Clinical Utility of Comprehensive Genomic Profiling in Patients with Unresectable Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Protocol
2.2. Clinical and Laboratory Data
2.3. F1CDx and F1LCDx
2.4. Expert Panel Discussion
2.5. Treatment after F1CDX or F1LCDX
3. Results
3.1. Patient Characteristics
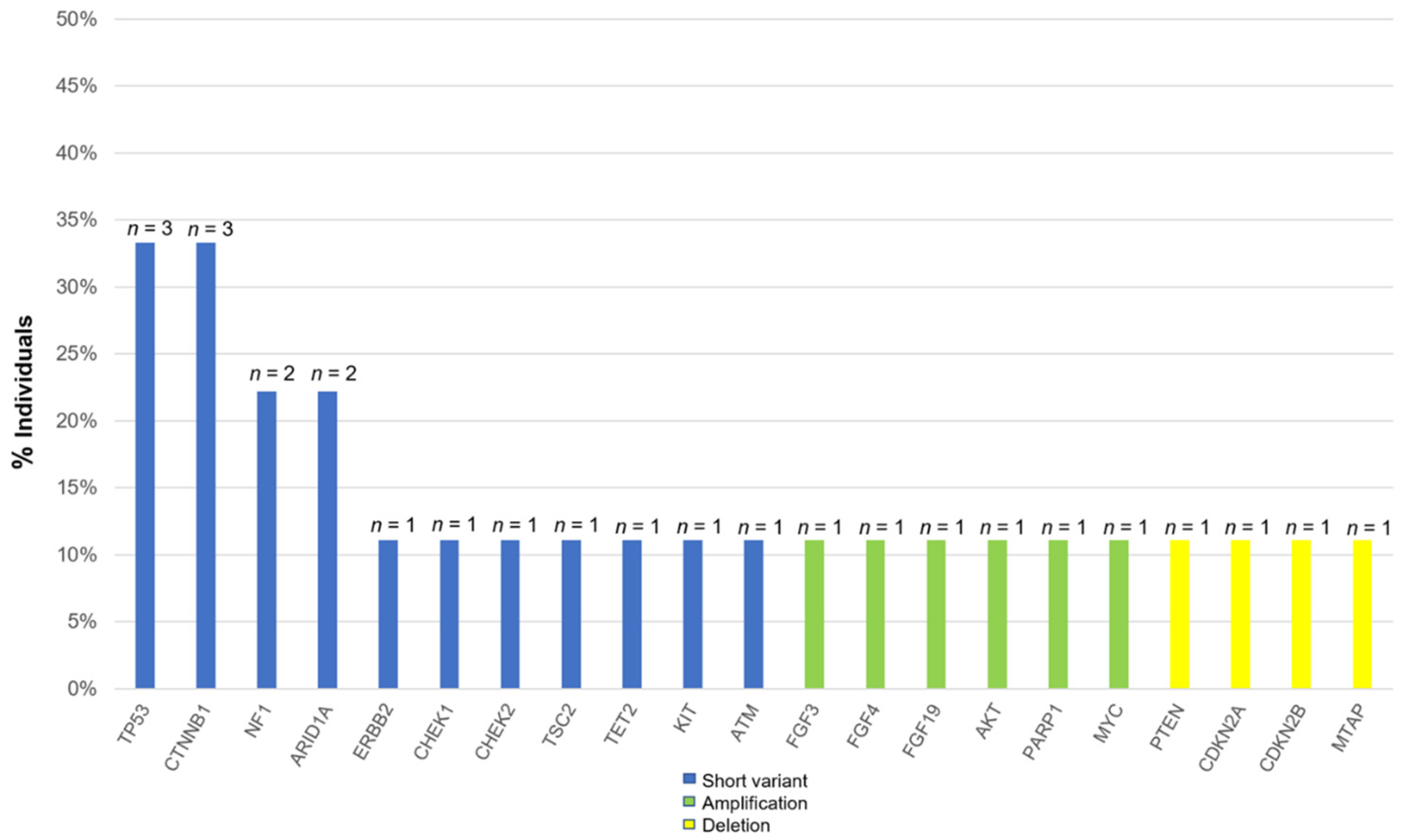
3.2. Common Alterations in Patients with HCC
3.3. Treatment after The Comprehensive Genomic Profiling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.-H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.-W.; Han, G.; Jassem, J.; et al. Lenvatinib versus Sorafenib in First-Line Treatment of Patients with Unresectable Hepatocellular Carcinoma: A Randomised Phase 3 Non-Inferiority Trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Kelley, R.K.; Rimassa, L.; Cheng, A.-L.; Kaseb, A.; Qin, S.; Zhu, A.X.; Chan, S.L.; Melkadze, T.; Sukeepaisarnjaroen, W.; Breder, V.; et al. Cabozantinib plus Atezolizumab versus Sorafenib for Advanced Hepatocellular Carcinoma (COSMIC-312): A Multicentre, Open-Label, Randomised, Phase 3 Trial. Lancet Oncol. 2022, 23, 995–1008. [Google Scholar] [CrossRef]
- Llovet, J.M.; Pinyol, R.; Kelley, R.K.; El-Khoueiry, A.; Reeves, H.L.; Wang, X.W.; Gores, G.J.; Villanueva, A. Molecular Pathogenesis and Systemic Therapies for Hepatocellular Carcinoma. Nat. Cancer 2022, 3, 386–401. [Google Scholar] [CrossRef]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular Therapies and Precision Medicine for Hepatocellular Carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Ding, X.; He, M.; Chan, A.W.H.; Song, Q.X.; Sze, S.C.; Chen, H.; Man, M.K.H.; Man, K.; Chan, S.L.; Lai, P.B.S.; et al. Genomic and Epigenomic Features of Primary and Recurrent Hepatocellular Carcinomas. Gastroenterology 2019, 157, 1630–1645.e6. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.-F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.-L.; Forner, A.; et al. Sorafenib in Advanced Hepatocellular Carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Cheng, A.-L.; Kang, Y.-K.; Chen, Z.; Tsao, C.-J.; Qin, S.; Kim, J.S.; Luo, R.; Feng, J.; Ye, S.; Yang, T.-S.; et al. Efficacy and Safety of Sorafenib in Patients in the Asia-Pacific Region with Advanced Hepatocellular Carcinoma: A Phase III Randomised, Double-Blind, Placebo-Controlled Trial. Lancet Oncol. 2009, 10, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Qin, S.; Merle, P.; Granito, A.; Huang, Y.-H.; Bodoky, G.; Pracht, M.; Yokosuka, O.; Rosmorduc, O.; Breder, V.; et al. Regorafenib for Patients with Hepatocellular Carcinoma Who Progressed on Sorafenib Treatment (RESORCE): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 389, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Kang, Y.-K.; Yen, C.-J.; Finn, R.S.; Galle, P.R.; Llovet, J.M.; Assenat, E.; Brandi, G.; Pracht, M.; Lim, H.Y.; et al. Ramucirumab after Sorafenib in Patients with Advanced Hepatocellular Carcinoma and Increased α-Fetoprotein Concentrations (REACH-2): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 2019, 20, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Abou-Alfa, G.K.; Meyer, T.; Cheng, A.-L.; El-Khoueiry, A.B.; Rimassa, L.; Ryoo, B.-Y.; Cicin, I.; Merle, P.; Chen, Y.; Park, J.-W.; et al. Cabozantinib in Patients with Advanced and Progressing Hepatocellular Carcinoma. N. Engl. J. Med. 2018, 379, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, Y.; Kano, Y.; Tohyama, K.; Matsudera, S.; Kumaki, Y.; Takahashi, K.; Mitsumura, T.; Harada, Y.; Sato, A.; Nakamura, H.; et al. Clinical Utility of Comprehensive Genomic Profiling in Japan: Result of PROFILE-F Study. PLoS ONE 2022, 17, e0266112. [Google Scholar] [CrossRef] [PubMed]
- Noji, R.; Tohyama, K.; Kugimoto, T.; Kuroshima, T.; Hirai, H.; Tomioka, H.; Michi, Y.; Tasaki, A.; Ohno, K.; Ariizumi, Y.; et al. Comprehensive Genomic Profiling Reveals Clinical Associations in Response to Immune Therapy in Head and Neck Cancer. Cancers 2022, 14, 3476. [Google Scholar] [CrossRef] [PubMed]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and Validation of a Clinical Cancer Genomic Profiling Test Based on Massively Parallel DNA Sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and Analytical Validation of FoundationOne Liquid CDx, a Novel 324-Gene CfDNA-Based Comprehensive Genomic Profiling Assay for Cancers of Solid Tumor Origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef]
- Ogawa, K.; Kanzaki, H.; Chiba, T.; Ao, J.; Qiang, N.; Ma, Y.; Zhang, J.; Yumita, S.; Ishino, T.; Unozawa, H.; et al. Effect of Atezolizumab plus Bevacizumab in Patients with Hepatocellular Carcinoma Harboring CTNNB1 Mutation in Early Clinical Experience. J. Cancer 2022, 13, 2656–2661. [Google Scholar] [CrossRef]
- Yamada, K.; Hori, Y.; Inoue, S.; Yamamoto, Y.; Iso, K.; Kamiyama, H.; Yamaguchi, A.; Kimura, T.; Uesugi, M.; Ito, J.; et al. E7386, a Selective Inhibitor of the Interaction between β-Catenin and CBP, Exerts Antitumor Activity in Tumor Models with Activated Canonical Wnt Signaling. Cancer Res. 2021, 81, 1052–1062. [Google Scholar] [CrossRef]
- Sun, E.J.; Wankell, M.; Palamuthusingam, P.; McFarlane, C.; Hebbard, L. Targeting the PI3K/Akt/MTOR Pathway in Hepatocellular Carcinoma. Biomedicines 2021, 9, 1639. [Google Scholar] [CrossRef]
- Matter, M.S.; Decaens, T.; Andersen, J.B.; Thorgeirsson, S.S. Targeting the MTOR Pathway in Hepatocellular Carcinoma: Current State and Future Trends. J. Hepatol. 2014, 60, 855–865. [Google Scholar] [CrossRef]
- Chen, J.-S.; Wang, Q.; Fu, X.-H.; Huang, X.-H.; Chen, X.-L.; Cao, L.-Q.; Chen, L.-Z.; Tan, H.-X.; Li, W.; Bi, J.; et al. Involvement of PI3K/PTEN/AKT/MTOR Pathway in Invasion and Metastasis in Hepatocellular Carcinoma: Association with MMP-9. Hepatol. Res. 2009, 39, 177–186. [Google Scholar] [CrossRef]
- Ocana, A.; Vera-Badillo, F.; Al-Mubarak, M.; Templeton, A.J.; Corrales-Sanchez, V.; Diez-Gonzalez, L.; Cuenca-Lopez, M.D.; Seruga, B.; Pandiella, A.; Amir, E. Activation of the PI3K/MTOR/AKT Pathway and Survival in Solid Tumors: Systematic Review and Meta-Analysis. PLoS ONE 2014, 9, e95219. [Google Scholar] [CrossRef]
- Zhu, A.X.; Abrams, T.A.; Miksad, R.; Blaszkowsky, L.S.; Meyerhardt, J.A.; Zheng, H.; Muzikansky, A.; Clark, J.W.; Kwak, E.L.; Schrag, D.; et al. Phase 1/2 Study of Everolimus in Advanced Hepatocellular Carcinoma. Cancer 2011, 117, 5094–5102. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Kudo, M.; Assenat, E.; Cattan, S.; Kang, Y.-K.; Lim, H.Y.; Poon, R.T.P.; Blanc, J.-F.; Vogel, A.; Chen, C.-L.; et al. Effect of Everolimus on Survival in Advanced Hepatocellular Carcinoma after Failure of Sorafenib: The EVOLVE-1 Randomized Clinical Trial. JAMA 2014, 312, 57–67. [Google Scholar] [CrossRef]
| n = 9 | |
| Age (Years), Median (IQR) | 65 (25–78) |
| Sex: Male/Female (%) | 7 (77.8%)/2 (22.2%) |
| Body Weight (kg), Median (IQR) | 57.7 (47.6–102.5) |
| Etiology HBV/HCV/Alcohol/Others (%) | 3 (33.3%)/3 (33.3%)/1 (11.1%)/2 (22.2%) |
| Platelets (10⁴/μL), Median (IQR) | 16.2 (6.2–31.4) |
| AST (U/L), Median (IQR) | 42 (22–65) |
| ALT (U/L), Median (IQR) | 34 (13–96) |
| Total Bilirubin (mg/dL), Median (IQR) | 0.6 (0.4–2.6) |
| Albumin (g/dL), Median (IQR) | 3.6 (2.2–4.0) |
| Prothrombin Time–International Normalized Ratio, Median (IQR) | 1.0 (0.9–1.2) |
| Creatinine (mg/dL), Median (IQR) | 1.0 (0.5–1.4) |
| eGFR (mL/min/1.73m2), Median (IQR) | 58.8 (42.1–149.3) |
| Urine Total Protein/Creatinine | 0.4 (0.1–7.9) |
| Child–Pugh A/B/C (%) | 6 (66.7%)/3 (33.3%)/0 |
| ECOG PS 0/1/2 (%) | 8 (88.9%)/1 (11.1%)/0 |
| BCLC Stage A/B/C (%) | 1 (11.1%)/0/8 (88.9%) |
| Macroscopic Vascular Invasion, Yes/No (%) | 2 (22.2%)/7 (77.8%) |
| Extrahepatic Metastasis, Yes/No (%) | 7 (77.8%)/2 (22.2%) |
| Baseline AFP Concentration (ng/mL), Median (IQR) | 6519.4 (1.7–19827.3) |
| Baseline AFP < 400 ng/mL, Yes/No (%) | 4 (44.4%)/5 (55.6%) |
| Baseline DCP Concentration (mAU/mL), Median (Range) | 2965.5 (14.9–36091.5) |
| Number of Past Systemic Therapies 1/2/3/4/5/6 (%) | 0/0/0/5 (55.6%)/3 (33.3%)/1 (11.1%) |
| Specimen Collection Liver/Bone/Blood | 2 (22.2%)/2 (22.2%)/5 (55.6%) |
| Types of CGP Foundation One® CDx Foundation One® Liquid CDx | 4 (44.4%) 5 (55.6%) |
| Initial HCC Treatment Resection/RFA/TACE/Systemic Therapy (%) | 4 (44.5%)/2 (22.2%)/0/3 (33.3%) |
| Observation Period from the Initial HCC Treatment (Months), Median (Range) | 57 (13–144) |
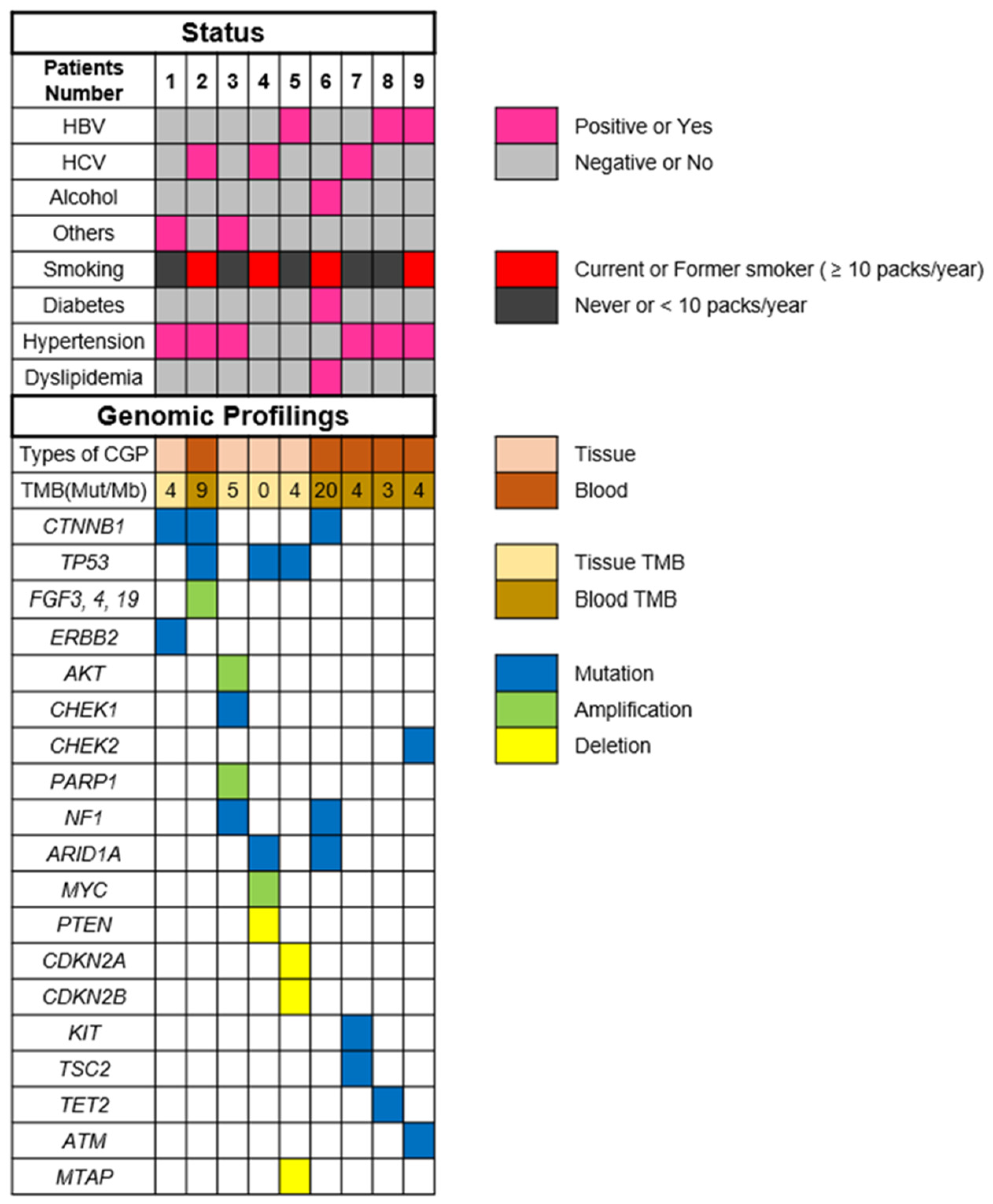
| Patients Number, Age, Sex | Etiology | Number of Prior Systemic Therapies | Metastatic Spread | CGP Method (Organ) | Mutations (VAF%) |
|---|---|---|---|---|---|
| ① 20s male | NBNC | 4 LEN + TAI→ATZ + BEV→SOR→REG | Nothing | F1CDx (liver) | CTNNB1 G34R (18.9%)/ D32Y (6.2%) ERBB2 P967Q (50%) |
| ② 70s female | HCV | 6 SOR→REG→LEN→ATZ + BEV→RAM→CAB | Lungs, bones | F1LCDx (blood) | CTNNB1 S33C (0.15%) FGF3 (present*) FGF4 (present*) FGF19 (present*) TP53 (0.53%) |
| ③ 30s male | NBNC | 5 SOR→REG→LEN→RAM→ATZ + BEV | Lungs, bones | F1CDx (bone) | AKT3 (amplification) CHEK1 K224* (54.6%) PARP1 (amplification) NF1 R2083C (present*) |
| ④ 70s male | HCV | 4 LEN→ATZ + BEV→CAB→SOR | Lungs, bones | F1CDx (bone) | TP53 S227fs*2 (46.9%) PTEN (loss exons) ARID1A splice site 2988 + 2T > G (62%) MYC (amplification) |
| ⑤ 50s male | HBV | 5 LEN→HAIC→ATZ + BEV→RAM→CAB | Nothing | F1CDx (liver) | CDKN2A/B (loss exsons) MTAP (loss exsons) TP53 R249S (80.2%) |
| ⑥ 60s male | ALD | 4 LEN + TAI→ATZ + BEV→SOR→REG | Lungs | F1LCDx (blood) | TMB-high (20 mutations/megabase) NF1 Y80fs*26 (1.4%) ARID1A K990fs*18 (18%) CTNNB1 K335T (16.4%) |
| ⑦ 60s male | HCV | 4 LEN→ATZ + BEV→CAB→REG | Lungs | F1LCDx (blood) | KIT D816E (0.29%) TSC2 splice site 2587_2639 + 36del89 (0.42%) |
| ⑧ 60s male | HBV | 4 LEN→ATZ + BEV→CAB→RAM | Lungs | F1LCDx (blood) | TET2 H1904R (0.55%) |
| ⑨ 60s male | HBV | 5 SOR→REG→ATZ + BEV→LEN→CAB | Peritoneal dissemination | F1LCDx (blood) | CHEK2 T43fs*15 (0.15%) ATM R1068fs*18 (1.5%) |
| Patients Number, Age, Sex | Reasons for Not Participating in Clinical Trials | After CGP | Survival Durationafter CGP |
|---|---|---|---|
| ① 20s male | Excluded from the clinical trial because of renal dysfunction | Treated with CAB (based on CGP) because CAB was reported to inhibit the beta-catenin pathway partially | 18 months |
| ② 70s female | irAE | Treated with REG (based on CGP). The expert panel recommended that retreatment of regorafenib would be an effective option | 6 months |
| ③ 30s male | Excluded from the clinical trial because of low platelet counts | Treated with CAB as 6th line | 17 months |
| ④ 70s male | Worsening PS | Treated with RAM as 5th line | 5 months |
| ⑤ 50s male | No applicable clinical trials | BSC | 4 months |
| ⑥ 60s male | The patient decided to receive pembrolizumab with health insurance by national health insurance | Treated with pembrolizumab (based on CGP) because the CGP found TMB-high | 3 months |
| ⑦ 60s male | Planned to participate in everolimus’s prospective trial of patient-proposed healthcare services with multiple targeted agents | The patient participated in the other clinical trial at National Cancer Center | 2 months |
| ⑧ 60s male | No applicable clinical trials | Treated with SOR as 5th line | 2 months |
| ⑨ 60s male | Excluded from the clinical trial because of HBV infection | Retreatment with ATZ + BEV | 3 months |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishido, S.; Tsuchiya, K.; Kano, Y.; Yasui, Y.; Takaura, K.; Uchihara, N.; Suzuki, K.; Tanaka, Y.; Miyamoto, H.; Yamada, M.; et al. Clinical Utility of Comprehensive Genomic Profiling in Patients with Unresectable Hepatocellular Carcinoma. Cancers 2023, 15, 719. https://doi.org/10.3390/cancers15030719
Ishido S, Tsuchiya K, Kano Y, Yasui Y, Takaura K, Uchihara N, Suzuki K, Tanaka Y, Miyamoto H, Yamada M, et al. Clinical Utility of Comprehensive Genomic Profiling in Patients with Unresectable Hepatocellular Carcinoma. Cancers. 2023; 15(3):719. https://doi.org/10.3390/cancers15030719
Chicago/Turabian StyleIshido, Shun, Kaoru Tsuchiya, Yoshihito Kano, Yutaka Yasui, Kenta Takaura, Naoki Uchihara, Keito Suzuki, Yuki Tanaka, Haruka Miyamoto, Michiko Yamada, and et al. 2023. "Clinical Utility of Comprehensive Genomic Profiling in Patients with Unresectable Hepatocellular Carcinoma" Cancers 15, no. 3: 719. https://doi.org/10.3390/cancers15030719
APA StyleIshido, S., Tsuchiya, K., Kano, Y., Yasui, Y., Takaura, K., Uchihara, N., Suzuki, K., Tanaka, Y., Miyamoto, H., Yamada, M., Matsumoto, H., Nobusawa, T., Keitoku, T., Tanaka, S., Maeyashiki, C., Tamaki, N., Takahashi, Y., Nakanishi, H., Sakurai, U., ... Izumi, N. (2023). Clinical Utility of Comprehensive Genomic Profiling in Patients with Unresectable Hepatocellular Carcinoma. Cancers, 15(3), 719. https://doi.org/10.3390/cancers15030719








