Patterns of Somatic Variants in Colorectal Adenoma and Carcinoma Tissue and Matched Plasma Samples from the Hungarian Oncogenome Program
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. DNA Isolation
2.3. Library Preparation and Next-Generation Sequencing
2.4. Bioinformatic Analyses
2.5. Validation of KRAS Mutation Status Using Digital PCR Technology
2.6. In Situ Hybridization
3. Results
3.1. Whole-Exome Sequencing Parameters of Tissue Samples
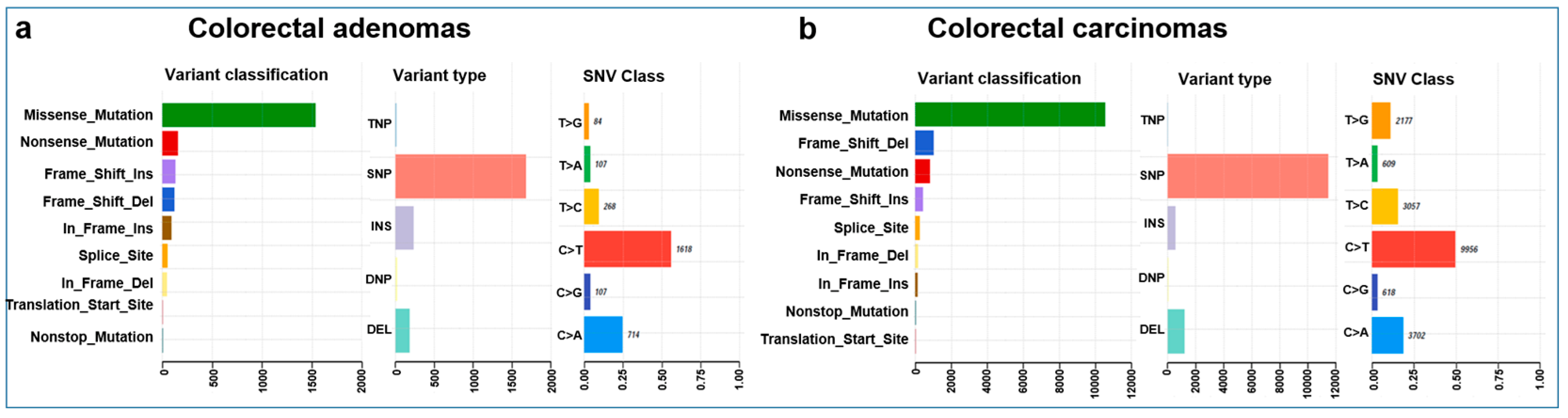
3.2. COSMIC Mutation Signatures
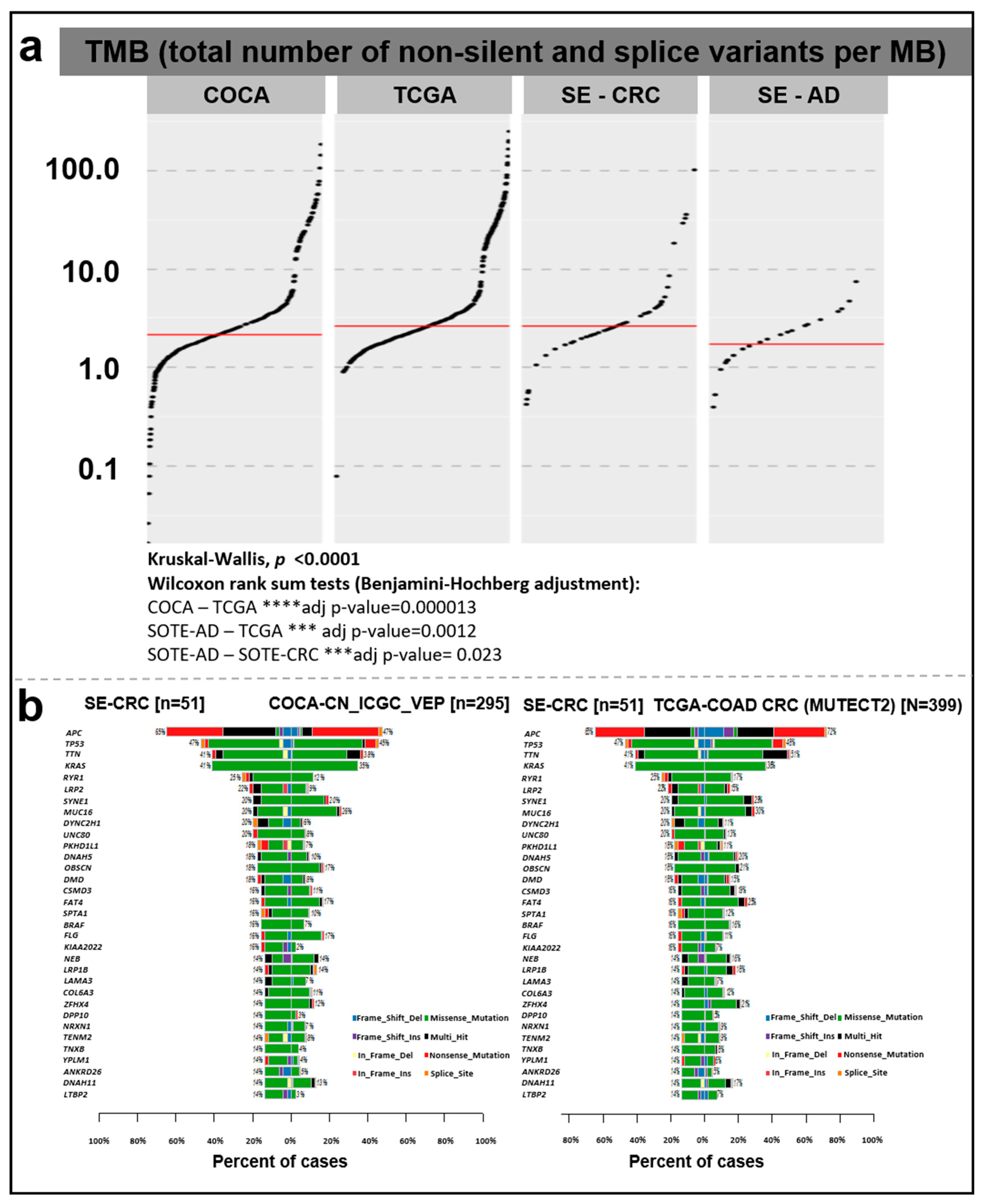
3.3. Tumor Mutation Burden Evaluation
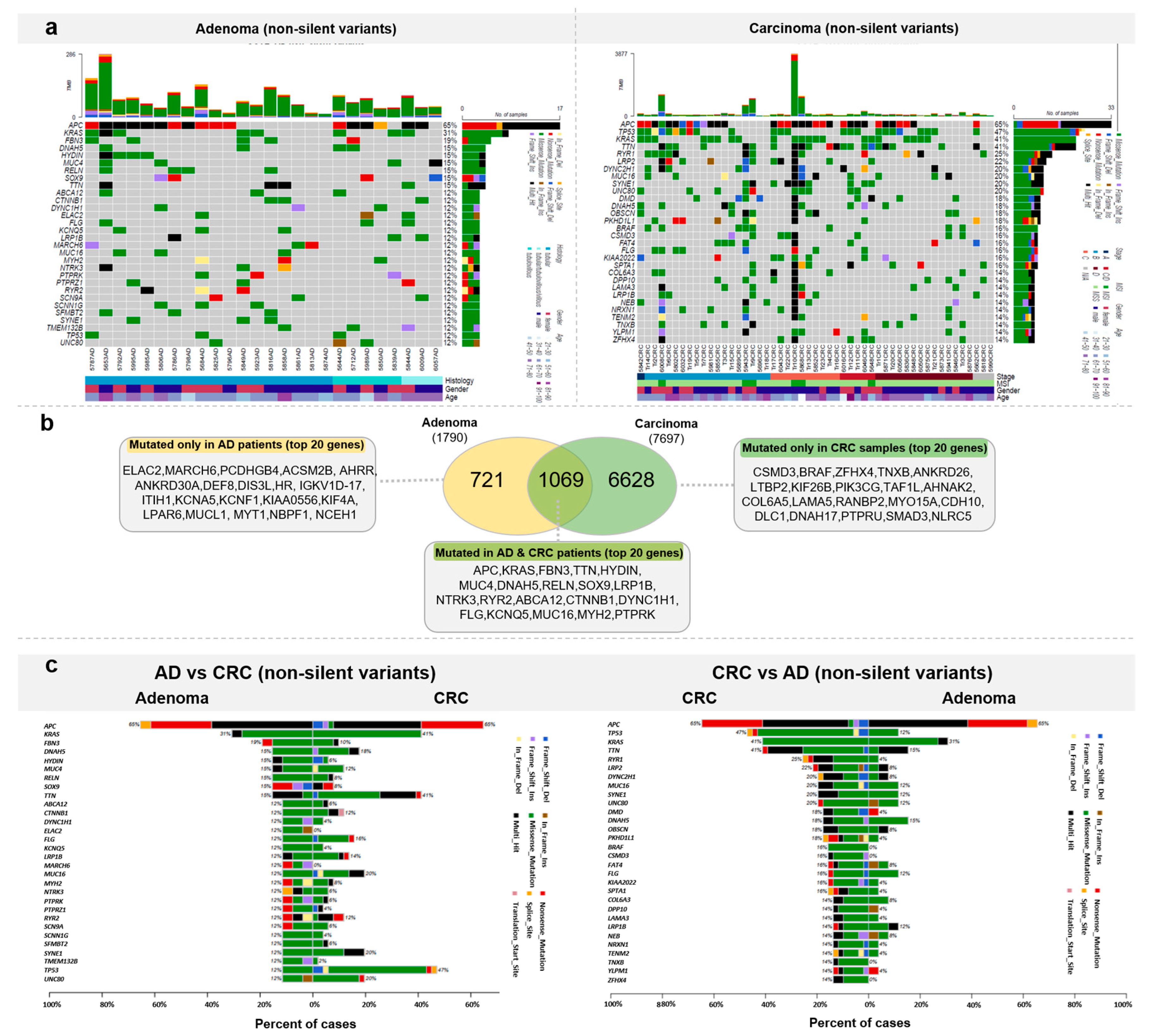
3.4. Somatic Mutation Landscape of Colorectal Tumors from a Hungarian Cohort
3.5. KRAS Mutation Landscape
3.6. ddPCR and In Situ Hybridization Validation of KRAS G12D
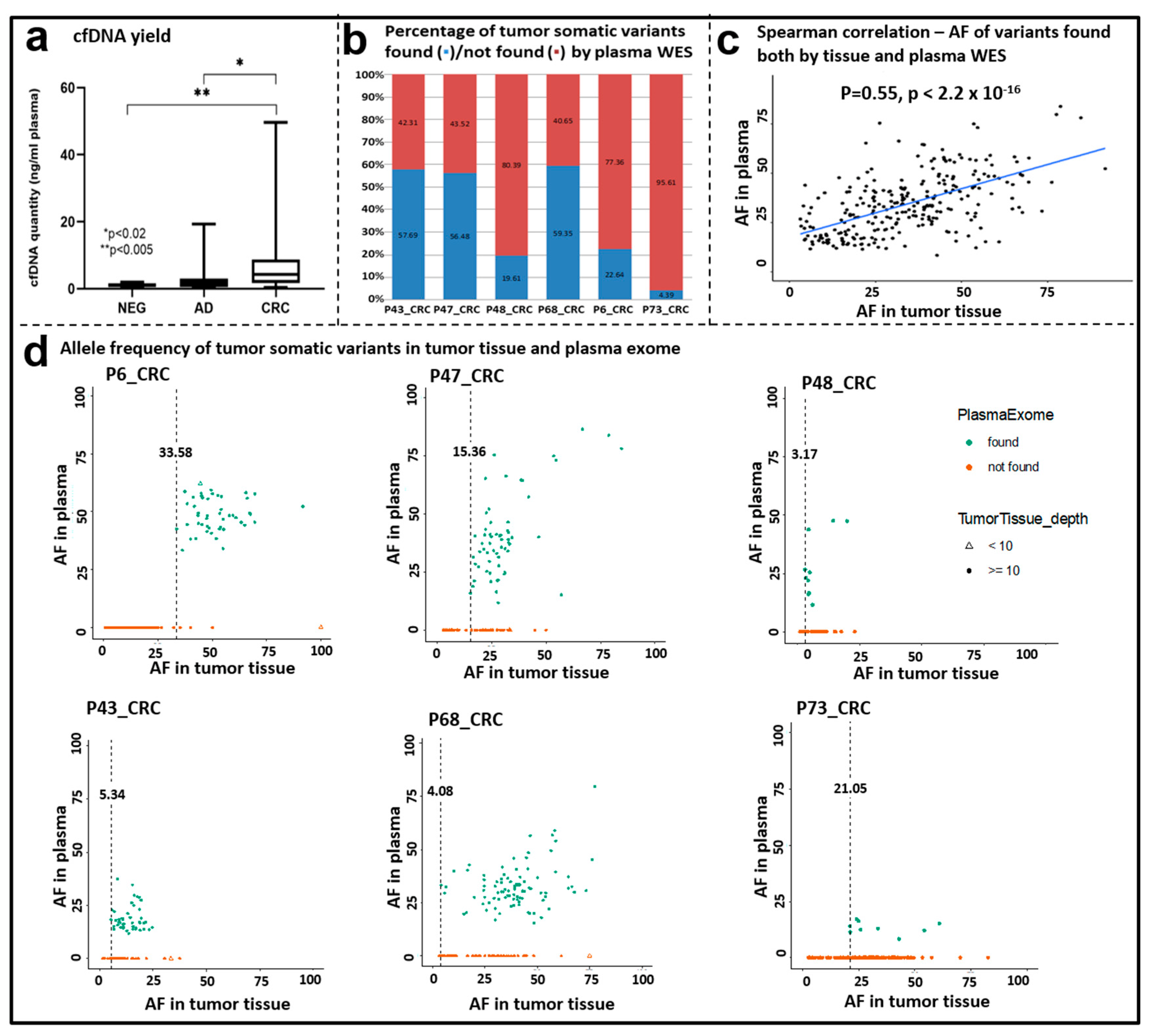
3.7. Whole Exome Sequencing of cfDNA Samples
3.8. CRC-Specific Targeted Sequencing
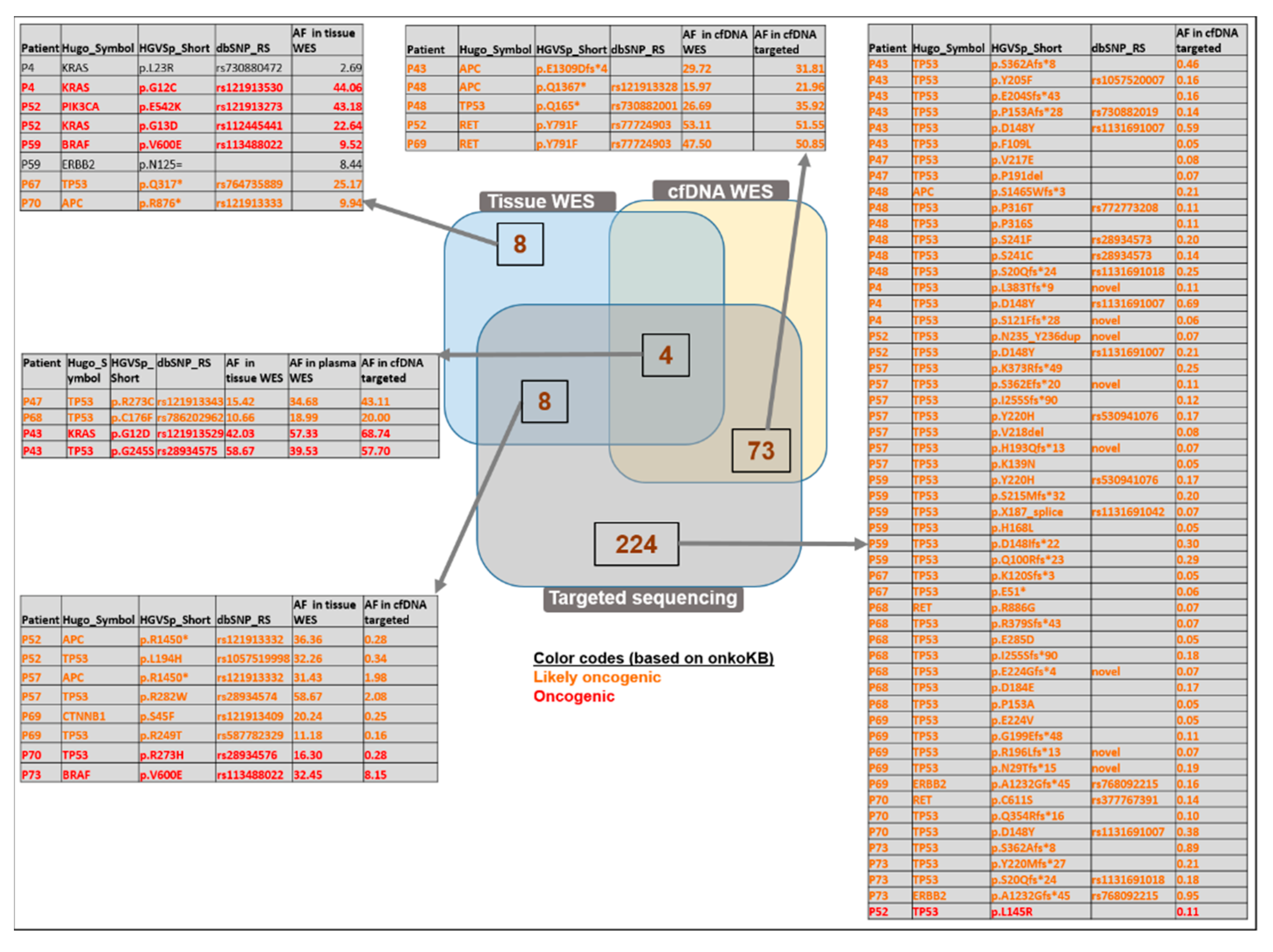
3.9. Comparison of WES and Targeted Panel Sequencing Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xi, Y.; Xu, P. Global Colorectal Cancer Burden in 2020 and Projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef]
- Boyle, P.; Langman, J.S. ABC of Colorectal Cancer: Epidemiology. BMJ 2000, 321, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Rutka, M.; Bor, R.; Molnár, T.; Farkas, K.; Pigniczki, D.; Fábián, A.; Györffy, M.; Bálint, A.; Milassin, Á.; Szücs, M.; et al. Efficacy of the Population-Based Pilot Colorectal Cancer Screening, Csongrád County, Hungary, 2015. Turk. J. Med. Sci. 2020, 50, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Csanádi, M.; Gini, A.; de Koning, H.; Széles, G.; Pitter, J.G.; Oroszi, B.; Pataki, P.; Fadgyas-Freyler, P.; Korponai, G.; Vokó, Z.; et al. Modeling Costs and Benefits of the Organized Colorectal Cancer Screening Programme and Its Potential Future Improvements in Hungary. J. Med. Screen. 2021, 28, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.W.; Krzyzanowska, M.K.; Serra, S.; Knox, J.J.; Dhani, N.C.; Mackay, H.; Hedley, D.; Moore, M.; Liu, G.; Burkes, R.L.; et al. Molecular Profiling of Patients with Advanced Colorectal Cancer: Princess Margaret Cancer Centre Experience. Clin. Color. Cancer 2018, 17, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Fuszek, P.; Horváth, H.C.; Speer, G.; Papp, J.; Haller, P.; Fischer, S.; Halasz, J.; Járay, B.; Székely, E.; Schaff, Z.; et al. Location and Age at Onset of Colorectal Cancer in Hungarian Patients between 1993 and 2004. Anticancer Res. 2006, 26, 527–532. [Google Scholar]
- Tusnády, G.; Gaudi, I.; Rejto, L.; Kásler, M.; Szentirmay, Z. Survival chances of Hungarian cancer patients in the National Cancer Registry. Magy. Onkol. 2008, 52, 339–349. [Google Scholar] [CrossRef]
- Kásler, M.; Ottó, S.; Kenessey, I. The current situation of cancer morbidity and mortality in the light of the National Cancer Registry. Orv. Hetil. 2017, 158, 84–89. [Google Scholar] [CrossRef]
- Inotai, A.; Abonyi-Tóth, Z.; Rokszin, G.; Vokó, Z. Prognosis, Cost, and Occurrence of Colorectal, Lung, Breast, and Prostate Cancer in Hungary. Value Health Reg. Issues 2015, 7, 1–8. [Google Scholar] [CrossRef]
- Vernia, F.; Longo, S.; Stefanelli, G.; Viscido, A.; Latella, G. Dietary Factors Modulating Colorectal Carcinogenesis. Nutrients 2021, 13, 143. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, F.; Kaaks, R.; Vainio, H. Weight Control and Physical Activity in Cancer Prevention. Obes. Rev. 2002, 3, 5–8. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A Genetic Model for Colorectal Tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Innocenti, F.; Ou, F.-S.; Qu, X.; Zemla, T.J.; Niedzwiecki, D.; Tam, R.; Mahajan, S.; Goldberg, R.M.; Bertagnolli, M.M.; Blanke, C.D.; et al. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J. Clin. Oncol. 2019, 37, 1217–1227. [Google Scholar] [CrossRef]
- Taieb, J.; Jung, A.; Sartore-Bianchi, A.; Peeters, M.; Seligmann, J.; Zaanan, A.; Burdon, P.; Montagut, C.; Laurent-Puig, P. The Evolving Biomarker Landscape for Treatment Selection in Metastatic Colorectal Cancer. Drugs 2019, 79, 1375–1394. [Google Scholar] [CrossRef] [PubMed]
- Thota, R.; Yang, M.; Pflieger, L.; Schell, M.J.; Rajan, M.; Davis, T.B.; Wang, H.; Presson, A.; Pledger, W.J.; Yeatman, T.J. APC and TP53 Mutations Predict Cetuximab Sensitivity across Consensus Molecular Subtypes. Cancers 2021, 13, 5394. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 174, 1034–1035. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Vacante, M.; Ciuni, R.; Basile, F.; Biondi, A. The Liquid Biopsy in the Management of Colorectal Cancer: An Overview. Biomedicines 2020, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid Biopsy and Tumor Heterogeneity in Metastatic Solid Tumors: The Potentiality of Blood Samples. J. Exp. Clin. Cancer Res. 2020, 39, 95. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual; Springer International Publishing: Cham, Switzerland, 2018; ISBN 9783319406176. [Google Scholar]
- Bosari, S.; Moneghini, L.; Graziani, D.; Lee, A.K.; Murray, J.J.; Coggi, G.; Viale, G. Bcl-2 oncoprotein in colorectal hyperplastic polyps, adenomas, and adenocarcinomas. Hum. Pathol. 1995, 26, 534–540. [Google Scholar] [CrossRef]
- Katkoori, V.R.; Shanmugam, C.; Jia, X.; Vitta, S.P.; Sthanam, M.; Callens, T.; Messiaen, L.; Chen, D.; Zhang, B.; Bumpers, H.L.; et al. Prognostic Significance and Gene Expression Profiles of p53 Mutations in Microsatellite-Stable Stage III Colorectal Adenocarcinomas. PLoS ONE 2012, 7, e30020. [Google Scholar] [CrossRef] [PubMed]
- Juárez, M.; Egoavil, C.; Rodríguez-Soler, M.; Hernández-Illán, E.; Guarinos, C.; García-Martínez, A.; Alenda, C.; Giner-Calabuig, M.; Murcia, O.; Mangas, C.; et al. KRAS and BRAF Somatic Mutations in Colonic Polyps and the Risk of Metachronous Neoplasia. PLoS ONE 2017, 12, e0184937. [Google Scholar] [CrossRef] [PubMed]
- Poulin, E.J.; Bera, A.K.; Lu, J.; Lin, Y.-J.; Strasser, S.D.; Paulo, J.A.; Huang, T.Q.; Morales, C.; Yan, W.; Cook, J.; et al. Tissue-Specific Oncogenic Activity of KRASA146T. Cancer Discov. 2019, 9, 738–755. [Google Scholar] [CrossRef]
- Liebs, S.; Eder, T.; Klauschen, F.; Schütte, M.; Yaspo, M.-L.; Keilholz, U.; Tinhofer, I.; Kidess-Sigal, E.; Braunholz, D. Applicability of Liquid Biopsies to Represent the Mutational Profile of Tumor Tissue from Different Cancer Entities. Oncogene 2021, 40, 5204–5212. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling Accurate Genetic Variant Discovery to Tens of Thousands of Samples. bioRxiv 2018. bioRxiv:201178. [Google Scholar]
- Zhao, H.; Sun, Z.; Wang, J.; Huang, H.; Kocher, J.-P.; Wang, L. CrossMap: A Versatile Tool for Coordinate Conversion between Genome Assemblies. Bioinformatics 2014, 30, 1006–1007. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving Access to Variant Interpretations and Supporting Evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP—Database for Single Nucleotide Polymorphisms and Other Classes of Minor Genetic Variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.-M.; Huang, W.; Wang, X.-M.M.; Jansen, M.; Ma, X.-J.; Kim, J.; Anderson, C.M.; Wu, X.; Pan, L.; Su, N.; et al. Robust RNA-Based in Situ Mutation Detection Delineates Colorectal Cancer Subclonal Evolution. Nat. Commun. 2017, 8, 1998. [Google Scholar] [CrossRef] [PubMed]
- Morris, V.K.; Lucas, F.A.S.; Overman, M.J.; Eng, C.; Morelli, M.P.; Jiang, Z.-Q.; Luthra, R.; Meric-Bernstam, F.; Maru, D.; Scheet, P.; et al. Clinicopathologic Characteristics and Gene Expression Analyses of Non-KRAS 12/13, RAS-Mutated Metastatic Colorectal Cancer. Ann. Oncol. 2014, 25, 2008–2014. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Perne, C.; Peters, S.; Cartolano, M.; Horpaopan, S.; Grimm, C.; Altmüller, J.; Sommer, A.K.; Hillmer, A.M.; Thiele, H.; Odenthal, M.; et al. Variant Profiling of Colorectal Adenomas from Three Patients of Two Families with MSH3-Related Adenomatous Polyposis. PLoS ONE 2021, 16, e0259185. [Google Scholar] [CrossRef]
- Reynolds, I.S.; Thomas, V.; O’Connell, E.; Fichtner, M.; McNamara, D.A.; Kay, E.W.; Prehn, J.H.M.; Burke, J.P.; Furney, S.J. Mucinous Adenocarcinoma of the Rectum: A Whole Genome Sequencing Study. Front. Oncol. 2020, 10, 1682. [Google Scholar] [CrossRef]
- Huang, H.; Deng, T.; Guo, Y.; Chen, H.; Cui, X.; Duan, J.; Yang, Y.; Guo, Z.; Ba, Y. Gene Mutational Clusters in the Tumors of Colorectal Cancer Patients With a Family History of Cancer. Front. Oncol. 2022, 12, 814397. [Google Scholar] [CrossRef]
- Beal, M.A.; Meier, M.J.; LeBlanc, D.P.; Maurice, C.; O’Brien, J.M.; Yauk, C.L.; Marchetti, F. Chemically Induced Mutations in a MutaMouse Reporter Gene Inform Mechanisms Underlying Human Cancer Mutational Signatures. Commun. Biol. 2020, 3, 438. [Google Scholar] [CrossRef]
- Xiao, J.; Li, W.; Huang, Y.; Huang, M.; Li, S.; Zhai, X.; Zhao, J.; Gao, C.; Xie, W.; Qin, H.; et al. A next-Generation Sequencing-Based Strategy Combining Microsatellite Instability and Tumor Mutation Burden for Comprehensive Molecular Diagnosis of Advanced Colorectal Cancer. BMC Cancer 2021, 21, 282. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor Mutational Burden Is Predictive of Response to Immune Checkpoint Inhibitors in MSI-High Metastatic Colorectal Cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef]
- Packer, L.M.; Williams, S.J.; Callaghan, S.; Gotley, D.C.; McGuckin, M.A. Expression of the cell surface mucin gene family in adenocarcinomas. Int. J. Oncol. 2004, 25, 1119–1126. [Google Scholar]
- Matsuyama, T.; Ishikawa, T.; Mogushi, K.; Yoshida, T.; Iida, S.; Uetake, H.; Mizushima, H.; Tanaka, H.; Sugihara, K. MUC12 mRNA expression is an independent marker of prognosis in stage II and stage III colorectal cancer. Int. J. Cancer 2010, 127, 2292–2299. [Google Scholar] [CrossRef]
- Lin, D.-L.; Wang, L.L.; Zhao, P.; Ran, W.W.; Wang, W.; Zhang, L.X.; Han, M.; Bao, H.; Liu, K.; Wu, X.; et al. Gastrointestinal Goblet Cell Adenocarcinomas Harbor Distinctive Clinicopathological, Immune, and Genomic Landscape. Front. Oncol. 2021, 11, 758643. [Google Scholar] [CrossRef]
- Kwong, K.Y.; Bloom, G.C.; Yang, I.; Boulware, D.; Coppola, D.; Haseman, J.; Chen, E.; McGrath, A.; Makusky, A.J.; Taylor, J.; et al. Synchronous global assessment of gene and protein expression in colorectal cancer progression. Genomics 2005, 86, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, M.S.; Bennett, W.P.; Hollstein, M.; Harris, C.C. Mutations in the p53 tumor suppressor gene: Clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994, 54, 4855–4878. [Google Scholar] [PubMed]
- Intarajak, T.; Udomchaiprasertkul, W.; Bunyoo, C.; Yimnoon, J.; Soonklang, K.; Wiriyaukaradecha, K.; Lamlertthon, W.; Sricharunrat, T.; Chaiwiriyawong, W.; Siriphongpreeda, B.; et al. Genetic Aberration Analysis in Thai Colorectal Adenoma and Early-Stage Adenocarcinoma Patients by Whole-Exome Sequencing. Cancers 2019, 11, 977. [Google Scholar] [CrossRef]
- Beuten, J.; Gelfond, J.A.L.; Franke, J.L.; Shook, S.; Johnson-Pais, T.L.; Thompson, I.M.; Leach, R.J. Multivariate Associations of MSR1, ELAC2, and RNASEL with Prostate Cancer in an Ethnic Diverse Cohort of Men. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Wolff, R.K.; Hoffman, M.D.; Wolff, E.C.; Herrick, J.S.; Sakoda, L.C.; Samowitz, W.S.; Slattery, M.L. Mutation analysis of adenomas and carcinomas of the colon: Early and late drivers. Genes Chromosomes Cancer 2018, 57, 366–376. [Google Scholar] [CrossRef]
- Tejpar, S.; Bertagnolli, M.; Bosman, F.; Lenz, H.J.; Garraway, L.; Waldman, F.; Warren, R.; Bild, A.; Collins-Brennan, D.; Hahn, H.; et al. Prognostic and predictive biomarkers in resected colon cancer: Current status and future perspectives for integrating genomics into biomarker discovery. Oncologist 2010, 15, 390–404. [Google Scholar] [CrossRef] [PubMed]
- Caputo, F.; Santini, C.; Bardasi, C.; Cerma, K.; Casadei-Gardini, A.; Spallanzani, A.; Andrikou, K.; Cascinu, S.; Gelsomino, F. BRAF-Mutated Colorectal Cancer: Clinical and Molecular Insights. Int. J. Mol. Sci. 2019, 20, 5369. [Google Scholar] [CrossRef]
- Yi, C.; Huang, Y.; Yu, X.; Li, X.; Zheng, S.; Ding, K.; Xu, J. Clinicopathologic distribution of KRAS and BRAF mutations in a Chinese population with colorectal cancer precursor lesions. Oncotarget 2016, 7, 17265–17274. [Google Scholar] [CrossRef]
- Lin, S.-H.; Raju, G.S.; Huff, C.; Ye, Y.; Gu, J.; Chen, J.S.; Hildebrandt, M.A.; Liang, H.; Menter, D.G.; Morris, J.; et al. The somatic mutation landscape of premalignant colorectal adenoma. Gut 2018, 67, 1299–1305. [Google Scholar] [CrossRef]
- Jensen, K.H.; Izarzugaza, J.M.G.; Juncker, A.S.; Hansen, R.B.; Hansen, T.F.; Timshel, P.; Blondal, T.; Jensen, T.S.; Rygaard-Hjalsted, E.; Mouritzen, P.; et al. Analysis of a Gene Panel for Targeted Sequencing of Colorectal Cancer Samples. Oncotarget 2018, 9, 9043–9060. [Google Scholar] [CrossRef]
- Zudaire, E.; Cuesta, N.; Murty, V.; Woodson, K.; Adams, L.; Gonzalez, N.; Martínez, A.; Narayan, G.; Kirsch, I.; Franklin, W.; et al. The Aryl Hydrocarbon Receptor Repressor Is a Putative Tumor Suppressor Gene in Multiple Human Cancers. J. Clin. Investig. 2008, 118, 640–650. [Google Scholar] [CrossRef]
- Hou, P.-F.; Jiang, T.; Chen, F.; Shi, P.-C.; Li, H.-Q.; Bai, J.; Song, J. KIF4A Facilitates Cell Proliferation via Induction of p21-Mediated Cell Cycle Progression and Promotes Metastasis in Colorectal Cancer. Cell Death Dis. 2018, 9, 477. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Saito, M.; Saito, K.; Kanke, Y.; Watanabe, Y.; Onozawa, H.; Hayase, S.; Sakamoto, W.; Ishigame, T.; Momma, T.; et al. Enhanced Expression of KIF4A in Colorectal Cancer Is Associated with Lymph Node Metastasis. Oncol. Lett. 2018, 15, 2188–2194. [Google Scholar] [CrossRef]
- Takahashi, K.; Fukushima, K.; Onishi, Y.; Inui, K.; Node, Y.; Fukushima, N.; Honoki, K.; Tsujiuchi, T. Lysophosphatidic Acid (LPA) Signaling via LPA4 and LPA6 Negatively Regulates Cell Motile Activities of Colon Cancer Cells. Biochem. Biophys. Res. Commun. 2017, 483, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Andries, V.; Vandepoele, K.; Staes, K.; Berx, G.; Bogaert, P.; Van Isterdael, G.; Ginneberge, D.; Parthoens, E.; Vandenbussche, J.; Gevaert, K.; et al. NBPF1, a Tumor Suppressor Candidate in Neuroblastoma, Exerts Growth Inhibitory Effects by Inducing a G1 Cell Cycle Arrest. BMC Cancer 2015, 15, 391. [Google Scholar] [CrossRef]
- Liu, Z.; Gomez, C.R.; Espinoza, I.; Le, T.P.T.; Shenoy, V.; Zhou, X. Correlation of Cholesteryl Ester Metabolism to Pathogenesis, Progression and Disparities in Colorectal Cancer. Lipids Health Dis. 2022, 21, 22. [Google Scholar] [CrossRef] [PubMed]
- Amato, C.M.; Hintzsche, J.D.; Wells, K.; Applegate, A.; Gorden, N.T.; Vorwald, V.M.; Tobin, R.P.; Nassar, K.; Shellman, Y.G.; Kim, J.; et al. Pre-Treatment Mutational and Transcriptomic Landscape of Responding Metastatic Melanoma Patients to Anti-PD1 Immunotherapy. Cancers 2020, 12, 1943. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.-H.; Mao, T.; Tao, Y.; Dong, T.; Tang, X.-X.; Ge, G.-H.; Xu, Z.-J. Pan-Cancer Analysis Identifies ITIH1 as a Novel Prognostic Indicator for Hepatocellular Carcinoma. Aging 2021, 13, 11096–11119. [Google Scholar] [CrossRef]
- Huang, Y.; Zhao, J.; Mao, G.; Lee, G.S.; Zhang, J.; Bi, L.; Gu, L.; Chang, Z.; Valentino, J.; Li, G.-M. Identification of Novel Genetic Variants Predisposing to Familial Oral Squamous Cell Carcinomas. Cell Discov. 2019, 5, 57. [Google Scholar] [CrossRef]
- Ma, R.; Jing, C.; Zhang, Y.; Cao, H.; Liu, S.; Wang, Z.; Chen, D.; Zhang, J.; Wu, Y.; Wu, J.; et al. The Somatic Mutation Landscape of Chinese Colorectal Cancer. J. Cancer 2020, 11, 1038–1046. [Google Scholar] [CrossRef]
- Yadamsuren, E.-A.; Nagy, S.; Pajor, L.; Lacza, A.; Bogner, B. Characteristics of Advanced- and Non Advanced Sporadic Polypoid Colorectal Adenomas: Correlation to KRAS Mutations. Pathol. Oncol. Res. 2012, 18, 1077–1084. [Google Scholar] [CrossRef]
- Meng, M.; Zhong, K.; Jiang, T.; Liu, Z.; Kwan, H.Y.; Su, T. The Current Understanding on the Impact of KRAS on Colorectal Cancer. Biomed. Pharmacother. 2021, 140, 111717. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.; Pilati, C.; Blons, H.; Laurent-Puig, P. Beyond KRAS Status and Response to Anti-EGFR Therapy in Metastatic Colorectal Cancer. Pharmacogenomics 2014, 15, 1043–1052. [Google Scholar] [CrossRef] [PubMed]
- Cefalì, M.; Epistolio, S.; Palmarocchi, M.C.; Frattini, M.; De Dosso, S. Research Progress on KRAS Mutations in Colorectal Cancer. J. Cancer Metastasis Treat. 2021, 7, 26. [Google Scholar] [CrossRef]
- Phipps, A.I.; Buchanan, D.D.; Makar, K.W.; Win, A.K.; Baron, J.A.; Lindor, N.M.; Potter, J.D.; Newcomb, P.A. KRAS-Mutation Status in Relation to Colorectal Cancer Survival: The Joint Impact of Correlated Tumour Markers. Br. J. Cancer 2013, 108, 1757–1764. [Google Scholar] [CrossRef]
- Roa, I.; Sánchez, T.; Majlis, A.; Schalper, K. KRAS gene mutation in colorectal cancer. Rev. Med. Chil. 2013, 141, 1166–1172. [Google Scholar] [CrossRef]
- Edkins, S.; O’Meara, S.; Parker, A.; Stevens, C.; Reis, M.; Jones, S.; Greenman, C.; Davies, H.; Dalgliesh, G.; Forbes, S.; et al. Recurrent KRAS Codon 146 Mutations in Human Colorectal Cancer. Cancer Biol. Ther. 2006, 5, 928–932. [Google Scholar] [CrossRef]
- Petit, J.; Carroll, G.; Gould, T.; Pockney, P.; Dun, M.; Scott, R.J. Cell-Free DNA as a Diagnostic Blood-Based Biomarker for Colorectal Cancer: A Systematic Review. J. Surg. Res. 2019, 236, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, S.; Zhan, T.; Betge, J.; Rauscher, B.; Belle, S.; Gutting, T.; Schulte, N.; Jesenofsky, R.; Härtel, N.; Gaiser, T.; et al. Detection of Mutational Patterns in Cell-Free DNA of Colorectal Cancer by Custom Amplicon Sequencing. Mol. Oncol. 2019, 13, 1669–1683. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhu, L.; Song, J.; Wang, G.; Li, P.; Li, W.; Luo, P.; Sun, X.; Wu, J.; Liu, Y.; et al. Liquid Biopsy at the Frontier of Detection, Prognosis and Progression Monitoring in Colorectal Cancer. Mol. Cancer 2022, 21, 86. [Google Scholar] [CrossRef] [PubMed]
- Basnet, S.; Zhang, Z.-Y.; Liao, W.-Q.; Li, S.-H.; Li, P.-S.; Ge, H.-Y. The Prognostic Value of Circulating Cell-Free DNA in Colorectal Cancer: A Meta-Analysis. J. Cancer 2016, 7, 1105–1113. [Google Scholar] [CrossRef]
- Bos, M.K.; Angus, L.; Nasserinejad, K.; Jager, A.; Jansen, M.P.; Martens, J.W.; Sleijfer, S. Whole Exome Sequencing of Cell-Free DNA–A Systematic Review and Bayesian Individual Patient Data Meta-Analysis. Cancer Treat. Res. 2020, 83, 101951. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, R.J.; Lee, J.H.; Strbenac, D.; Yang, J.Y.H.; Menzies, A.M.; Carlino, M.S.; Long, G.V.; Spillane, A.J.; Stretch, J.R.; Saw, R.P.M.; et al. Analysis of the Whole-Exome Sequencing of Tumor and Circulating Tumor DNA in Metastatic Melanoma. Cancers 2019, 11, 1905. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.-Y.; Ho, J.Y.; Kang, J.; Hur, S.Y.; Kim, S.; Choi, Y.J.; Han, M.-R. Whole-Exome Sequencing Reveals Clinical Potential of Circulating Tumor DNA from Peritoneal Fluid and Plasma in Endometrial Cancer. Cancers 2022, 14, 2506. [Google Scholar] [CrossRef]
- Chae, Y.K.; Davis, A.A.; Carneiro, B.A.; Chandra, S.; Mohindra, N.; Kalyan, A.; Kaplan, J.; Matsangou, M.; Pai, S.; Costa, R.; et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget 2016, 7, 65364–65373. [Google Scholar] [CrossRef]
- Lebofsky, R.; Decraene, C.; Bernard, V.; Kamal, M.; Blin, A.; Leroy, Q.; Frio, T.R.; Pierron, G.; Callens, C.; Bieche, I.; et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol. Oncol. 2015, 9, 783–790. [Google Scholar] [CrossRef]
- Liebs, S.; Keilholz, U.; Kehler, I.; Schweiger, C.; Haybäck, J.; Nonnenmacher, A. Detection of mutations in circulating cell-free DNA in relation to disease stage in colorectal cancer. Cancer Med. 2019, 8, 3761–3769. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Kidess, E.; Heirich, K.; Wiggin, M.; Vysotskaia, V.; Visser, B.C.; Marziali, A.; Wiedenmann, B.; Norton, J.A.; Lee, M.; Jeffrey, S.S.; et al. Mutation profiling of tumor DNA from plasma and tumor tissue of colorectal cancer patients with a novel, high-sensitivity multiplexed mutation detection platform. Oncotarget 2015, 6, 2549–2561. [Google Scholar] [CrossRef]
- Chan, R.-H.; Lin, P.-C.; Chen, S.-H.; Lin, S.-C.; Chen, P.-C.; Lin, B.-W.; Shen, M.-R.; Yeh, Y.-M. Clinical Utility of a Cell-Free DNA Assay in Patients With Colorectal Cancer. Front. Oncol. 2021, 11, 589673. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-S.; Lee, C.-C.; Ke, T.-W.; Chang, C.-M.; Chao, D.-S.; Huang, H.-Y.; Chang, J.-G. Molecular Characterization of Colorectal Cancer Using Whole-Exome Sequencing in a Taiwanese Population. Cancer Med. 2019, 8, 3738–3747. [Google Scholar] [CrossRef]
- Samowitz, W.S.; Curtin, K.; Ma, K.-N.; Edwards, S.; Schaffer, D.; Leppert, M.F.; Slattery, M.L. Prognostic Significance of p53 Mutations in Colon Cancer at the Population Level. Int. J. Cancer 2002, 99, 597–602. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, M.; Fang, Y.; Li, J.; Chen, Y.; Jiao, S. TP53 R273C Mutation Is Associated With Poor Prognosis in LGG Patients. Front. Genet. 2022, 13, 720651. [Google Scholar] [CrossRef]
- Li, J.; Yang, L.; Gaur, S.; Zhang, K.; Wu, X.; Yuan, Y.-C.; Li, H.; Hu, S.; Weng, Y.; Yen, Y. Mutants TP53 p.R273H and p.R273C but Not p.R273G Enhance Cancer Cell Malignancy. Hum. Mutat. 2014, 35, 575–584. [Google Scholar] [CrossRef]


| Characteristics | Tissue WES | Plasma WES | Plasma Targeted Panel | |
|---|---|---|---|---|
| Age | n (Years, (Median)) | n (Years, (Median)) | n (Years, (Median)) | |
| N | 18 [21] | 7 [22] | ||
| AD | 27 [23] | 17 [23] | ||
| CRC | 51 [24] | 33 [69.5] | 11 [25] | |
| Sex | n = 92 | n = 57 | n = 11 | |
| Male | 48 | 29 | 7 | |
| Female | 44 | 28 | 4 | |
| Anatomic location of the primary lesion | ||||
| AD | n = 27 | n = 17 | ||
| Cecum | 1 | 1 | ||
| Ascending colon | 1 | |||
| Descending colon | 1 | 1 | ||
| Sigmoid colon | 6 | 3 | ||
| Rectosigmoid | 4 | |||
| Rectum | 2 | 1 | ||
| Multiple locations | 12 | 11 | ||
| CRC | n = 51 | n = 33 | n = 11 | |
| Cecum | 10 | 6 | 2 | |
| Ascending colon | 5 | 5 | 2 | |
| Transverse colon | 3 | 4 | 2 | |
| Descending colon | 1 | |||
| Sigmoid colon | 11 | 8 | 2 | |
| Rectosigmoid | 2 | 1 | ||
| Rectum | 16 | 7 | 2 | |
| Multiple location | 6 | 3 | 1 | |
| Adenoma histology | n = 27 | n = 17 | ||
| Tubular | 19 | 13 | ||
| Tubulovillous | 5 | 3 | ||
| Tubular–tubulovillous | 2 | 1 | ||
| CRC AJCC stage | n = 51 | n = 33 | n = 11 | |
| I | 5 | 5 | 1 | |
| lla | 7 | 8 | 2 | |
| llb | 1 | 1 | ||
| llla | 9 | 2 | ||
| IIIb | 5 | 3 | ||
| lllc | 4 | 4 | 1 | |
| IV | 17 | 10 | 7 | |
| Unkown | 3 | |||
| Hugo_Symbol | SE-CRC | COCA | p-Value | Odds Ratio |
|---|---|---|---|---|
| ZNF717 | 1 | 73 | 5.70 × 10−5 | 6.11 × 10−2 |
| FRG1 | 0 | 55 | 1.06 × 10−4 | 0.00 × 100 |
| MUC3A | 0 | 51 | 1.93 × 10−4 | 0 |
| MUC6 | 3 | 82 | 3.43 × 10−4 | 1.63 × 10−1 |
| KIAA2022 | 8 | 7 | 3.76 × 10−4 | 7.58 × 100 |
| DBF4 | 4 | 0 | 4.26 × 10−4 | Inf |
| KCNJ12 | 0 | 46 | 5.57 × 10−4 | 0 |
| ZSCAN5A | 5 | 2 | 9.68 × 10−4 | 1.57 × 101 |
| FNDC3A | 4 | 1 | 1.90 × 10−3 | 2.46 × 101 |
| ITGB2 | 4 | 1 | 1.90 × 10−3 | 2.46 × 101 |
| OR1M1 | 4 | 1 | 1.90 × 10−3 | 2.46 × 101 |
| SMAD3 | 6 | 5 | 2.00 × 10−3 | 7.66 × 100 |
| CCDC171 | 5 | 3 | 2.29 × 10−3 | 1.05 × 101 |
| LTBP2 | 7 | 8 | 2.56 × 10−3 | 5.66 × 100 |
| MUC2 | 2 | 61 | 2.69 × 10−3 | 1.57 × 10−1 |
| RP11-764K9.4 | 0 | 39 | 2.82 × 10−3 | 0 |
| ALG11 | 3 | 0 | 3.04 × 10−3 | Inf |
| CCDC86 | 3 | 0 | 3.04 × 10−3 | Inf |
| OC90 | 3 | 0 | 3.04 × 10−3 | Inf |
| PRKAR1B | 3 | 0 | 3.04 × 10−3 | Inf |
| SLC25A44 | 3 | 0 | 3.04 × 10−3 | Inf |
| UBOX5 | 3 | 0 | 3.04 × 10−3 | Inf |
| DYNC2H1 | 10 | 18 | 3.30 × 10−3 | 3.73 × 100 |
| HLA-DRB1 | 0 | 34 | 4.59 × 10−3 | 0 |
| ALDH3A2 | 4 | 2 | 5.06 × 10−3 | 1.23 × 101 |
| ALPL | 4 | 2 | 5.06 × 10−3 | 1.23 × 101 |
| MFN2 | 4 | 2 | 5.06 × 10−3 | 1.23 × 101 |
| SEMG2 | 4 | 2 | 5.06 × 10−3 | 1.23 × 101 |
| PTPRU | 6 | 7 | 5.84 × 10−3 | 5.44 × 100 |
| RANBP2 | 6 | 7 | 5.84 × 10−3 | 5.44 × 100 |
| DPP10 | 7 | 10 | 6.07 × 10−3 | 4.50 × 100 |
| FRG2C | 0 | 32 | 7.50 × 10−3 | 0 |
| FRG1B | 0 | 31 | 7.66 × 10−3 | 0 |
| TNXB | 7 | 11 | 8.81 × 10−3 | 4.08 × 100 |
| Hugo_Symbol | SE-CRC | TCGA | p-Value | Odds Ratio |
| MUC12 | 4 | 0 | 5.10 × 10−5 | Inf |
| ZNF729 | 4 | 0 | 5.10 × 10−5 | Inf |
| ENTPD5 | 3 | 0 | 6.21 × 10−4 | Inf |
| TRIM49B | 3 | 0 | 6.21 × 10−4 | Inf |
| BCL2 | 4 | 2 | 6.70 × 10−4 | 2.24 × 101 |
| B4GALNT3 | 5 | 5 | 7.38 × 10−4 | 1.14 × 101 |
| NMBR | 5 | 5 | 7.38 × 10−4 | 1.14 × 101 |
| FOXD3 | 4 | 3 | 1.46 × 10−3 | 1.50 × 101 |
| SLC7A7 | 4 | 3 | 1.46 × 10−3 | 1.50 × 101 |
| ZSCAN5A | 5 | 9 | 4.47 × 10−3 | 6.33 × 100 |
| ALDH3A2 | 4 | 5 | 4.61 × 10−3 | 8.97 × 100 |
| PIK3CA | 4 | 132 | 5.00 × 10−3 | 2.61 × 10−1 |
| C1orf43 | 3 | 2 | 5.47 × 10−3 | 1.65 × 101 |
| CDT1 | 3 | 2 | 5.47 × 10−3 | 1.65 × 101 |
| GPR27 | 3 | 2 | 5.47 × 10−3 | 1.65 × 101 |
| KRTAP13-4 | 3 | 2 | 5.47 × 10−3 | 1.65 × 101 |
| STMN2 | 3 | 2 | 5.47 × 10−3 | 1.65 × 101 |
| ZFHX2 | 3 | 2 | 5.47 × 10−3 | 1.65 × 101 |
| SHCBP1L | 5 | 10 | 6.26 × 10−3 | 5.69 × 100 |
| AL033381.1 | 2 | 0 | 7.41 × 10−3 | Inf |
| ANHX | 2 | 0 | 7.41 × 10−3 | Inf |
| ANKRD31 | 2 | 0 | 7.41 × 10−3 | Inf |
| C11orf85 | 2 | 0 | 7.41 × 10−3 | Inf |
| ERVMER34-1 | 2 | 0 | 7.41 × 10−3 | Inf |
| ERVV-1 | 2 | 0 | 7.41 × 10−3 | Inf |
| FAM153C | 2 | 0 | 7.41 × 10−3 | Inf |
| FAM188B2 | 2 | 0 | 7.41 × 10−3 | Inf |
| FBXL8 | 2 | 0 | 7.41 × 10−3 | Inf |
| FREM3 | 2 | 0 | 7.41 × 10−3 | Inf |
| GCGR | 2 | 0 | 7.41 × 10−3 | Inf |
| IZUMO3 | 2 | 0 | 7.41 × 10−3 | Inf |
| KRTAP9-1 | 2 | 0 | 7.41 × 10−3 | Inf |
| LRRC53 | 2 | 0 | 7.41 × 10−3 | Inf |
| MUC22 | 2 | 0 | 7.41 × 10−3 | Inf |
| OR56B3P | 2 | 0 | 7.41 × 10−3 | Inf |
| PP2D1 | 2 | 0 | 7.41 × 10−3 | Inf |
| RP11-10J21.3 | 2 | 0 | 7.41 × 10−3 | Inf |
| RP11-766F14.2 | 2 | 0 | 7.41 × 10−3 | Inf |
| SAR1B | 2 | 0 | 7.41 × 10−3 | Inf |
| STARD9 | 2 | 0 | 7.41 × 10−3 | Inf |
| TDRD15 | 2 | 0 | 7.41 × 10−3 | Inf |
| TRAV9-1 | 2 | 0 | 7.41 × 10−3 | Inf |
| TRIM64C | 2 | 0 | 7.41 × 10−3 | Inf |
| YRDC | 2 | 0 | 7.41 × 10−3 | Inf |
| ZSCAN5C | 2 | 0 | 7.41 × 10−3 | Inf |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalmár, A.; Galamb, O.; Szabó, G.; Pipek, O.; Medgyes-Horváth, A.; Barták, B.K.; Nagy, Z.B.; Szigeti, K.A.; Zsigrai, S.; Csabai, I.; et al. Patterns of Somatic Variants in Colorectal Adenoma and Carcinoma Tissue and Matched Plasma Samples from the Hungarian Oncogenome Program. Cancers 2023, 15, 907. https://doi.org/10.3390/cancers15030907
Kalmár A, Galamb O, Szabó G, Pipek O, Medgyes-Horváth A, Barták BK, Nagy ZB, Szigeti KA, Zsigrai S, Csabai I, et al. Patterns of Somatic Variants in Colorectal Adenoma and Carcinoma Tissue and Matched Plasma Samples from the Hungarian Oncogenome Program. Cancers. 2023; 15(3):907. https://doi.org/10.3390/cancers15030907
Chicago/Turabian StyleKalmár, Alexandra, Orsolya Galamb, Gitta Szabó, Orsolya Pipek, Anna Medgyes-Horváth, Barbara K. Barták, Zsófia B. Nagy, Krisztina A. Szigeti, Sára Zsigrai, István Csabai, and et al. 2023. "Patterns of Somatic Variants in Colorectal Adenoma and Carcinoma Tissue and Matched Plasma Samples from the Hungarian Oncogenome Program" Cancers 15, no. 3: 907. https://doi.org/10.3390/cancers15030907
APA StyleKalmár, A., Galamb, O., Szabó, G., Pipek, O., Medgyes-Horváth, A., Barták, B. K., Nagy, Z. B., Szigeti, K. A., Zsigrai, S., Csabai, I., Igaz, P., Molnár, B., & Takács, I. (2023). Patterns of Somatic Variants in Colorectal Adenoma and Carcinoma Tissue and Matched Plasma Samples from the Hungarian Oncogenome Program. Cancers, 15(3), 907. https://doi.org/10.3390/cancers15030907









