Plasma nanoDSF Denaturation Profile at Baseline Is Predictive of Glioblastoma EGFR Status
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Patient Cohort
2.2. EGFR Amplification Determination by Next Generation Sequencing
2.3. MGMT Promoter Methylation Determination by Pyrosequencing
2.4. Sample Analysis by NanoDSF
2.5. AI Analyses
3. Results
3.1. Patient Characteristics
3.2. Molecular Profile
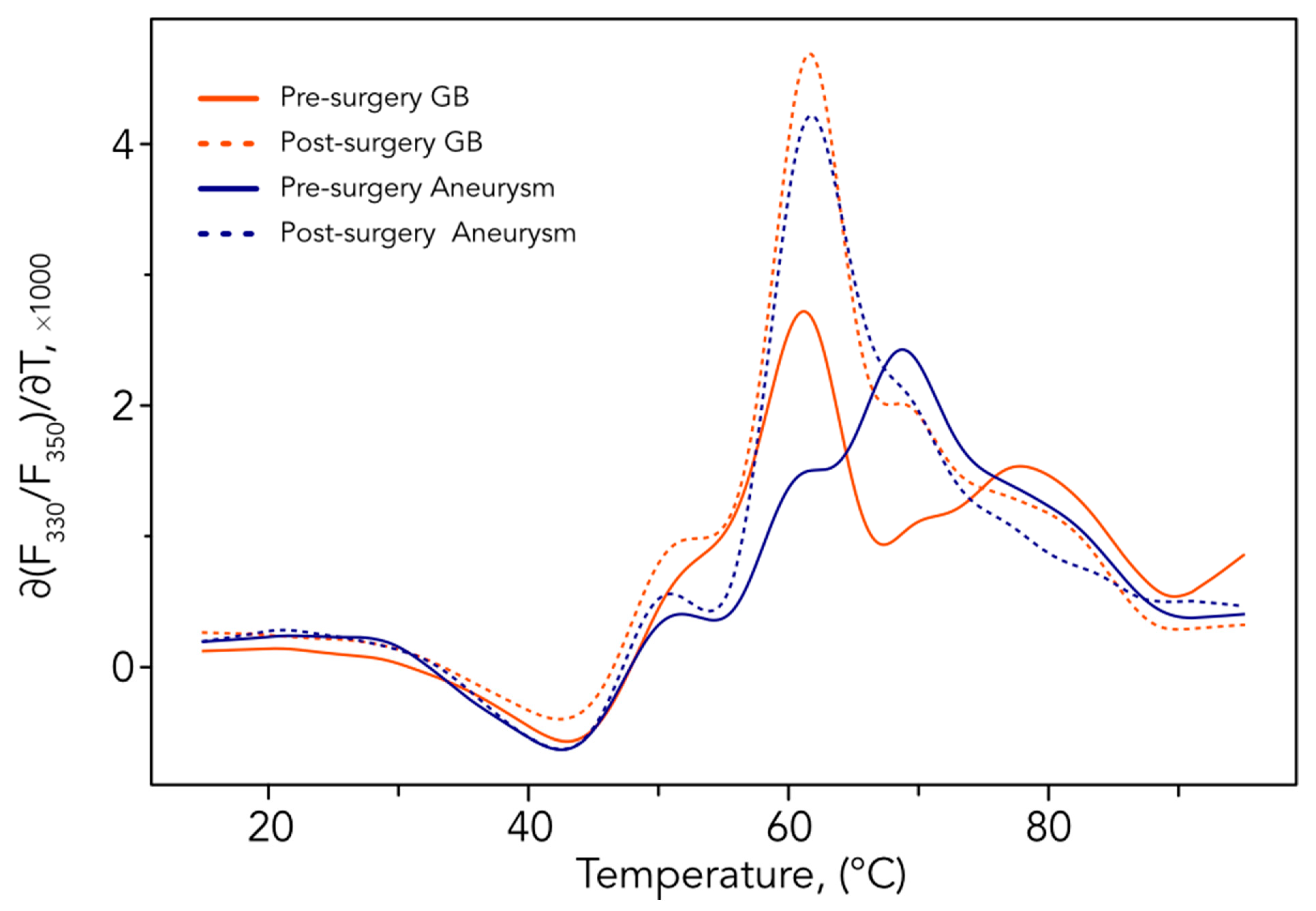
3.3. Comparative Analysis before and after Surgery
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary Brain Tumours in Adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Wesseling, P.; Aldape, K.; Brat, D.J.; Capper, D.; Cree, I.A.; Eberhart, C.; Figarella-Branger, D.; Fouladi, M.; Fuller, G.N.; et al. cIMPACT-NOW Update 6: New Entity and Diagnostic Principle Recommendations of the cIMPACT-Utrecht Meeting on Future CNS Tumor Classification and Grading. Brain Pathol. 2020, 30, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European Association for Neuro-Oncology (EANO) Guideline on the Diagnosis and Treatment of Adult Astrocytic and Oligodendroglial Gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef]
- Tsvetkov, P.O.; Devred, F. Plasmatic Signature of Disease by Differential Scanning Calorimetry (DSC). Methods Mol. Biol. 2019, 1964, 45–57. [Google Scholar]
- Garbett, N.C.; Mekmaysy, C.S.; DeLeeuw, L.; Chaires, J.B. Clinical Application of Plasma Thermograms. Utility, Practical Approaches and Considerations. Methods 2015, 76, 41–50. [Google Scholar] [CrossRef]
- Garbett, N.C.; Mekmaysy, C.S.; Helm, C.W.; Jenson, A.B.; Chaires, J.B. Differential Scanning Calorimetry of Blood Plasma for Clinical Diagnosis and Monitoring. Exp. Mol. Pathol. 2009, 86, 186–191. [Google Scholar] [CrossRef]
- Tsvetkov, P.O.; Tabouret, E.; Roman, A.Y.; Romain, S.; Bequet, C.; Ishimbaeva, O.; Honoré, S.; Figarella-Branger, D.; Chinot, O.; Devred, F. Differential Scanning Calorimetry of Plasma in Glioblastoma: Toward a New Prognostic / Monitoring Tool. Oncotarget 2018, 9, 9391–9399. [Google Scholar] [CrossRef]
- Schneider, G.; Kaliappan, A.; Nguyen, T.Q.; Buscaglia, R.; Brock, G.N.; Hall, M.B.; DeSpirito, C.; Wilkey, D.W.; Merchant, M.L.; Klein, J.B.; et al. The Utility of Differential Scanning Calorimetry Curves of Blood Plasma for Diagnosis, Subtype Differentiation and Predicted Survival in Lung Cancer. Cancers 2021, 13, 5326. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.O.; Eyraud, R.; Ayache, S.; Bougaev, A.A.; Malesinski, S.; Benazha, H.; Gorokhova, S.; Buffat, C.; Dehais, C.; Sanson, M.; et al. An AI-Powered Blood Test to Detect Cancer Using NanoDSF. Cancers 2021, 13, 1294. [Google Scholar] [CrossRef] [PubMed]
- Quillien, V.; Lavenu, A.; Sanson, M.; Legrain, M.; Dubus, P.; Karayan-Tapon, L.; Mosser, J.; Ichimura, K.; Figarella-Branger, D. Outcome-Based Determination of Optimal Pyrosequencing Assay for MGMT Methylation Detection in Glioblastoma Patients. J. Neurooncol. 2014, 116, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 9781493938438. [Google Scholar]
- Yi, Z.; Qu, C.; Zeng, Y.; Liu, Z. Liquid Biopsy: Early and Accurate Diagnosis of Brain Tumor. J. Cancer Res. Clin. Oncol. 2022, 148, 2347–2373. [Google Scholar] [CrossRef] [PubMed]
- Terrier, L.; Gilard, V.; Marguet, F.; Fontanilles, M.; Derrey, S. Stereotactic Brain Biopsy: Evaluation of Robot-Assisted Procedure in 60 Patients. Acta Neurochir. 2019, 161, 545–552. [Google Scholar] [CrossRef]
- Yi, Z.; Long, L.; Zeng, Y.; Liu, Z. Current Advances and Challenges in Radiomics of Brain Tumors. Front. Oncol. 2021, 11, 732196. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid Biopsy and Minimal Residual Disease—Latest Advances and Implications for Cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro. Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Fontanilles, M.; Marguet, F.; Beaussire, L.; Magne, N.; Pépin, L.-F.; Alexandru, C.; Tennevet, I.; Hanzen, C.; Langlois, O.; Jardin, F.; et al. Cell-Free DNA and Circulating TERT Promoter Mutation for Disease Monitoring in Newly-Diagnosed Glioblastoma. Acta Neuropathol. Commun. 2020, 8, 179. [Google Scholar] [CrossRef]
- Shankar, G.M.; Balaj, L.; Stott, S.L.; Nahed, B.; Carter, B.S. Liquid Biopsy for Brain Tumors. Expert Rev. Mol. Diagn. 2017, 17, 943–947. [Google Scholar] [CrossRef]
- Müller Bark, J.; Kulasinghe, A.; Chua, B.; Day, B.W.; Punyadeera, C. Circulating Biomarkers in Patients with Glioblastoma. Br. J. Cancer 2020, 122, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Sarkaria, J.N.; Hu, L.S.; Parney, I.F.; Pafundi, D.H.; Brinkmann, D.H.; Laack, N.N.; Giannini, C.; Burns, T.C.; Kizilbash, S.H.; Laramy, J.K.; et al. Is the Blood-Brain Barrier Really Disrupted in All Glioblastomas? A Critical Assessment of Existing Clinical Data. Neuro. Oncol. 2018, 20, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Solar, P.; Hendrych, M.; Barak, M.; Valekova, H.; Hermanova, M.; Jancalek, R. Blood-Brain Barrier Alterations and Edema Formation in Different Brain Mass Lesions. Front. Cell Neurosci. 2022, 16, 922181. [Google Scholar] [CrossRef]
- Sharma, G.; Jain, S.K.; Sinha, V.D. Peripheral Inflammatory Blood Markers in Diagnosis of Glioma and IDH Status. J. Neurosci. Rural Pract. 2021, 12, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, J.; Tan, D.; Cheng, P.; Wu, A. Relationship between Circulating Inflammatory Factors and Glioma Risk and Prognosis: A Meta-Analysis. Cancer Med. 2019, 8, 7454–7468. [Google Scholar] [CrossRef] [PubMed]
- Dharmajaya, R.; Sari, D.K. Role and Value of Inflammatory Markers in Brain Tumors: A Case Controlled Study. Ann. Med. Surg. (Lond.) 2021, 63, 102107. [Google Scholar] [CrossRef]
- He, Q.; Li, L.; Ren, Q. The Prognostic Value of Preoperative Systemic Inflammatory Response Index (SIRI) in Patients With High-Grade Glioma and the Establishment of a Nomogram. Front. Oncol. 2021, 11, 671811. [Google Scholar] [CrossRef]
- Jarmuzek, P.; Kot, M.; Defort, P.; Stawicki, J.; Komorzycka, J.; Nowak, K.; Tylutka, A.; Zembron-Lacny, A. Prognostic Values of Combined Ratios of White Blood Cells in Glioblastoma: A Retrospective Study. J. Clin. Med. Res. 2022, 11, 3397. [Google Scholar] [CrossRef]
- Joshkon, A.; Tabouret, E.; Traboulsi, W.; Bachelier, R.; Simoncini, S.; Roffino, S.; Jiguet-Jiglaire, C.; Badran, B.; Guillet, B.; Foucault-Bertaud, A.; et al. Soluble CD146, a Biomarker and a Target for Preventing Resistance to Anti-Angiogenic Therapy in Glioblastoma. Acta Neuropathol. Commun. 2022, 10, 151. [Google Scholar] [CrossRef]
- Jiguet-Jiglaire, C.; Boissonneau, S.; Denicolai, E.; Hein, V.; Lasseur, R.; Garcia, J.; Romain, S.; Appay, R.; Graillon, T.; Mason, W.; et al. Plasmatic MMP9 Released from Tumor-Infiltrating Neutrophils Is Predictive for Bevacizumab Efficacy in Glioblastoma Patients: An AVAglio Ancillary Study. Acta Neuropathol. Commun. 2022, 10, 1. [Google Scholar] [CrossRef]
- Vasunilashorn, S.M.; Ngo, L.H.; Dillon, S.T.; Fong, T.G.; Carlyle, B.C.; Kivisäkk, P.; Trombetta, B.A.; Vlassakov, K.V.; Kunze, L.J.; Arnold, S.E.; et al. Plasma and Cerebrospinal Fluid Inflammation and the Blood-Brain Barrier in Older Surgical Patients: The Role of Inflammation after Surgery for Elders (RISE) Study. J. Neuroinflammation 2021, 18, 103. [Google Scholar] [CrossRef] [PubMed]

| Factors | Prospective Cohort | |
|---|---|---|
| N = 38 | % | |
| Age (median, rage) | 65.5 (42.1–85.4) | |
| Gender (Women/Men) | 27/11 | 71/29 |
| Initial KPS (median, range) | 70 (40–100) | |
| Cognitive symptom | 9 | 24 |
| Steroid doses | 40 (10–120) | |
| Surgery types | ||
| Gross total resection | 27 | 75 |
| Partial resection | 9 | 25 |
| First-line treatment | ||
| Radio-chemotherapy | 28 | 76 |
| Radio-chemotherapy + bevacizumab | ||
| Chemotherapy alone | 9 | 24 |
| N | LR | SVM | RF | Adaboost | |
|---|---|---|---|---|---|
| EGFR amplification (False positive/False negative) | 16/11 ** | 59.3% (10/1) | 55.6% (1/11) | 63.0% (4/6) | 81.5% (1/4) |
| MGMT promoter (False positive/False negative) | 18/7 ** | 48.0% (12/1) | 76.0% (1/5) | 72.0% (1/6) | 60.0% (4/6) |
|
post-/pre-surgery (GBM only) (False post-surgery/False pre-surgery) | 28/33 * | 77.0% (3/11) | 80.3% (6/6) | 82.0% (4/7) | 80.3% (5/7) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eyraud, R.; Ayache, S.; Tsvetkov, P.O.; Kalidindi, S.S.; Baksheeva, V.E.; Boissonneau, S.; Jiguet-Jiglaire, C.; Appay, R.; Nanni-Metellus, I.; Chinot, O.; et al. Plasma nanoDSF Denaturation Profile at Baseline Is Predictive of Glioblastoma EGFR Status. Cancers 2023, 15, 760. https://doi.org/10.3390/cancers15030760
Eyraud R, Ayache S, Tsvetkov PO, Kalidindi SS, Baksheeva VE, Boissonneau S, Jiguet-Jiglaire C, Appay R, Nanni-Metellus I, Chinot O, et al. Plasma nanoDSF Denaturation Profile at Baseline Is Predictive of Glioblastoma EGFR Status. Cancers. 2023; 15(3):760. https://doi.org/10.3390/cancers15030760
Chicago/Turabian StyleEyraud, Rémi, Stéphane Ayache, Philipp O. Tsvetkov, Shanmugha Sri Kalidindi, Viktoriia E. Baksheeva, Sébastien Boissonneau, Carine Jiguet-Jiglaire, Romain Appay, Isabelle Nanni-Metellus, Olivier Chinot, and et al. 2023. "Plasma nanoDSF Denaturation Profile at Baseline Is Predictive of Glioblastoma EGFR Status" Cancers 15, no. 3: 760. https://doi.org/10.3390/cancers15030760
APA StyleEyraud, R., Ayache, S., Tsvetkov, P. O., Kalidindi, S. S., Baksheeva, V. E., Boissonneau, S., Jiguet-Jiglaire, C., Appay, R., Nanni-Metellus, I., Chinot, O., Devred, F., & Tabouret, E. (2023). Plasma nanoDSF Denaturation Profile at Baseline Is Predictive of Glioblastoma EGFR Status. Cancers, 15(3), 760. https://doi.org/10.3390/cancers15030760






