Gene-Transcript Expression in Urine Supernatant and Urine Cell-Sediment Are Different but Equally Useful for Detecting Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Cohort
2.2. Cell and EV Fractions
2.3. NanoString Data: Feature Selection and Analysis
2.4. RT-PCR Analysis
3. Results
3.1. Gene-Transcript Expression in Urine EV and Urine Cell-Sediment Are Different
3.1.1. No Differences Were Observed in the Expression of Housekeeper Genes between Cancer and Non-Cancer in Both Urine Fractions
3.1.2. EV and Cell Fractions Have a Different Profile of Tissue of Origin
3.1.3. Most Gene-Probes Are Differentially Expressed between EV and Cell Fractions
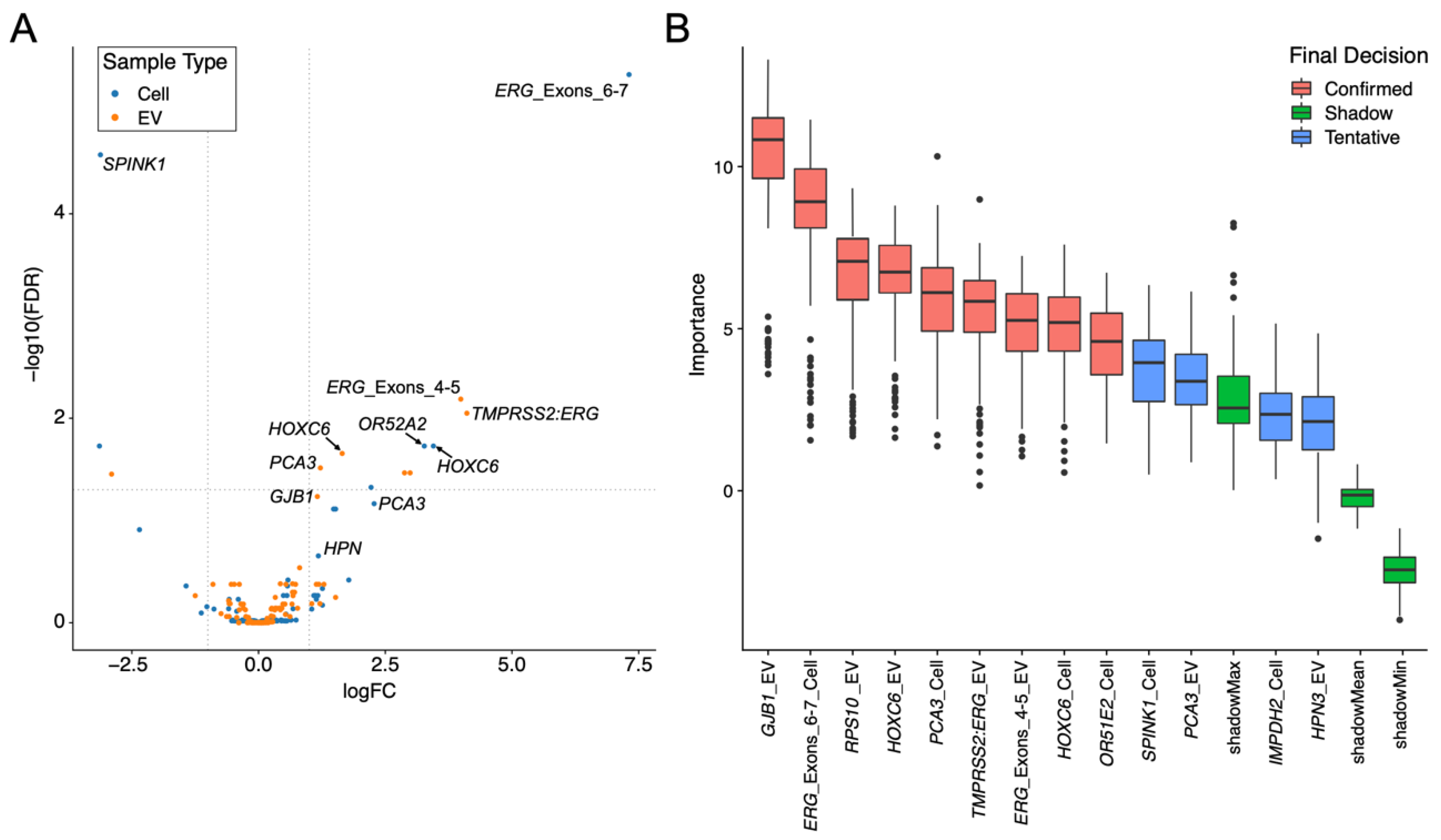
3.1.4. Expression Changes between Non-Cancer and Cancer Are Different in the EV and Cell Fractions
3.2. Expression Levels from RT-PCR and NanoString Are Strongly Correlated for Both EV and Cell Urine Fractions
3.3. Each Urine Fraction Has Different Genes That Are Important in Predicting the Presence of Prostate Cancer
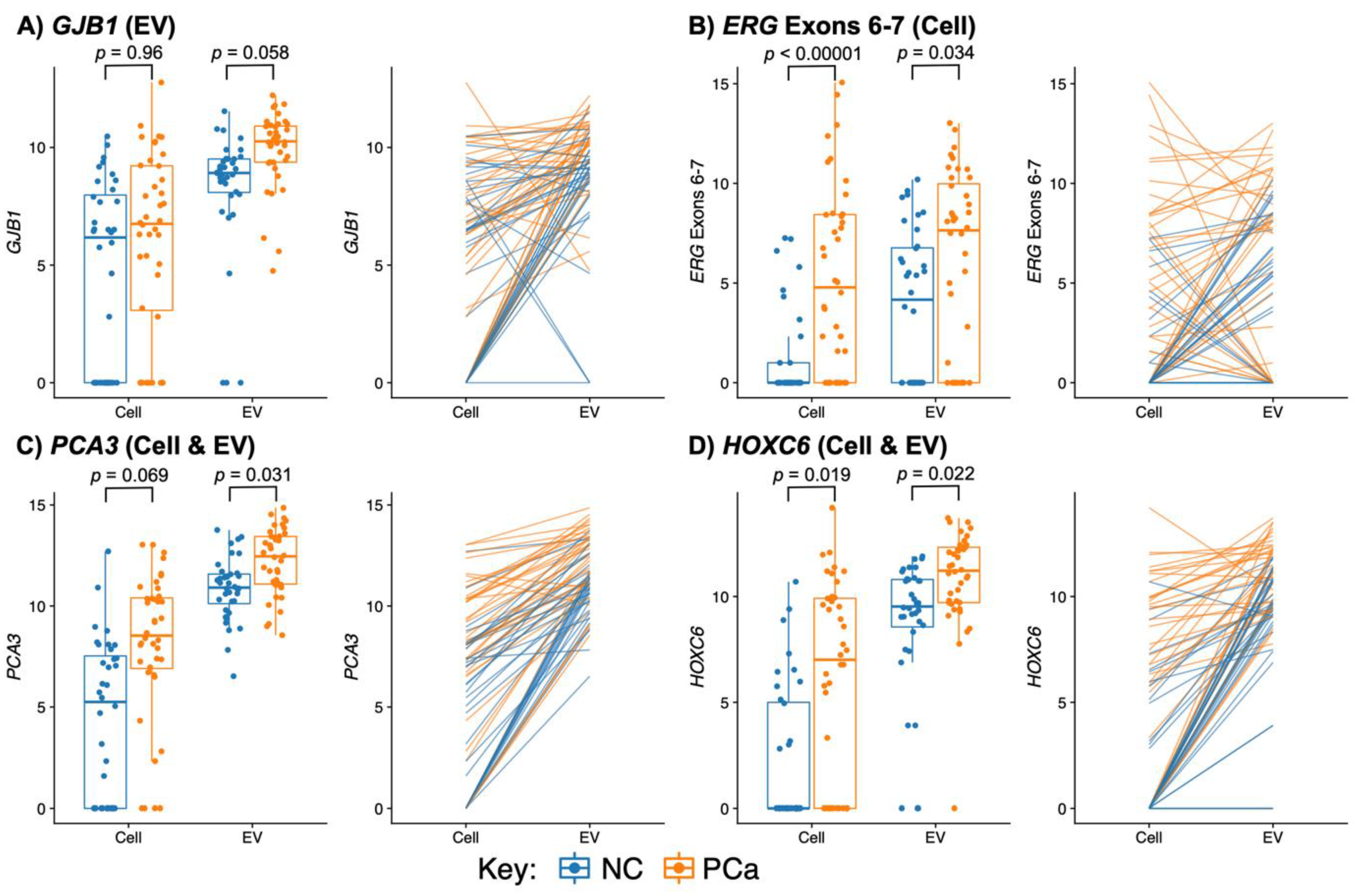
3.3.1. PCA3 and HOXC6 Were Useful in Both EV and Cell Sediment Fractions
3.3.2. EV Fraction Genes Useful for Prostate Cancer Detection
3.3.3. Cellular Genes Useful for PCa Detection
3.3.4. TMPRSS2:ERG and ERG Probes Are Useful Biomarkers in Urine
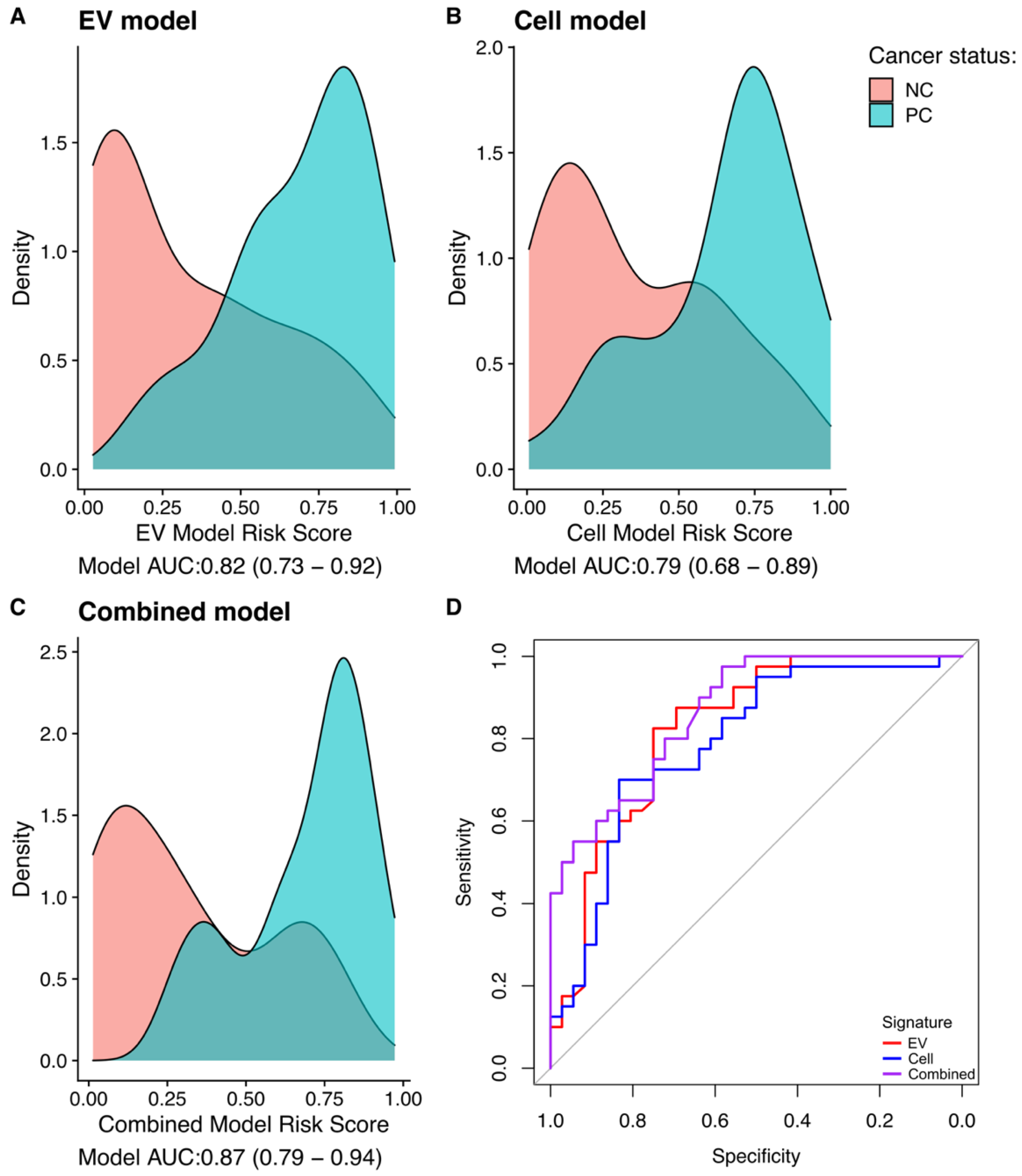
3.4. Gene-Transcript Expression in Urine EV and Urine Cell-Sediment Are Equally Useful for Detecting Prostate Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- National Institute for Health and Care Excellence (NICE). Prostate Cancer: Diagnosis and Management; National Institute for Health and Care Excellence (NICE): London, UK, 2021; pp. 1–58. [Google Scholar]
- Bernemann, C.; Schnoeller, T.J.; Luedeke, M.; Steinestel, K.; Boegemann, M.; Schrader, A.J.; Steinestel, J. Expression of AR-V7 in Circulating Tumour Cells Does Not Preclude Response to Next Generation Androgen Deprivation Therapy in Patients with Castration Resistant Prostate Cancer. Eur. Urol. 2017, 71, 1–3. [Google Scholar] [CrossRef]
- Hennigan, S.T.; Trostel, S.Y.; Terrigino, N.T.; Voznesensky, O.S.; Schaefer, R.J.; Whitlock, N.C.; Wilkinson, S.; Carrabba, N.V.; Atway, R.; Shema, S.; et al. Low Abundance of Circulating Tumor DNA in Localized Prostate Cancer. JCO Precis. Oncol. 2019, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Herbut, P.A.; Lubin, E. Cancer Cells in Prostatic Secretions. J. Urol. 1947, 57, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.J.; Welton, J.; Staffurth, J.; Court, J.; Mason, M.D.; Tabi, Z.; Clayton, A. Can urinary exosomes act as treatment response markers in prostate cancer? J. Transl. Med. 2009, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Hessels, D.; Gunnewiek, J.M.K.; van Oort, I.; Karthaus, H.F.; van Leenders, G.J.; van Balken, B.; Kiemeney, L.; Witjes, J.; Schalken, J.A. DD3PCA3-based Molecular Urine Analysis for the Diagnosis of Prostate Cancer. Eur. Urol. 2003, 44, 8–16. [Google Scholar] [CrossRef]
- Hessels, D.; Smit, F.P.; Verhaegh, G.W.; Witjes, J.A.; Cornel, E.B.; Schalken, J.A. Detection of TMPRSS2-ERG Fusion Transcripts and Prostate Cancer Antigen 3 in Urinary Sediments May Improve Diagnosis of Prostate Cancer. Clin. Cancer Res. 2007, 13, 5103–5108. [Google Scholar] [CrossRef]
- Miranda, K.C.; Bond, D.T.; McKee, M.; Skog, J.; Păunescu, T.G.; Da Silva, N.; Brown, D.; Russo, L.M. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010, 78, 191–199. [Google Scholar] [CrossRef]
- Connell, S.P.; Yazbek-Hanna, M.; McCarthy, F.; Hurst, R.; Webb, M.; Curley, H.; Walker, H.; Mills, R.; Ball, R.Y.; Sanda, M.G.; et al. A four-group urine risk classifier for predicting outcomes in patients with prostate cancer. BJU Int. 2019, 124, 609–620. [Google Scholar] [CrossRef]
- Dijkstra, S.; Birker, I.L.; Smit, F.P.; Leyten, G.H.J.M.; De Reijke, T.M.; Van Oort, I.M.; Mulders, P.F.A.; Jannink, S.A.; Schalken, J.A. Prostate Cancer Biomarker Profiles in Urinary Sediments and Exosomes. J. Urol. 2014, 191, 1132–1138. [Google Scholar] [CrossRef]
- Pellegrini, K.L.; Patil, D.; Douglas, K.J.; Lee, G.; Wehrmeyer, K.; Torlak, M.; Clark, J.; Cooper, C.S.; Moreno, C.S.; Sanda, M.G. Detection of prostate cancer-specific transcripts in extracellular vesicles isolated from post-DRE urine. Prostate 2017, 77, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, R.J.; Dijkstra, S.; Jannink, S.A.; Steffens, M.G.; van Oort, I.M.; Mulders, P.F.; Schalken, J.A. Comparative analysis of prostate cancer specific biomarkers PCA3 and ERG in whole urine, urinary sediments and exosomes. Clin. Chem. Lab. Med. 2016, 54, 483–492. [Google Scholar] [CrossRef] [PubMed]
- DeAntoni, E.P.; Crawford, E.D.; Oesterling, J.E.; Ross, C.A.; Berger, E.R.; McLeod, D.G.; Staggers, F.; Stone, N.N. Age- and race-specific reference ranges for prostate-specific antigen from a large community-based study. Urology 1996, 48, 234–239. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Connell, S.P.; O’Reilly, E.; Tuzova, A.; Webb, M.; Hurst, R.; Mills, R.; Zhao, F.; Bapat, B.; Cooper, C.S.; Perry, A.S.; et al. Development of a multivariable risk model integrating urinary cell DNA methylation and cell-free RNA data for the detection of significant prostate cancer. Prostate 2020, 80, 547–558. [Google Scholar] [CrossRef]
- Sequeiros, T.; Bastarós, J.M.; Sánchez, M.; Rigau, M.; Montes, M.; Placer, J.; Planas, J.; de Torres, I.; Reventós, J.; Pegtel, D.M.; et al. Urinary biomarkers for the detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia. Prostate 2015, 75, 1102–1113. [Google Scholar] [CrossRef]
- Clark, J.; Merson, S.; Jhavar, S.; Flohr, P.; Edwards, S.; Foster, C.S.; Eeles, R.; Martin, F.L.; Phillips, D.H.; Crundwell, M.; et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene 2006, 26, 2667–2673. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Routledge: New York, NY, USA, 2013; ISBN 0-8058-0283-5. [Google Scholar] [CrossRef]
- Wen, S.; Wei, Y.; Zen, C.; Xiong, W.; Niu, Y.; Zhao, Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer 2020, 19, 171. [Google Scholar] [CrossRef]
- Gervais, F.G.; Singaraja, R.; Xanthoudakis, S.; Gutekunst, C.-A.; Leavitt, B.R.; Metzler, M.; Hackam, A.S.; Tam, J.; Vaillancourt, J.P.; Houtzager, V.; et al. Recruitment and activation of caspase-8 by the Huntingtin-interacting protein Hip-1 and a novel partner Hippi. Nature 2002, 4, 95–105. [Google Scholar] [CrossRef]
- De Jonge, H.J.M.; Fehrmann, R.; De Bont, E.S.J.M.; Hofstra, R.; Gerbens, F.; Kamps, W.A.; de Vries, E.; Van Der Zee, A.G.J.; Meerman, G.J.T.; Ter Elst, A. Evidence Based Selection of Housekeeping Genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Zhang, Z.; Wang, G. BTG2: A rising star of tumor suppressors (Review). Int. J. Oncol. 2014, 46, 459–464. [Google Scholar] [CrossRef]
- Shabani, S.; Elahi, E.; Bahraniasl, M.; Babaheidarian, P.; Sadeghpour, A.; Majidzadeh, T.; Talebi, A.; Mahjoubi, F. Multi-stage analysis of FOXM1, PYROXD1, hTERT, PPARA, PIM3, BMI1 and MCTP1 expression patterns in colorectal cancer. Carcinogenesis 2022, 9, 10. [Google Scholar]
- Lu, Z.; Qi, L.; Bo, X.J.; Liu, G.D.; Wang, J.M.; Li, G. Expression of CD26 and CXCR4 in prostate carcinoma and its relationship with clinical parameters. J. Res. Med. Sci. 2013, 18, 647–652. [Google Scholar] [PubMed]
- Su, W.-P.; Sun, L.-N.; Yang, S.-L.; Zhao, H.; Zeng, T.-Y.; Wu, W.-Z.; Wang, D. Apolipoprotein C1 promotes prostate cancer cell proliferation in vitro. J. Biochem. Mol. Toxicol. 2018, 32, e22158. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Shi, Y. Histone H1.2 as a trigger for apoptosis. Nat. Struct. Mol. Biol. 2003, 10, 983–985. [Google Scholar] [CrossRef] [PubMed]
- Itkonen, H.M.; Brown, M.; Urbanucci, A.; Tredwell, G.; Lau, C.H.; Barfeld, S.; Hart, C.; Guldvik, I.J.; Takhar, M.; Heemers, H.V.; et al. Lipid degradation promotes prostate cancer cell survival. Oncotarget 2017, 8, 38264–38275. [Google Scholar] [CrossRef]
- Yu, C.; Cao, H.; He, X.; Sun, P.; Feng, Y.; Chen, L.; Gong, H. Cyclin-dependent kinase inhibitor 3 (CDKN3) plays a critical role in prostate cancer via regulating cell cycle and DNA replication signaling. Biomed. Pharmacother. 2017, 96, 1109–1118. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.-W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent Fusion of TMPRSS2 and ETS Transcription Factor Genes in Prostate Cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef]
- Maraj, B.; Markham, A. Prostate-specific membrane antigen (FOLH1): Recent advances in characterising this putative prostate cancer gene. Prostate Cancer Prostatic Dis. 1999, 2, 180–185. [Google Scholar] [CrossRef]
- Hamid, A.R.A.H.; Hoogland, A.M.; Smit, F.; Jannink, S.; van Rijt-van de Westerlo, C.; Jansen, C.F.J.; van Leenders, G.J.L.H.; Verhaegh, G.W.; Schalken, J.A. The role of HOXC6 in prostate cancer development. Prostate 2015, 75, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Rigau, M.; Morote, J.; Mir, M.C.; Ballesteros, C.; Ortega, I.; Sanchez, A.; Colás, E.; Garcia, M.; Ruiz, A.; Abal, M.; et al. PSGR and PCA3 as biomarkers for the detection of prostate cancer in urine. Prostate 2010, 70, 1760–1767. [Google Scholar] [CrossRef] [PubMed]
- Haas, M. The Na-K-Cl cotransporters. Am. J. Physiol. Physiol. 1994, 267, C869–C885. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Rhodes, D.R.; Yu, J.; Varambally, S.; Mehra, R.; Perner, S.; Demichelis, F.; Helgeson, B.E.; Laxman, B.; Morris, D.S.; et al. The Role of SPINK1 in ETS Rearrangement-Negative Prostate Cancers. Cancer Cell 2008, 13, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Lobban, E.D.; Smith, B.A.; Hall, G.D.; Harnden, P.; Roberts, P.; Selby, P.J.; Trejdosiewicz, L.K.; Southgate, J. Uroplakin Gene Expression by Normal and Neoplastic Human Urothelium. Am. J. Pathol. 1998, 153, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Groskopf, J.; Aubin, S.M.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. APTIMA PCA3 Molecular Urine Test: Development of a Method to Aid in the Diagnosis of Prostate Cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef]
- De Kok, J.B.; Verhaegh, G.W.; Roelofs, R.W.; Hessels, D.; Kiemeney, L.A.; Aalders, T.W.; Swinkels, D.W.; Schalken, J.A. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002, 62, 2695–2698. [Google Scholar] [PubMed]
- Heidenreich, A.; Bellmunt, J.; Bolla, M.; Joniau, S.; Mason, M.; Matveev, V.; Mottet, N.; Schmid, H.-P.; van der Kwast, T.; Wiegel, T.; et al. EAU Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Treatment of Clinically Localised Disease. Eur. Urol. 2011, 59, 61–71. [Google Scholar] [CrossRef]
- Alshalalfa, M.; Verhaegh, G.W.; Gibb, E.A.; Santiago-Jiménez, M.; Erho, N.; Jordan, J.; Yousefi, K.; Lam, L.L.; Kolisnik, T.; Chelissery, J.; et al. Low PCA3 expression is a marker of poor differentiation in localized prostate tumors: Exploratory analysis from 12,076 patients. Oncotarget 2017, 8, 50804–50813. [Google Scholar] [CrossRef]
- Lee, G.L.; Dobi, A.; Srivastava, S. Diagnostic performance of the PCA3 urine test. Nat. Rev. Urol. 2011, 8, 123–124. [Google Scholar] [CrossRef]
- Gou, X.; Huang, P.; Mou, C.; Luo, Y. The PCA3 test for guiding repeat biopsy of prostate cancer and its cut-off score: A systematic review and meta-analysis. Asian J. Androl. 2014, 16, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Liu, P.; Young, A.N.; Yin-Goen, Q.; Lim, S.D.; Laycock, N.; Amin, M.B.; Carney, J.K.; Marshall, F.F.; Petros, A.J.; et al. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene 2005, 24, 188–198. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, X.; Song, P.; Wang, H.; Wang, X. HOXC6 in the prognosis of prostate cancer. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2715–2720. [Google Scholar] [CrossRef] [PubMed]
- Pepe, P.; Dibenedetto, G.; Pepe, L.; Pennisi, M. Multiparametric MRI Versus SelectMDx Accuracy in the Diagnosis of Clinically Significant PCa in Men Enrolled in Active Surveillance. In Vivo 2019, 34, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, R.J.; van der Leest, M.M.G.; Israël, B.; Hannink, G.; YantiSetiasti, A.; Cornel, E.B.; de Kaa, C.A.H.-V.; Klaver, O.S.; Sedelaar, J.P.M.; Van Criekinge, W.; et al. Clinical use of the SelectMDx urinary-biomarker test with or without mpMRI in prostate cancer diagnosis: A prospective, multicenter study in biopsy-naïve men. Prostate Cancer Prostatic Dis. 2021, 24, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Annamalai, L.; Chakraborty, S.; Johnson, K.; Song, X.-H.; Batra, S.K.; Mehta, P.P. Androgen-regulated Formation and Degradation of Gap Junctions in Androgen-responsive Human Prostate Cancer Cells. Mol. Biol. Cell 2006, 17, 5400–5416. [Google Scholar] [CrossRef]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 3698. [Google Scholar] [CrossRef]
- Erdem, H.; Çırakoğlu, A.; Benli, E.; Çankaya, S. Association of Connexin 32 with Prostate Volume and PSA Level in Prostatic Adenocarcinoma and Adenomyomatous Hyperplasia. J. Urol. Surg. 2020, 7, 103–108. [Google Scholar] [CrossRef]
- Luna-Coronell, J.A.; Vierlinger, K.; Gamperl, M.; Hofbauer, J.; Berger, I.; Weinhäusel, A. The prostate cancer immunome: In silico functional analysis of antigenic proteins from microarray profiling with IgG. Proteomics 2016, 16, 1204–1214. [Google Scholar] [CrossRef]
- Wu, Q. Hepsin and prostate cancer. Front. Biosci. 2007, 12, 5052–5059. [Google Scholar] [CrossRef]
- Xia, C.; Ma, W.; Wang, F.; Hua, S.-B.; Liu, M. Identification of a prostate-specific G-protein coupled receptor in prostate cancer. Oncogene 2001, 20, 5903–5907. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.-D.; Zhang, Y.-Q.; He, H.-C.; Dai, Q.-S.; Qin, G.-Q.; Chen, J.-H.; Cai, C.; Fu, X.; Bi, X.-C.; Zhu, J.-G.; et al. Identification of novel serological tumor markers for human prostate cancer using integrative transcriptome and proteome analysis. Med. Oncol. 2012, 29, 2877–2888. [Google Scholar] [CrossRef] [PubMed]
- Laxman, B.; Morris, D.S.; Yu, J.; Siddiqui, J.; Cao, J.; Mehra, R.; Lonigro, R.J.; Tsodikov, A.; Wei, J.T.; Tomlins, S.A.; et al. A First-Generation Multiplex Biomarker Analysis of Urine for the Early Detection of Prostate Cancer. Cancer Res 2008, 68, 645–649. [Google Scholar] [CrossRef]
- Attard, G.; Clark, J.; Ambroisine, L.; Fisher, G.; Kovacs, G.; Flohr, P.; Berney, D.; Foster, C.S.; Fletcher, A.; Gerald, W.L.; et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene 2008, 27, 253–263. [Google Scholar] [CrossRef]
- Clark, J.P.; Cooper, C.S. ETS gene fusions in prostate cancer. Nat. Rev. Urol. 2009, 6, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.; Attard, G.; Jhavar, S.; Flohr, P.; Reid, A.; De-Bono, J.; Eeles, R.; Scardino, P.; Cuzick, J.; Fisher, G.; et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene 2007, 27, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Mehra, R.; Han, B.; Tomlins, S.A.; Wang, L.; Menon, A.; Wasco, M.J.; Shen, R.; Montie, J.E.; Chinnaiyan, A.M.; Shah, R.B. Heterogeneity of TMPRSS2 Gene Rearrangements in Multifocal Prostate Adenocarcinoma: Molecular Evidence for an Independent Group of Diseases. Cancer Res. 2007, 67, 7991–7995. [Google Scholar] [CrossRef] [PubMed]
- Young, A.; Palanisamy, N.; Siddiqui, J.; Wood, D.P.; Wei, J.T.; Chinnaiyan, A.M.; Kunju, L.P.; Tomlins, S.A. Correlation of Urine TMPRSS2:ERG and PCA3 to ERG+ and Total Prostate Cancer Burden. Am. J. Clin. Pathol. 2012, 138, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2015, 70, 45–53. [Google Scholar] [CrossRef]
- Fujita, K.; Pavlovich, C.P.; Netto, G.J.; Konishi, Y.; Isaacs, W.B.; Ali, S.; De Marzo, A.; Meeker, A.K. Specific detection of prostate cancer cells in urine by multiplex immunofluorescence cytology. Hum. Pathol. 2009, 40, 924–933. [Google Scholar] [CrossRef]
- Quek, S.-I.; Wong, O.M.; Chen, A.; Borges, G.T.; Ellis, W.J.; Salvanha, D.M.; Vêncio, R.Z.; Weaver, B.; Ench, Y.M.; Leach, R.J.; et al. Processing of voided urine for prostate cancer RNA biomarker analysis. Prostate 2015, 75, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Leyten, G.H.; Hessels, D.; Smit, F.P.; Jannink, S.A.; de Jong, H.; Melchers, W.J.; Cornel, E.B.; de Reijke, T.M.; Vergunst, H.; Kil, P.; et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin. Cancer Res. 2015, 21, 3061–3070. [Google Scholar] [CrossRef] [PubMed]
- Donovan, M.J.; Noerholm, M.; Bentink, S.; Belzer, S.; Skog, J.; O’Neill, V.; Cochran, J.S.; Brown, G.A. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015, 18, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Motamedinia, P.; Scott, A.N.; Bate, K.L.; Sadeghi, N.; Salazar, G.; Shapiro, E.; Ahn, J.; Lipsky, M.; Lin, J.; Hruby, G.W.; et al. Urine Exosomes for Non-Invasive Assessment of Gene Expression and Mutations of Prostate Cancer. PLoS ONE 2016, 11, e0154507. [Google Scholar] [CrossRef]
- Ahmed, H.U.; El-Shater Bosaily, A.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef]
- Kim, S.; Schroeder, C.M.; Xie, X.S. Single-Molecule Study of DNA Polymerization Activity of HIV-1 Reverse Transcriptase on DNA Templates. J. Mol. Biol. 2010, 395, 995–1006. [Google Scholar] [CrossRef]
- Bracht, J.W.P.; Gimenez-Capitan, A.; Huang, C.-Y.; Potie, N.; Pedraz-Valdunciel, C.; Warren, S.; Rosell, R.; Molina-Vila, M.A. Analysis of extracellular vesicle mRNA derived from plasma using the nCounter platform. Sci. Rep. 2021, 11, 3712. [Google Scholar] [CrossRef]
- David, A.; Mabjeesh, N.; Azar, I.; Biton, S.; Engel, S.; Bernstein, J.; Romano, J.; Avidor, Y.; Waks, T.; Eshhar, Z.; et al. Unusual Alternative Splicing within the Human Kallikrein Genes KLK2 and KLK3 Gives Rise to Novel Prostate-specific Proteins. J. Biol. Chem. 2002, 277, 18084–18090. [Google Scholar] [CrossRef]
- Hopkins, J.F.; Sabelnykova, V.Y.; Weischenfeldt, J.; Simon, R.; Aguiar, J.A.; Alkallas, R.; Heisler, L.E.; Zhang, J.; Watson, J.D.; Chua, M.L.K.; et al. Mitochondrial mutations drive prostate cancer aggression. Nat. Commun. 2017, 8, 656. [Google Scholar] [CrossRef]

| Characteristic | Non-Cancer | Prostate Cancer |
|---|---|---|
| Number of Samples | 36 | 40 |
| Age (IQR) | 66 (12.3) | 70.0 (9.5) |
| PSA (ng/mL) (IQR) | 6.3 (4.0) | 9.1 (5.4) |
| Biopsy results (N, %) | No Bx (7, 19%) Neg Bx (29, 79%) | GG1 (6, 26%) GG2 (17, 74%) GG3 (6, 35 %) GG ≥ 4 (11, 65%) |
| Gene Name | Log2 FC | p-Value | Expression | Gene Function/Link to PCa |
|---|---|---|---|---|
| NEAT1 | −8.65 | <0.00001 | Lower in EVs | Bone metastasis [21] |
| MIR4435.1HG lOC541471 | −4.43 | <0.00001 | Lower in EVs | No publications on PCa |
| IFT57 | 2.81 | <0.00001 | Higher in EVs | Pro-apoptotic [22] |
| B2M | −3.9 | <0.00001 | Lower in EVs | Housekeeper [23] |
| BTG2 | −3.67 | <0.00001 | Lower in EVs | Tumor-suppressor [24] |
| MCTP1 | −9.42 | <0.00001 | Lower in EVs | Calcium signaling [25] |
| DPP4 | 3.05 | <0.00001 | Higher in EVs | Overexpressed in PCa [26] |
| APOC1 | −8.16 | <0.00001 | Lower in EVs | Overexpressed in PCa [27] |
| H1.2 | 1.68 | <0.00001 | Higher in EVs | Apoptotic response to DNA damage [28] |
| ECI2 | 2.09 | <0.00001 | Higher in EVs | Knock-out may have a therapeutic response in PCa [29] |
| Gene Name | Fraction | log FC | p Value | Expression | Gene Function/Link to Cancer |
|---|---|---|---|---|---|
| CDKN3 | EVs | −2.9 | 0.0352 | Lower in PCa | Overexpressed in PCa [30] |
| ERG_Exons_4-5 | EVs | 3.99 | 0.00650 | Higher in PCa | Overexpression due to TMPRSS2:ERG translocation [31] |
| ERG_Exons_6-7 | Cell | 7.31 | 4.40 × 10−6 | Higher in PCa | As above |
| ERG_Exons_6-7 | EVs | 2.88 | 0.0342 | Higher in PCa | As above |
| FOLH1 | Cell | 2.22 | 0.0474 | Higher in PCa | Overexpressed in PCa [32] |
| HOXC6 | Cell | 3.45 | 0.0187 | Higher in PCa | Overexpressed in PCa urine sediment [33] |
| HOXC6 | EVs | 1.65 | 0.0221 | Higher in PCa | Overexpressed in PCa urine sediment [33] |
| OR51E2 | Cell | 3.27 | 0.0187 | Higher in PCa | Overexpressed in PCa urine sediment [34] |
| PCA3 | EVs | 1.22 | 0.0306 | Higher in PCa | Overexpressed in PCa urine cell sediment [7] |
| SLC12A1 | EVs | 2.99 | 0.0342 | Higher in PCa | Kidney-specific [35] |
| SPINK1 | Cell | −3.12 | 0.0187 | Lower in PCa | Overexpressed in TMPRSS2:ERG-negative PCa [36] |
| TMPRSS2:ERG | EVs | 4.11 | 0.0893 | Higher in PCa | Translocation in 50% of PCa [31] |
| UPK2 | Cell | −3.14 | 0.0187 | Lower in PCa | bladder-specific expression [37] |
| Gene | Experiment | Spearman Correlation Coefficient | p-Value |
|---|---|---|---|
| FOLH1 | Cells | 0.71 | <0.00001 |
| FOLH1 | EV | 0.68 | <0.00001 |
| KLK3 | Cells | 0.7 | <0.00001 |
| KLK3 | EV | 0.51 | <0.00001 |
| OR51E2 | Cells | 0.77 | <0.00001 |
| OR51E2 | EV | 0.74 | <0.00001 |
| PCA3 | Cells | 0.88 | <0.00001 |
| PCA3 | EV | 0.95 | <0.00001 |
| RPLP2 | Cells | 0.86 | <0.00001 |
| RPLP2 | EV | 0.79 | <0.00001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yazbek Hanna, M.; Winterbone, M.; O’Connell, S.P.; Olivan, M.; Hurst, R.; Mills, R.; Cooper, C.S.; Brewer, D.S.; Clark, J. Gene-Transcript Expression in Urine Supernatant and Urine Cell-Sediment Are Different but Equally Useful for Detecting Prostate Cancer. Cancers 2023, 15, 789. https://doi.org/10.3390/cancers15030789
Yazbek Hanna M, Winterbone M, O’Connell SP, Olivan M, Hurst R, Mills R, Cooper CS, Brewer DS, Clark J. Gene-Transcript Expression in Urine Supernatant and Urine Cell-Sediment Are Different but Equally Useful for Detecting Prostate Cancer. Cancers. 2023; 15(3):789. https://doi.org/10.3390/cancers15030789
Chicago/Turabian StyleYazbek Hanna, Marcelino, Mark Winterbone, Shea P. O’Connell, Mireia Olivan, Rachel Hurst, Rob Mills, Colin S. Cooper, Daniel S. Brewer, and Jeremy Clark. 2023. "Gene-Transcript Expression in Urine Supernatant and Urine Cell-Sediment Are Different but Equally Useful for Detecting Prostate Cancer" Cancers 15, no. 3: 789. https://doi.org/10.3390/cancers15030789
APA StyleYazbek Hanna, M., Winterbone, M., O’Connell, S. P., Olivan, M., Hurst, R., Mills, R., Cooper, C. S., Brewer, D. S., & Clark, J. (2023). Gene-Transcript Expression in Urine Supernatant and Urine Cell-Sediment Are Different but Equally Useful for Detecting Prostate Cancer. Cancers, 15(3), 789. https://doi.org/10.3390/cancers15030789








