HO-1089 and HO-1197, Novel Herbal Formulas, Have Antitumor Effects via Suppression of PLK1 (Polo-like Kinase 1) Expression in Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Background
2. Materials and Methods
2.1. Preparation of HO-1089 and HO-1197
2.2. Cell Lines and Culture Conditions
2.3. Cell Cycle Analysis
2.4. Dose–Response Curves
2.5. Protein Separation and Immunoblot Analysis
2.6. Sorafenib Combination Assay
2.7. Reactive Oxygen Species Detection
2.8. Cell Migration Assay
2.9. Microarray Analysis
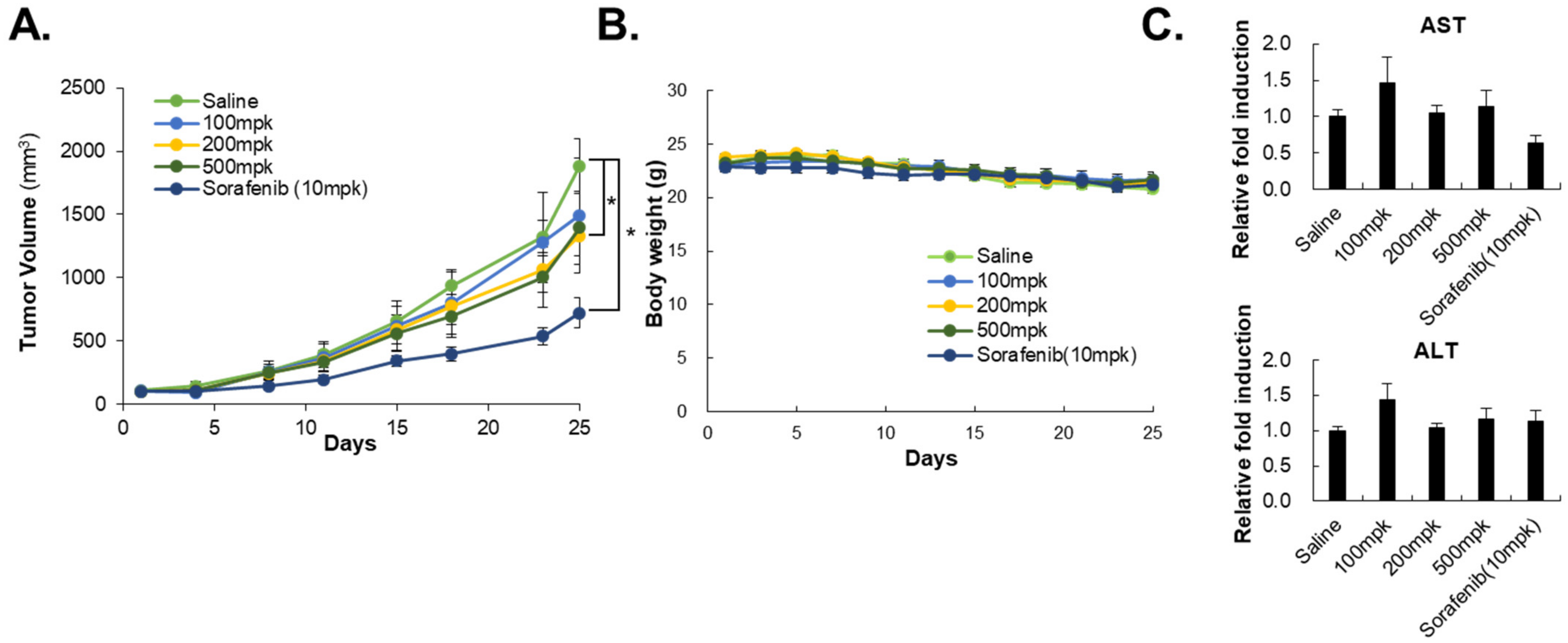
2.10. In Vivo Model of a Tumor Xenograft
2.11. Statistical Analysis
3. Results
3.1. HO-1089 Induces Apoptosis in HCC
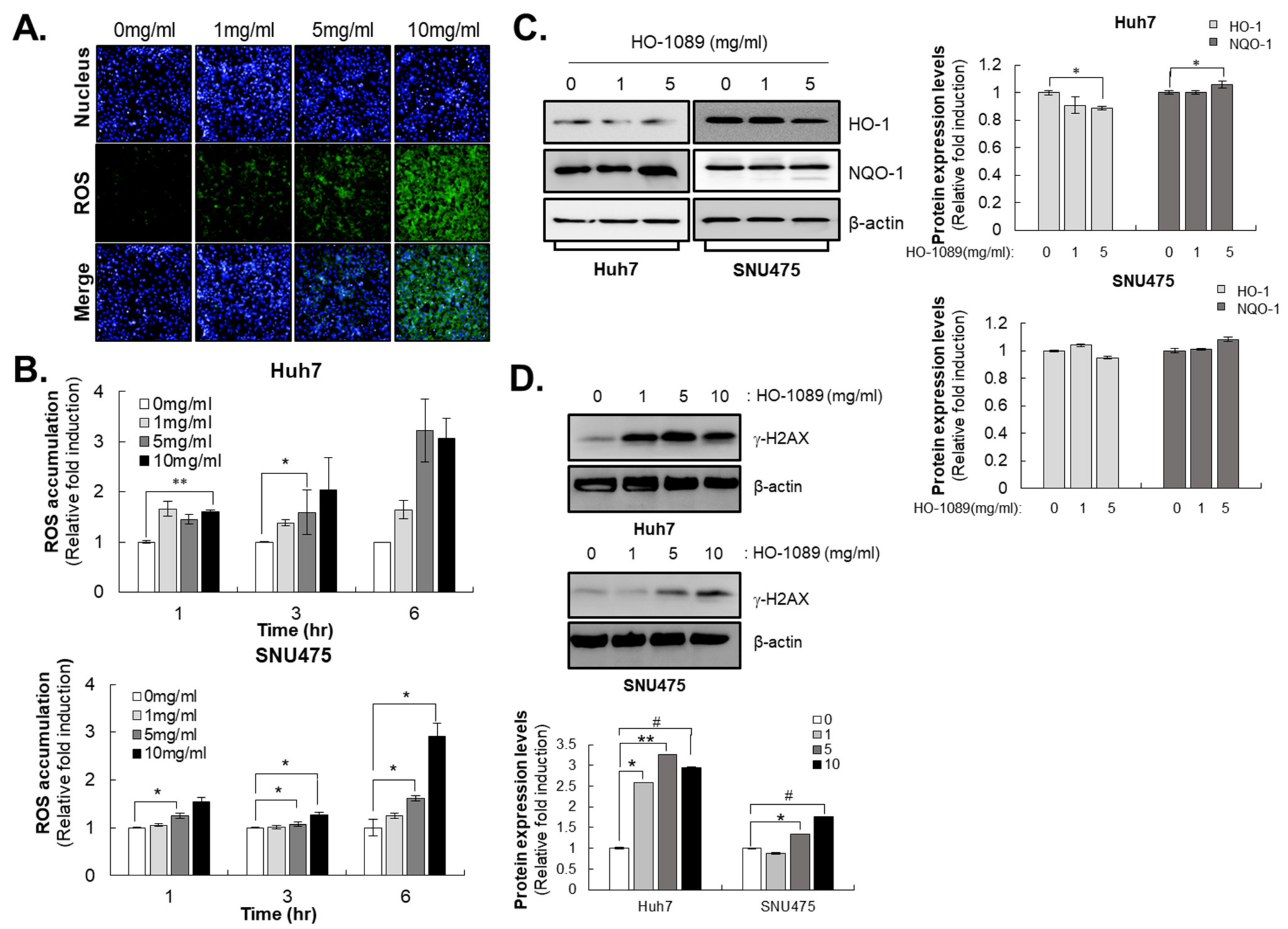
3.2. HO-1089 Induces DNA Damage by ROS Accumulation in HCC
3.3. HO-1089 Attenuates Migration of HCC Cells
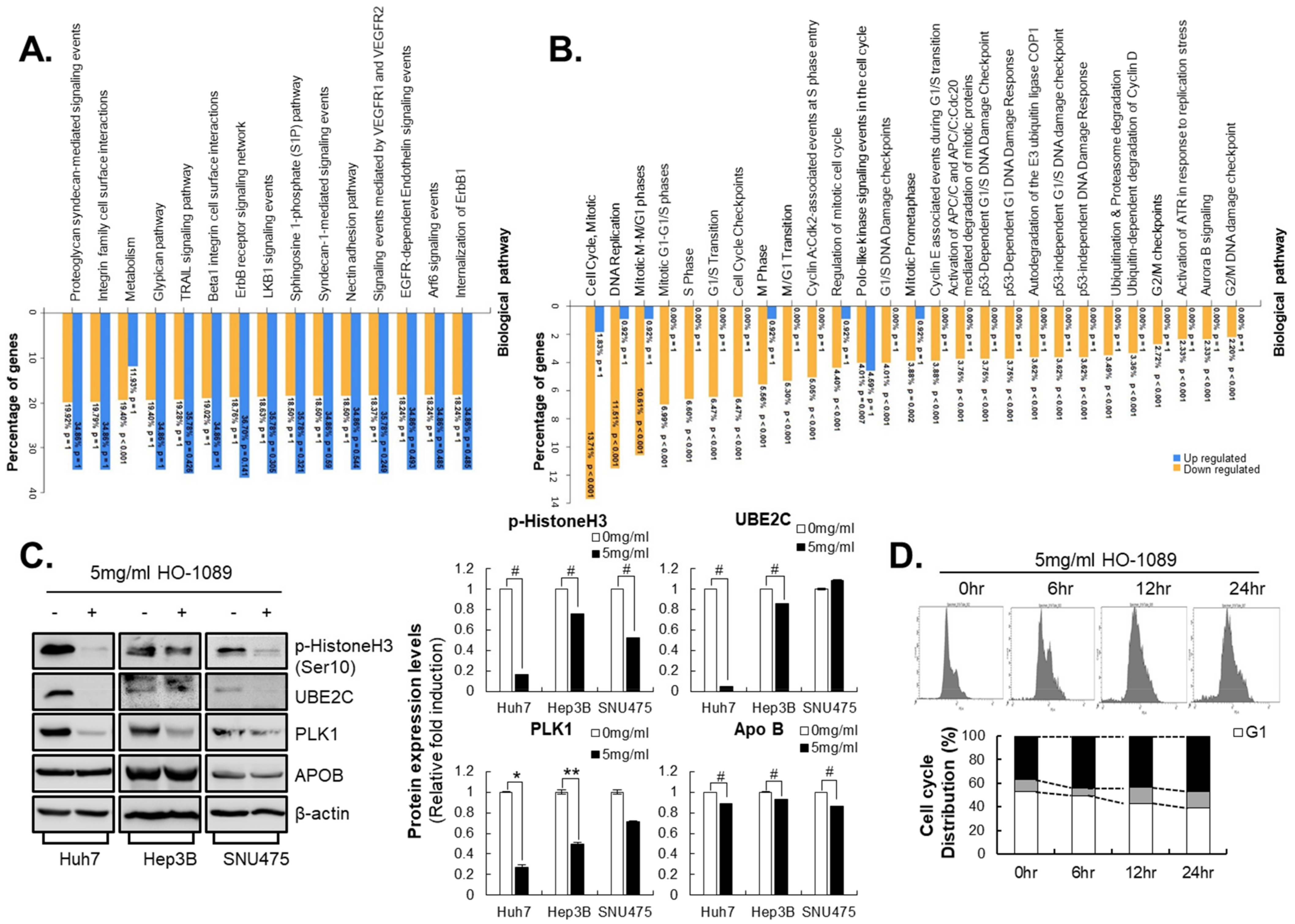
3.4. HO-1089 Induces Cell Cycle Arrest via Suppression of Mitosis-Related Proteins in HCC
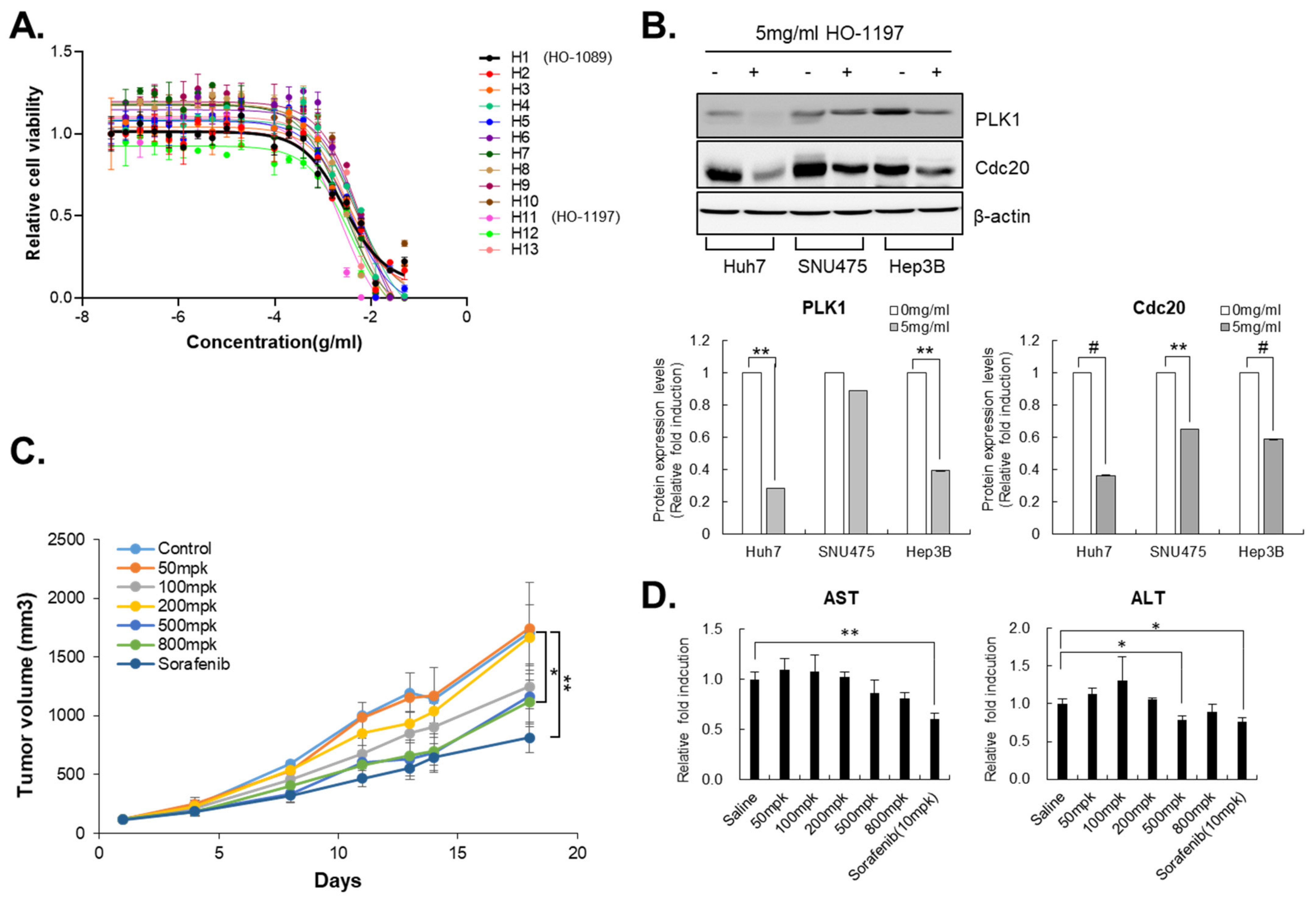
3.5. HO-1197, a Novel Herbal Formula, Has a Better Improved Anticancer Efficacy Than HO-1089
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PLK1 | polo-like kinase 1 |
| HCC | hepatocellular carcinoma |
| HCS | high content screening |
| ROS | reactive oxygen species |
References
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Kedar Mukthinuthalapati, V.V.P.; Sewram, V.; Ndlovu, N.; Kimani, S.; Abdelaziz, A.O.; Chiao, E.Y.; Abou-Alfa, G.K. Hepatocellular Carcinoma in Sub-Saharan Africa. JCO Glob. Oncol. 2021, 7, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Hilmi, M.; Neuzillet, C.; Calderaro, J.; Lafdil, F.; Pawlotsky, J.M.; Rousseau, B. Angiogenesis and immune checkpoint inhibitors as therapies for hepatocellular carcinoma: Current knowledge and future research directions. J. ImmunoTherapy Cancer 2019, 7, 333. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef]
- Eso, Y.; Taura, K.; Seno, H. Does immune checkpoint inhibitor exhibit limited efficacy against non-viral hepatocellular carcinoma?: A review of clinical trials. Hepatol. Res. 2022, 52, 67–74. [Google Scholar] [CrossRef]
- Shek, D.; Read, S.A.; Nagrial, A.; Carlino, M.S.; Gao, B.; George, J.; Ahlenstiel, G. Immune-Checkpoint Inhibitors for Advanced Hepatocellular Carcinoma: A Synopsis of Response Rates. Oncologist 2021, 26, e1216–e1225. [Google Scholar] [CrossRef]
- Huang, M.Y.; Zhang, L.L.; Ding, J.; Lu, J.J. Anticancer drug discovery from Chinese medicinal herbs. Chin. Med. 2018, 13, 35. [Google Scholar] [CrossRef]
- Zhang, Y.; Lou, Y.; Wang, J.; Yu, C.; Shen, W. Research Status and Molecular Mechanism of the Traditional Chinese Medicine and Antitumor Therapy Combined Strategy Based on Tumor Microenvironment. Front. Immunol. 2020, 11, 609705. [Google Scholar] [CrossRef]
- Xie, Q.; Yang, K.M.; Heo, G.E.; Song, M. Literature based discovery of alternative TCM medicine for adverse reactions to depression drugs. BMC Bioinform. 2020, 21 (Suppl. 5), 405. [Google Scholar] [CrossRef]
- Fan, Y.; Ma, Z.; Zhao, L.; Wang, W.; Gao, M.; Jia, X.; Ouyang, H.; He, J. Anti-tumor activities and mechanisms of Traditional Chinese medicines formulas: A review. Biomed. Pharmacother. 2020, 132, 110820. [Google Scholar] [CrossRef]
- Huang, L.; Xu, H.; Wu, T.; Li, G. Hedyotis diffusa Willd. Suppresses Hepatocellular Carcinoma via Downregulating AKT/mTOR Pathways. Evid. Based Complement. Altern. Med. 2021, 2021, 5210152. [Google Scholar] [CrossRef]
- Yeh, Y.C.; Chen, H.Y.; Yang, S.H.; Lin, Y.H.; Chiu, J.H.; Lin, Y.H.; Chen, J.L. Hedyotis diffusa Combined with Scutellaria barbata Are the Core Treatment of Chinese Herbal Medicine Used for Breast Cancer Patients: A Population-Based Study. Evid. Based Complement. Altern. Med. 2014, 2014, 202378. [Google Scholar] [CrossRef]
- Lu, W.L.; Yang, T.; Song, Q.J.; Fang, Z.Q.; Pan, Z.Q.; Liang, C.; Jia, D.W.; Peng, P.K. Akebia trifoliata (Thunb.) Koidz Seed Extract inhibits human hepatocellular carcinoma cell migration and invasion in vitro. J. Ethnopharmacol. 2019, 234, 204–215. [Google Scholar] [CrossRef]
- Lu, W.L.; Ren, H.Y.; Liang, C.; Zhang, Y.Y.; Xu, J.; Pan, Z.Q.; Liu, X.M.; Wu, Z.H.; Fang, Z.Q. Akebia trifoliate (Thunb.) Koidz Seed Extract Inhibits the Proliferation of Human Hepatocellular Carcinoma Cell Lines via Inducing Endoplasmic Reticulum Stress. Evid. Based Complement. Altern. Med. 2014, 2014, 192749. [Google Scholar] [CrossRef]
- Song, Y.G.; Kang, L.; Tian, S.; Cui, L.L.; Li, Y.; Bai, M.; Fang, X.Y.; Cao, L.H.; Coleman, K.; Miao, M.S. Study on the anti-hepatocarcinoma effect and molecular mechanism of Prunella vulgaris total flavonoids. J. Ethnopharmacol. 2021, 273, 113891. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, L.; Feng, J.; Lin, W.; Cai, Q.; Peng, J. Spica Prunellae extract suppresses the growth of human colon carcinoma cells by targeting multiple oncogenes via activating miR-34a. Oncol. Rep. 2017, 38, 1895–1901. [Google Scholar] [CrossRef]
- Seo, W.G.; Hwang, J.C.; Kang, S.K.; Jin, U.H.; Suh, S.J.; Moon, S.K.; Kim, C.H. Suppressive effect of Zedoariae rhizoma on pulmonary metastasis of B16 melanoma cells. J. Ethnopharmacol. 2005, 101, 249–257. [Google Scholar] [CrossRef]
- Pan, Z.; Zhuang, J.; Ji, C.; Cai, Z.; Liao, W.; Huang, Z. Curcumin inhibits hepatocellular carcinoma growth by targeting VEGF expression. Oncol. Lett. 2018, 15, 4821–4826. [Google Scholar] [CrossRef]
- Li, J.; Wei, H.; Liu, Y.; Li, Q.; Guo, H.; Guo, Y.; Chang, Z. Curcumin Inhibits Hepatocellular Carcinoma via Regulating miR-21/TIMP3 Axis. Evid. Based Complement. Altern. Med. 2020, 2020, 2892917. [Google Scholar] [CrossRef]
- Jang, J.W.; Song, Y.; Kim, S.H.; Kim, J.S.; Kim, K.M.; Choi, E.K.; Kim, J.; Seo, H.R. CD133 confers cancer stem-like cell properties by stabilizing EGFR-AKT signaling in hepatocellular carcinoma. Cancer Lett. 2017, 389, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.C.; Li, Y.X.; Cheng, J.T. The use of herbal medicine in cancer-related anorexia/ cachexia treatment around the world. Curr. Pharm. Des. 2012, 18, 4819–4826. [Google Scholar] [CrossRef] [PubMed]
- Damery, S.; Gratus, C.; Grieve, R.; Warmington, S.; Jones, J.; Routledge, P.; Greenfield, S.; Dowswell, G.; Sherriff, J.; Wilson, S. The use of herbal medicines by people with cancer: A cross-sectional survey. Br. J. Cancer 2011, 104, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Ernst, E. Prevalence of use of complementary/alternative medicine: A systematic review. Bull. World Health Organ. 2000, 78, 252–257. [Google Scholar]
- Yeh, M.H.; Wu, H.C.; Lin, N.W.; Hsieh, J.J.; Yeh, J.W.; Chiu, H.P.; Wu, M.C.; Tsai, T.Y.; Yeh, C.C.; Li, T.M. Long-term use of combined conventional medicine and Chinese herbal medicine decreases the mortality risk of patients with lung cancer. Complement. Ther. Med. 2020, 52, 102427. [Google Scholar] [CrossRef]
- Li, Y.; Martin, R.C., 2nd. Herbal medicine and hepatocellular carcinoma: Applications and challenges. Evid. Based Complement. Altern. Med. 2011, 2011, 541209. [Google Scholar] [CrossRef]
- Barr, F.A.; Sillje, H.H.; Nigg, E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004, 5, 429–440. [Google Scholar] [CrossRef]
- Gheghiani, L.; Loew, D.; Lombard, B.; Mansfeld, J.; Gavet, O. PLK1 Activation in Late G2 Sets Up Commitment to Mitosis. Cell Rep. 2017, 19, 2060–2073. [Google Scholar] [CrossRef]
- Takai, N.; Hamanaka, R.; Yoshimatsu, J.; Miyakawa, I. Polo-like kinases (Plks) and cancer. Oncogene 2005, 24, 287–291. [Google Scholar] [CrossRef]
- Mok, W.C.; Wasser, S.; Tan, T.; Lim, S.G. Polo-like kinase 1, a new therapeutic target in hepatocellular carcinoma. World J. Gastroenterol. 2012, 18, 3527–3536. [Google Scholar] [CrossRef]
- Eckerdt, F.; Yuan, J.; Strebhardt, K. Polo-like kinases and oncogenesis. Oncogene 2005, 24, 267–276. [Google Scholar] [CrossRef]
- Driscoll, D.L.; Chakravarty, A.; Bowman, D.; Shinde, V.; Lasky, K.; Shi, J.; Vos, T.; Stringer, B.; Amidon, B.; D’Amore, N.; et al. Plk1 inhibition causes post-mitotic DNA damage and senescence in a range of human tumor cell lines. PLoS ONE 2014, 9, e111060. [Google Scholar] [CrossRef]
- Hikichi, Y.; Honda, K.; Hikami, K.; Miyashita, H.; Kaieda, I.; Murai, S.; Uchiyama, N.; Hasegawa, M.; Kawamoto, T.; Sato, T.; et al. TAK-960, a novel, orally available, selective inhibitor of polo-like kinase 1, shows broad-spectrum preclinical antitumor activity in multiple dosing regimens. Mol. Cancer Ther. 2012, 11, 700–709. [Google Scholar] [CrossRef]
- Ieta, K.; Ojima, E.; Tanaka, F.; Nakamura, Y.; Haraguchi, N.; Mimori, K.; Inoue, H.; Kuwano, H.; Mori, M. Identification of overexpressed genes in hepatocellular carcinoma, with special reference to ubiquitin-conjugating enzyme E2C gene expression. Int. J. Cancer 2007, 121, 33–38. [Google Scholar] [CrossRef]
- Mo, C.H.; Gao, L.; Zhu, X.F.; Wei, K.L.; Zeng, J.J.; Chen, G.; Feng, Z.B. The clinicopathological significance of UBE2C in breast cancer: A study based on immunohistochemistry, microarray and RNA-sequencing data. Cancer Cell Int. 2017, 17, 83. [Google Scholar] [CrossRef]
- Shen, Z.; Jiang, X.; Zeng, C.; Zheng, S.; Luo, B.; Zeng, Y.; Ding, R.; Jiang, H.; He, Q.; Guo, J.; et al. High expression of ubiquitin-conjugating enzyme 2C (UBE2C) correlates with nasopharyngeal carcinoma progression. BMC Cancer 2013, 13, 192. [Google Scholar] [CrossRef]
- Fujita, T.; Ikeda, H.; Taira, N.; Hatoh, S.; Naito, M.; Doihara, H. Overexpression of UbcH10 alternates the cell cycle profile and accelerate the tumor proliferation in colon cancer. BMC Cancer 2009, 9, 87. [Google Scholar] [CrossRef]
- Okamoto, Y.; Ozaki, T.; Miyazaki, K.; Aoyama, M.; Miyazaki, M.; Nakagawara, A. UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res. 2003, 63, 4167–4173. [Google Scholar]


| No | Log2Ratio1 | Gene Name | Protein |
|---|---|---|---|
| 1 | −4.7104928 | HIST1H2AI | histone cluster 1, H2ai |
| 2 | −4.235694 | ID1 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| 3 | −4.0174427 | KNG1 | kininogen 1 |
| 4 | −3.903392 | ZNF93 | zinc finger protein 93 |
| 5 | −3.821343 | ORM2 | orosomucoid 2 |
| 6 | −3.7330516 | HIST1H2BM | histone cluster 1, H2bm |
| 7 | −3.505795 | APOB | apolipoprotein B |
| 8 | −3.503174 | F8A1 | coagulation factor VIII-associated 1 |
| 9 | −3.465386 | HIST1H3I | histone cluster 1, H3i |
| 10 | −3.3550105 | AGMAT | agmatinase |
| 11 | −3.288377 | UBE2C | ubiquitin-conjugating enzyme E2C |
| 12 | −3.236855 | IFITM2 | interferon induced transmembrane protein 2 |
| 13 | −3.230311 | PLK1 | polo-like kinase 1 |
| 14 | −3.194956 | NAR-E | small ILF3/NF90-associated RNA E |
| 15 | −3.133499 | HABP2 | hyaluronan binding protein 2 |
| 16 | −3.1148475 | NCOA5 | nuclear receptor coactivator 5 |
| 17 | −3.097753 | APOA2 | apolipoprotein A-II |
| 18 | −3.097341 | CDCA8 | cell division cycle associated 8 |
| 19 | −3.094738 | APOH | apolipoprotein H |
| 20 | −3.073547 | CDC20 | cell division cycle 20 |
| Name of Cell Line | IC50 (mg/mL) |
|---|---|
| Huh7 | 2.75 |
| H460 | 8.92 |
| A549 | 5.49 |
| AGS | 1.5 |
| HT29 | 2.27 |
| MDA-MB231 | 2.18 |
| SNU213 | 14.5 |
| A375 | 1.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Lee, S.-Y.; Kim, S.; Choi, I.; Kim, N.; Park, J.; Seo, H.R. HO-1089 and HO-1197, Novel Herbal Formulas, Have Antitumor Effects via Suppression of PLK1 (Polo-like Kinase 1) Expression in Hepatocellular Carcinoma. Cancers 2023, 15, 851. https://doi.org/10.3390/cancers15030851
Song Y, Lee S-Y, Kim S, Choi I, Kim N, Park J, Seo HR. HO-1089 and HO-1197, Novel Herbal Formulas, Have Antitumor Effects via Suppression of PLK1 (Polo-like Kinase 1) Expression in Hepatocellular Carcinoma. Cancers. 2023; 15(3):851. https://doi.org/10.3390/cancers15030851
Chicago/Turabian StyleSong, Yeonhwa, Su-Yeon Lee, Sanghwa Kim, Inhee Choi, Namjeong Kim, Jongmin Park, and Haeng Ran Seo. 2023. "HO-1089 and HO-1197, Novel Herbal Formulas, Have Antitumor Effects via Suppression of PLK1 (Polo-like Kinase 1) Expression in Hepatocellular Carcinoma" Cancers 15, no. 3: 851. https://doi.org/10.3390/cancers15030851
APA StyleSong, Y., Lee, S.-Y., Kim, S., Choi, I., Kim, N., Park, J., & Seo, H. R. (2023). HO-1089 and HO-1197, Novel Herbal Formulas, Have Antitumor Effects via Suppression of PLK1 (Polo-like Kinase 1) Expression in Hepatocellular Carcinoma. Cancers, 15(3), 851. https://doi.org/10.3390/cancers15030851





