | = |
| Ovarian serous cystadenocarcinoma (OV) | = | = | = | = | = | ↑ | = | = | = | = | = | ↑ | = |
| Pancreatic adenocarcinoma (PAAD) | = | ↑ | = | ↑ | = | ↑ | ↑ | ↓ | = | = | ↑ | = | = |
| Pheochromocytoma and paraganglioma (PCPG) | = | = | = | = | = | = | ↑ | ↓ | = | = | = | = | = |
| Prostate adenocarcinoma (PRAD) | = | = | = | = | = | ↑ | = | = | = | = | = | = | = |
| Rectum adenocarcinoma (READ) | = | ↓ | = | = | = | ↑ | = | = | = | ↑ | ↑ | = | = |
| Sarcoma (SARC) | = | = | = | = | = | = | = | = | ↓ | ↓ | ↓ | = | = |
| Skin cutaneous melanoma (SKCM) | = | = | = | = | = | ↓ | ↑ | ↓ | = | = | ↓ | ↓ | = |
| Stomach adenocarcinoma (STAD) | = | = | ↓ | ↓ | = | ↑ | = | = | = | = | ↑ | ↓ | = |
| Testicular germ cell tumors (TGCT) | = | = | ↓ | = | = | ↓ | = | = | = | ↓ | = | = | = |
| Thyroid carcinoma (THCA) | = | ↑ | = | = | = | = | = | = | = | = | = | ↓ | = |
| Thymoma (THYM) | = | ↑ | = | = | = | = | ↑ | = | = | = | ↓ | = | = |
| Uterine corpus endometrial carcinoma (UCEC) | = | ↓ | = | = | = | ↑ | ↓ | = | = | = | = | = | = |
| Uterine carcinosarcoma (UCS) | = | ↓ | = | = | = | ↑ | ↓ | = | = | = | = | = | = |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korbecki, J.; Rębacz-Maron, E.; Kupnicka, P.; Chlubek, D.; Baranowska-Bosiacka, I. Synthesis and Significance of Arachidonic Acid, a Substrate for Cyclooxygenases, Lipoxygenases, and Cytochrome P450 Pathways in the Tumorigenesis of Glioblastoma Multiforme, Including a Pan-Cancer Comparative Analysis. Cancers 2023, 15, 946. https://doi.org/10.3390/cancers15030946
Korbecki J, Rębacz-Maron E, Kupnicka P, Chlubek D, Baranowska-Bosiacka I. Synthesis and Significance of Arachidonic Acid, a Substrate for Cyclooxygenases, Lipoxygenases, and Cytochrome P450 Pathways in the Tumorigenesis of Glioblastoma Multiforme, Including a Pan-Cancer Comparative Analysis. Cancers. 2023; 15(3):946. https://doi.org/10.3390/cancers15030946
Chicago/Turabian StyleKorbecki, Jan, Ewa Rębacz-Maron, Patrycja Kupnicka, Dariusz Chlubek, and Irena Baranowska-Bosiacka. 2023. "Synthesis and Significance of Arachidonic Acid, a Substrate for Cyclooxygenases, Lipoxygenases, and Cytochrome P450 Pathways in the Tumorigenesis of Glioblastoma Multiforme, Including a Pan-Cancer Comparative Analysis" Cancers 15, no. 3: 946. https://doi.org/10.3390/cancers15030946
APA StyleKorbecki, J., Rębacz-Maron, E., Kupnicka, P., Chlubek, D., & Baranowska-Bosiacka, I. (2023). Synthesis and Significance of Arachidonic Acid, a Substrate for Cyclooxygenases, Lipoxygenases, and Cytochrome P450 Pathways in the Tumorigenesis of Glioblastoma Multiforme, Including a Pan-Cancer Comparative Analysis. Cancers, 15(3), 946. https://doi.org/10.3390/cancers15030946






