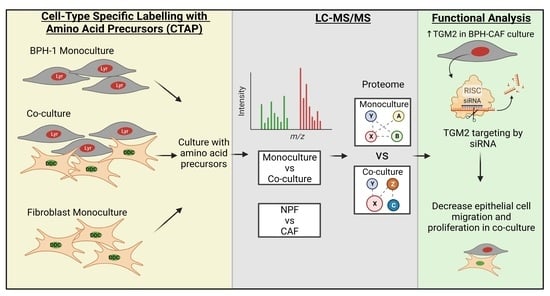
Cell-Type-Specific Signalling Networks Impacted by Prostate Epithelial-Stromal Intercellular Communication
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Nonmalignant Prostate Fibroblasts (NPF) and Cancer-Associated Fibroblasts (CAF) from Primary Prostate Tissue
2.2. Transfection and Cell-Type Labelling with Amino Acid Precursors
2.3. Phosphopeptide Enrichment
2.4. Mass Spectrometry Analysis
2.5. Mass Spectrometry Statistical Analysis
2.6. Functional Annotation Analysis
2.7. Western Blot Analysis
2.8. siRNA Knockdown
2.9. MTS Proliferation Assay for Cells in Monoculture
2.10. Scratch Assay
2.11. Co-Culture Proliferation Assay
2.12. Co-Culture Random Cell Migration Assay
3. Results
3.1. Global Proteomic Analysis of NPF or CAF Co-Cultures with BPH-1 Prostate Epithelial Cells Using CTAP
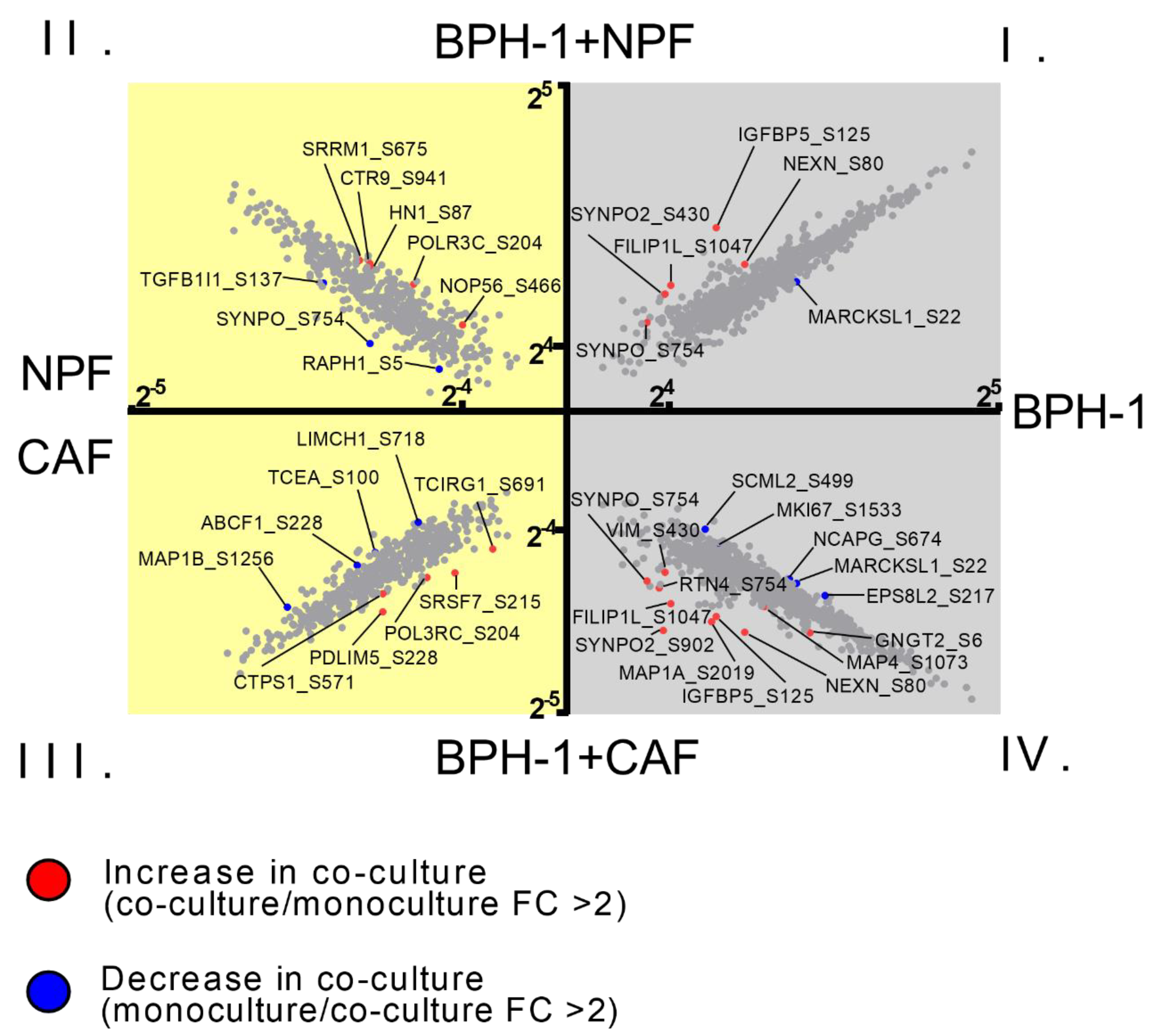
3.2. Global Phosphoproteomic Analysis of NPF or CAF Co-Cultures with BPH-1 Prostate Epithelial Cells Using CTAP
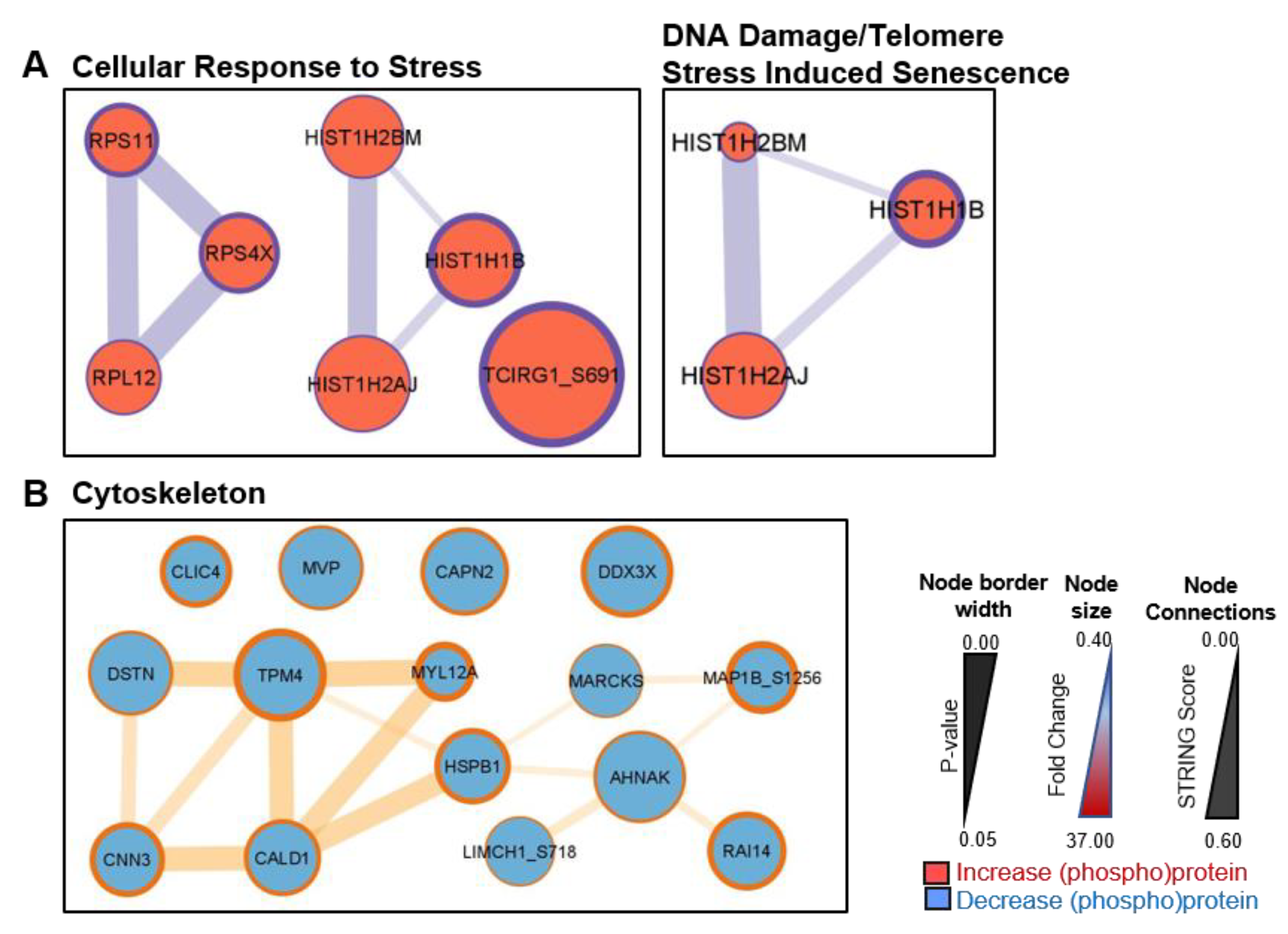
3.3. Signalling between BPH-1 Epithelial Cells and CAFs in Co-Culture
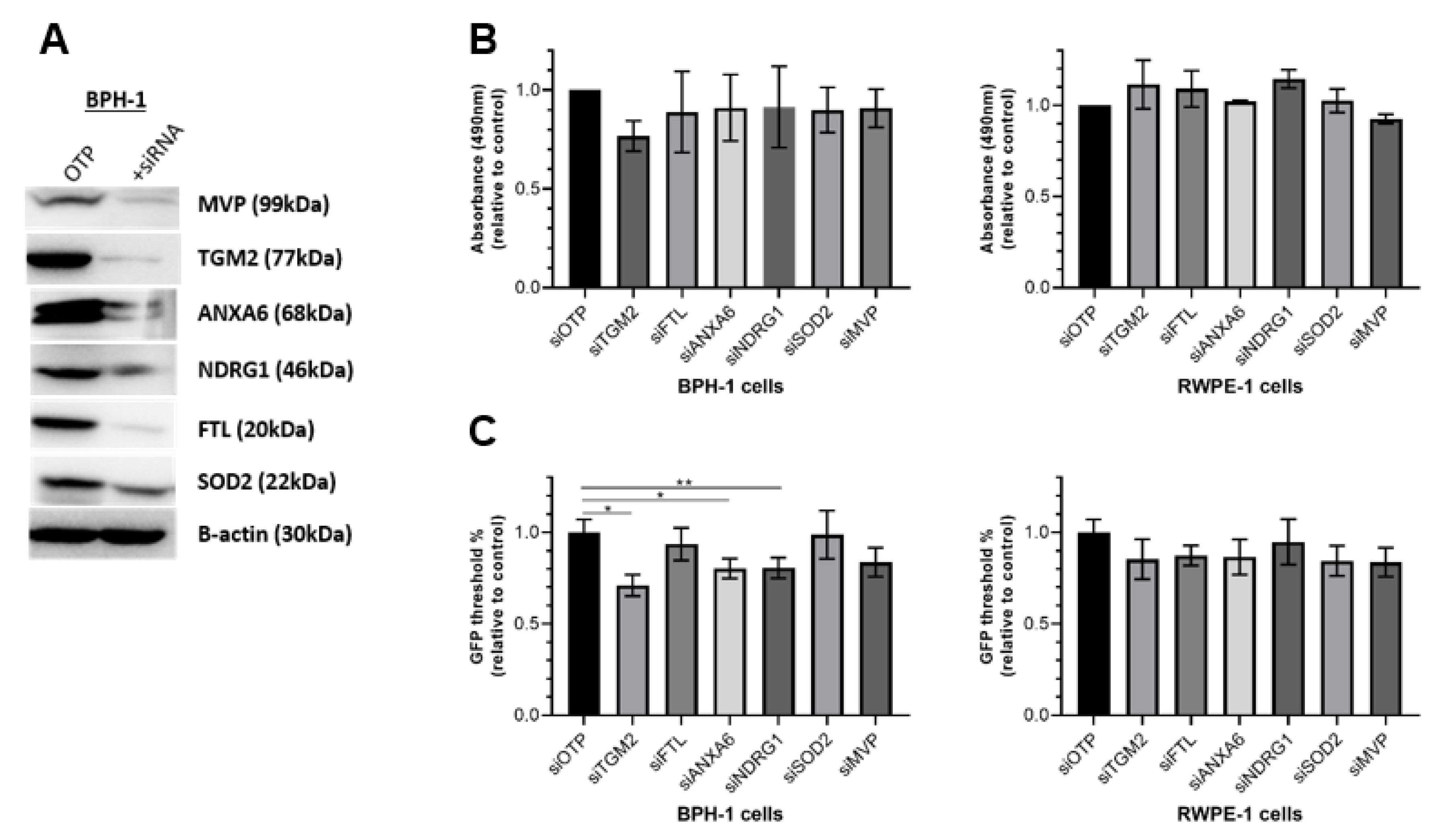
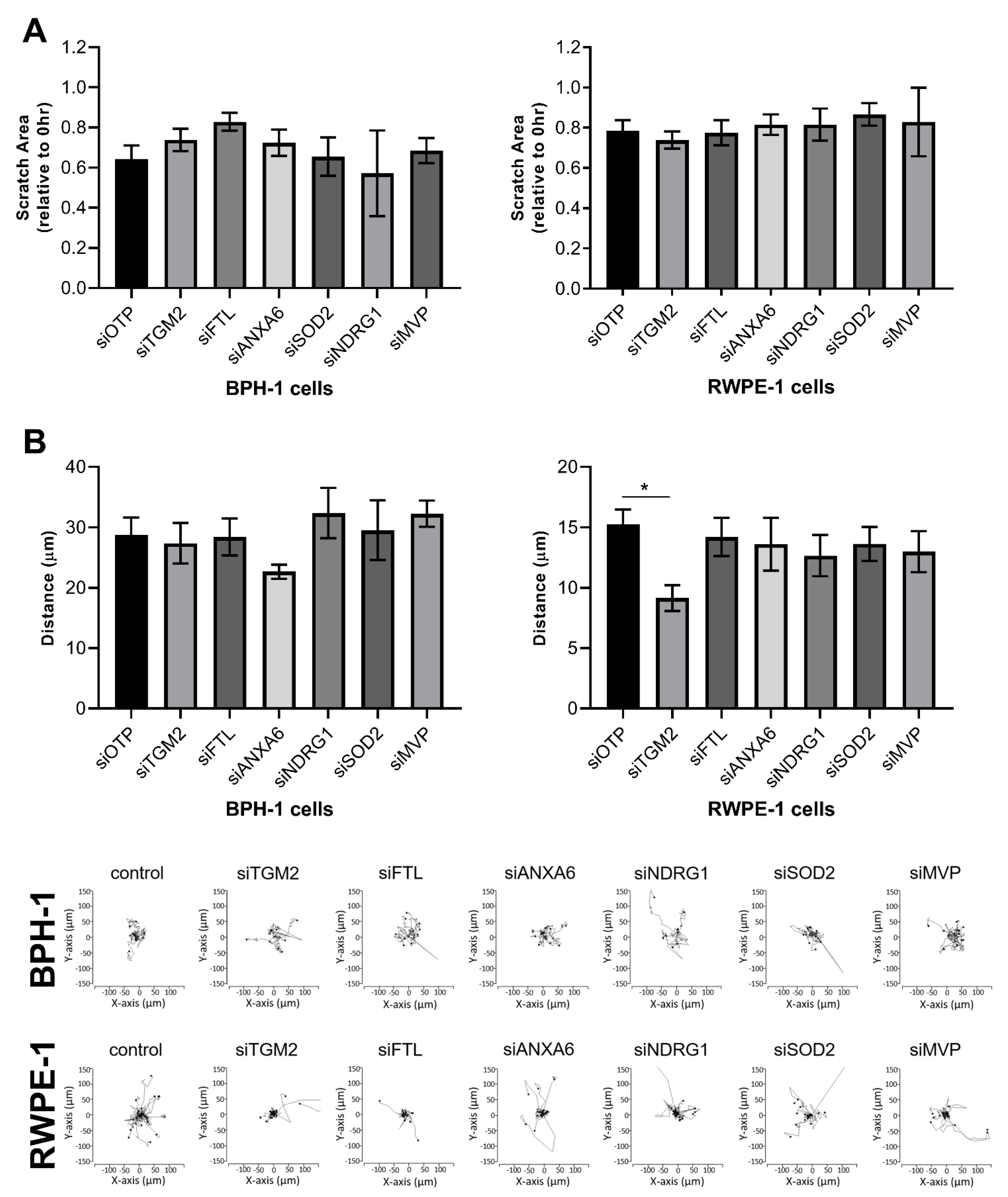
3.4. Functional Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bonollo, F.; Thalmann, G.N.; Kruithof-de Julio, M.; Karkampouna, S. The Role of Cancer-Associated Fibroblasts in Prostate Cancer Tumorigenesis. Cancers 2020, 12, 1887. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.K.; Taubenberger, A.V.; Taylor, R.A.; Niranjan, B.; Chea, Z.Y.; Zotenko, E.; Sieh, S.; Pedersen, J.S.; Norden, S.; Frydenberg, M. A bioengineered microenvironment to quantitatively measure the tumorigenic properties of cancer-associated fibroblasts in human prostate cancer. Biomaterials 2013, 34, 4777–4785. [Google Scholar] [CrossRef]
- Ayala, G.; Tuxhorn, J.A.; Wheeler, T.M.; Frolov, A.; Scardino, P.T.; Ohori, M.; Wheeler, M.; Spitler, J.; Rowley, D.R. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin. Cancer Res. 2003, 9, 4792–4801. [Google Scholar]
- Wu, Z.; Shi, J.; Lai, C.; Li, K.; Li, K.; Li, Z.; Tang, Z.; Liu, C.; Xu, K. Clinicopathological significance and prognostic value of cancer-associated fibroblasts in prostate cancer patients. Urol. Oncol. 2021, 39, 433.e417–433.e423. [Google Scholar] [CrossRef] [PubMed]
- Olumi, A.F.; Grossfeld, G.D.; Hayward, S.W.; Carroll, P.R.; Tlsty, T.D.; Cunha, G.R. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999, 59, 5002–5011. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, E.V.; Pereira, B.A.; Lawrence, M.G.; Ma, X.; Rebello, R.J.; Chan, H.; Niranjan, B.; Wu, Y.; Ellem, S.; Guan, X.; et al. Proteomic Profiling of Human Prostate Cancer-associated Fibroblasts (CAF) Reveals LOXL2-dependent Regulation of the Tumor Microenvironment. Mol. Cell. Proteom. MCP 2019, 18, 1410–1427. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, N.P.; Soufi, B.; Walkowicz, W.E.; Pedicord, V.A.; Mavrakis, K.J.; Macek, B.; Gin, D.Y.; Sander, C.; Miller, M.L. Cell-selective labeling using amino acid precursors for proteomic studies of multicellular environments. Nat. Methods 2013, 10, 768–773. [Google Scholar] [CrossRef]
- Tape, C.J.; Ling, S.; Dimitriadi, M.; McMahon, K.M.; Worboys, J.D.; Leong, H.S.; Norrie, I.C.; Miller, C.J.; Poulogiannis, G.; Lauffenburger, D.A.; et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell 2016, 165, 910–920. [Google Scholar] [CrossRef]
- Tape, C.J.; Norrie, I.C.; Worboys, J.D.; Lim, L.; Lauffenburger, D.A.; Jørgensen, C. Cell-specific labeling enzymes for analysis of cell-cell communication in continuous co-culture. Mol. Cell. Proteom. MCP 2014, 13, 1866–1876. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Taylor, R.A.; Toivanen, R.; Pedersen, J.; Norden, S.; Pook, D.W.; Frydenberg, M.; Australian Prostate Cancer, B.; Papargiris, M.M.; Niranjan, B.; et al. A preclinical xenograft model of prostate cancer using human tumors. Nat. Protoc. 2013, 8, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, S.J.; Karayel, O.; James, D.E.; Mann, M. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat. Protoc. 2018, 13, 1897–1916. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Chan, C.; Naseer, M.; Yau, M.; Lo, R.; Sribnaia, A.; Ring, G.; Que, J.; Wee, K.; Winsor, G.L.; et al. Curating the innate immunity interactome. BMC Syst. Biol. 2010, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Brummer, T.; Schramek, D.; Hayes, V.M.; Bennett, H.L.; Caldon, C.E.; Musgrove, E.A.; Daly, R.J. Increased proliferation and altered growth factor dependence of human mammary epithelial cells overexpressing the Gab2 docking protein. J. Biol. Chem. 2006, 281, 626–637. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Kai, F.; Fawcett, J.P.; Duncan, R. Synaptopodin-2 induces assembly of peripheral actin bundles and immature focal adhesions to promote lamellipodia formation and prostate cancer cell migration. Oncotarget 2015, 6, 11162–11174. [Google Scholar] [CrossRef]
- Quiros-Gonzalez, I.; Gonzalez-Menendez, P.; Mayo, J.C.; Hevia, D.; Artime-Naveda, F.; Fernandez-Vega, S.; Fernandez-Fernandez, M.; Rodriguez-Gonzalez, P.; Garcia-Alonso, J.I.; Sainz, R.M. Androgen-Dependent Prostate Cancer Cells Reprogram Their Metabolic Signature upon GLUT1 Upregulation by Manganese Superoxide Dismutase. Antioxidants 2022, 11, 313. [Google Scholar] [CrossRef]
- Kim, Y.S.; Gupta Vallur, P.; Phaëton, R.; Mythreye, K.; Hempel, N. Insights into the Dichotomous Regulation of SOD2 in Cancer. Antioxidants 2017, 6, 86. [Google Scholar] [CrossRef]
- Eckert, R.L.; Kaartinen, M.T.; Nurminskaya, M.; Belkin, A.M.; Colak, G.; Johnson, G.V.; Mehta, K. Transglutaminase regulation of cell function. Physiol. Rev. 2014, 94, 383–417. [Google Scholar] [CrossRef] [PubMed]
- Tatsukawa, H.; Furutani, Y.; Hitomi, K.; Kojima, S. Transglutaminase 2 has opposing roles in the regulation of cellular functions as well as cell growth and death. Cell Death Dis. 2016, 7, e2244. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.S.; Greenberg, C.S. TGM2 and implications for human disease: Role of alternative splicing. Front. Biosci.-Landmark 2013, 18, 504–519. [Google Scholar] [CrossRef]
- Berg, T.J.; Marques, C.; Pantazopoulou, V.; Johansson, E.; von Stedingk, K.; Lindgren, D.; Jeannot, P.; Pietras, E.J.; Bergström, T.; Swartling, F.J.; et al. The Irradiated Brain Microenvironment Supports Glioma Stemness and Survival via Astrocyte-Derived Transglutaminase 2. Cancer Res. 2021, 81, 2101–2115. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.; Rheinstein, P.H. Druggable genetic targets in endometrial cancer. Cancer Treat. Res. Commun. 2021, 30, 100502. [Google Scholar] [CrossRef]
- Malkomes, P.; Lunger, I.; Oppermann, E.; Abou-El-Ardat, K.; Oellerich, T.; Günther, S.; Canbulat, C.; Bothur, S.; Schnütgen, F.; Yu, W.; et al. Transglutaminase 2 promotes tumorigenicity of colon cancer cells by inactivation of the tumor suppressor p53. Oncogene 2021, 40, 4352–4367. [Google Scholar] [CrossRef]
- Ahluwalia, P.; Ahluwalia, M.; Mondal, A.K.; Sahajpal, N.; Kota, V.; Rojiani, M.V.; Rojiani, A.M.; Kolhe, R. Prognostic and therapeutic implications of extracellular matrix associated gene signature in renal clear cell carcinoma. Sci. Rep. 2021, 11, 7561. [Google Scholar] [CrossRef]
- Urick, M.E.; Yu, E.J.; Bell, D.W. High-risk endometrial cancer proteomic profiling reveals that FBXW7 mutation alters L1CAM and TGM2 protein levels. Cancer 2021, 127, 2905–2915. [Google Scholar] [CrossRef]
- Sun, X.; Shu, Y.; Xu, M.; Jiang, J.; Wang, L.; Wang, J.; Huang, D.; Zhang, J. ANXA6 suppresses the tumorigenesis of cervical cancer through autophagy induction. Clin. Transl. Med. 2020, 10, e208. [Google Scholar] [CrossRef]
- Korolkova, O.Y.; Widatalla, S.E.; Williams, S.D.; Whalen, D.S.; Beasley, H.K.; Ochieng, J.; Grewal, T.; Sakwe, A.M. Diverse Roles of Annexin A6 in Triple-Negative Breast Cancer Diagnosis, Prognosis and EGFR-Targeted Therapies. Cells 2020, 9, 1855. [Google Scholar] [CrossRef]
- Grewal, T.; Hoque, M.; Conway, J.R.W.; Reverter, M.; Wahba, M.; Beevi, S.S.; Timpson, P.; Enrich, C.; Rentero, C. Annexin A6-A multifunctional scaffold in cell motility. Cell Adhes. Migr. 2017, 11, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Lomnytska, M.I.; Becker, S.; Bodin, I.; Olsson, A.; Hellman, K.; Hellström, A.C.; Mints, M.; Hellman, U.; Auer, G.; Andersson, S. Differential expression of ANXA6, HSP27, PRDX2, NCF2, and TPM4 during uterine cervix carcinogenesis: Diagnostic and prognostic value. Br. J. Cancer 2011, 104, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Leca, J.; Martinez, S.; Lac, S.; Nigri, J.; Secq, V.; Rubis, M.; Bressy, C.; Sergé, A.; Lavaut, M.N.; Dusetti, N.; et al. Cancer-associated fibroblast-derived annexin A6+ extracellular vesicles support pancreatic cancer aggressiveness. J. Clin. Investig. 2016, 126, 4140–4156. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Guo, A.; Zhou, C. Breast Cancer Stem Cell-Derived ANXA6-Containing Exosomes Sustain Paclitaxel Resistance and Cancer Aggressiveness in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 718721. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Liu, S.; Guo, C.; Wang, J.; Greenaway, F.T.; Sun, M.Z. Role of annexin A6 in cancer. Oncol. Lett. 2015, 10, 1947–1952. [Google Scholar] [CrossRef]
- Xin, W.; Rhodes, D.R.; Ingold, C.; Chinnaiyan, A.M.; Rubin, M.A. Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am. J. Pathol. 2003, 162, 255–261. [Google Scholar] [CrossRef]
- Lim, S.C.; Geleta, B.; Maleki, S.; Richardson, D.R.; Kovačević, Ž. The metastasis suppressor NDRG1 directly regulates androgen receptor signaling in prostate cancer. J. Biol. Chem. 2021, 297, 101414. [Google Scholar] [CrossRef]
- Quan, Y.; Cui, Y.; Wahafu, W.; Liu, Y.; Ping, H.; Zhang, X. MLL5α activates AR/NDRG1 signaling to suppress prostate cancer progression. Am. J. Cancer Res. 2020, 10, 1608–1629. [Google Scholar]
- Quan, Y.; Zhang, X.; Butler, W.; Du, Z.; Wang, M.; Liu, Y.; Ping, H. The role of N-cadherin/c-Jun/NDRG1 axis in the progression of prostate cancer. Int. J. Biol. Sci. 2021, 17, 3288–3304. [Google Scholar] [CrossRef]
- Ledet, R.J.; Ruff, S.E.; Wang, Y.; Nayak, S.; Schneider, J.A.; Ueberheide, B.; Logan, S.K.; Garabedian, M.J. Identification of PIM1 substrates reveals a role for NDRG1 phosphorylation in prostate cancer cellular migration and invasion. Commun. Biol. 2021, 4, 36. [Google Scholar] [CrossRef]
- Lage-Vickers, S.; Bizzotto, J.; Valacco, M.P.; Sanchis, P.; Nemirovsky, S.; Labanca, E.; Scorticati, C.; Mazza, O.; Mitrofanova, A.; Navone, N.; et al. The expression of YWHAZ and NDRG1 predicts aggressive outcome in human prostate cancer. Commun. Biol. 2021, 4, 103. [Google Scholar] [CrossRef]
- Du, A.; Jiang, Y.; Fan, C. NDRG1 Downregulates ATF3 and Inhibits Cisplatin-Induced Cytotoxicity in Lung Cancer A549 Cells. Int. J. Med. Sci. 2018, 15, 1502–1507. [Google Scholar] [CrossRef]
- Kim, S.C.; Shin, Y.K.; Kim, Y.A.; Jang, S.G.; Ku, J.L. Identification of genes inducing resistance to ionizing radiation in human rectal cancer cell lines: Re-sensitization of radio-resistant rectal cancer cells through down regulating NDRG1. BMC Cancer 2018, 18, 594. [Google Scholar] [CrossRef]
- Villodre, E.S.; Gong, Y.; Hu, X.; Huo, L.; Yoon, E.C.; Ueno, N.T.; Woodward, W.A.; Tripathy, D.; Song, J.; Debeb, B.G. NDRG1 Expression Is an Independent Prognostic Factor in Inflammatory Breast Cancer. Cancers 2020, 12, 3711. [Google Scholar] [CrossRef] [PubMed]
- Villodre, E.S.; Hu, X.; Eckhardt, B.L.; Larson, R.; Huo, L.; Yoon, E.C.; Gong, Y.; Song, J.; Liu, S.; Ueno, N.T.; et al. NDRG1 in Aggressive Breast Cancer Progression and Brain Metastasis. J. Natl. Cancer Inst. 2022, 114, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Yamamoto, Y.; Shiota, M.; Kuruma, H.; Beraldi, E.; Matsuyama, H.; Zoubeidi, A.; Gleave, M. Cotargeting Androgen Receptor and Clusterin Delays Castrate-Resistant Prostate Cancer Progression by Inhibiting Adaptive Stress Response and AR Stability. Cancer Res. 2013, 73, 5206–5217. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xie, S. Therapeutic targeting of cellular stress responses in cancer. Thorac. Cancer 2018, 9, 1575–1582. [Google Scholar] [CrossRef]
- Nguyen, H.G.; Conn, C.S.; Kye, Y.; Xue, L.; Forester, C.M.; Cowan, J.E.; Hsieh, A.C.; Cunningham, J.T.; Truillet, C.; Tameire, F.; et al. Development of a stress response therapy targeting aggressive prostate cancer. Sci. Transl. Med. 2018, 10, eaar2036. [Google Scholar] [CrossRef]
- Verginadis, I.I.; Avgousti, H.; Monslow, J.; Skoufos, G.; Chinga, F.; Kim, K.; Leli, N.M.; Karagounis, I.V.; Bell, B.I.; Velalopoulou, A.; et al. A stromal Integrated Stress Response activates perivascular cancer-associated fibroblasts to drive angiogenesis and tumour progression. Nat. Cell Biol. 2022, 24, 940–953. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]

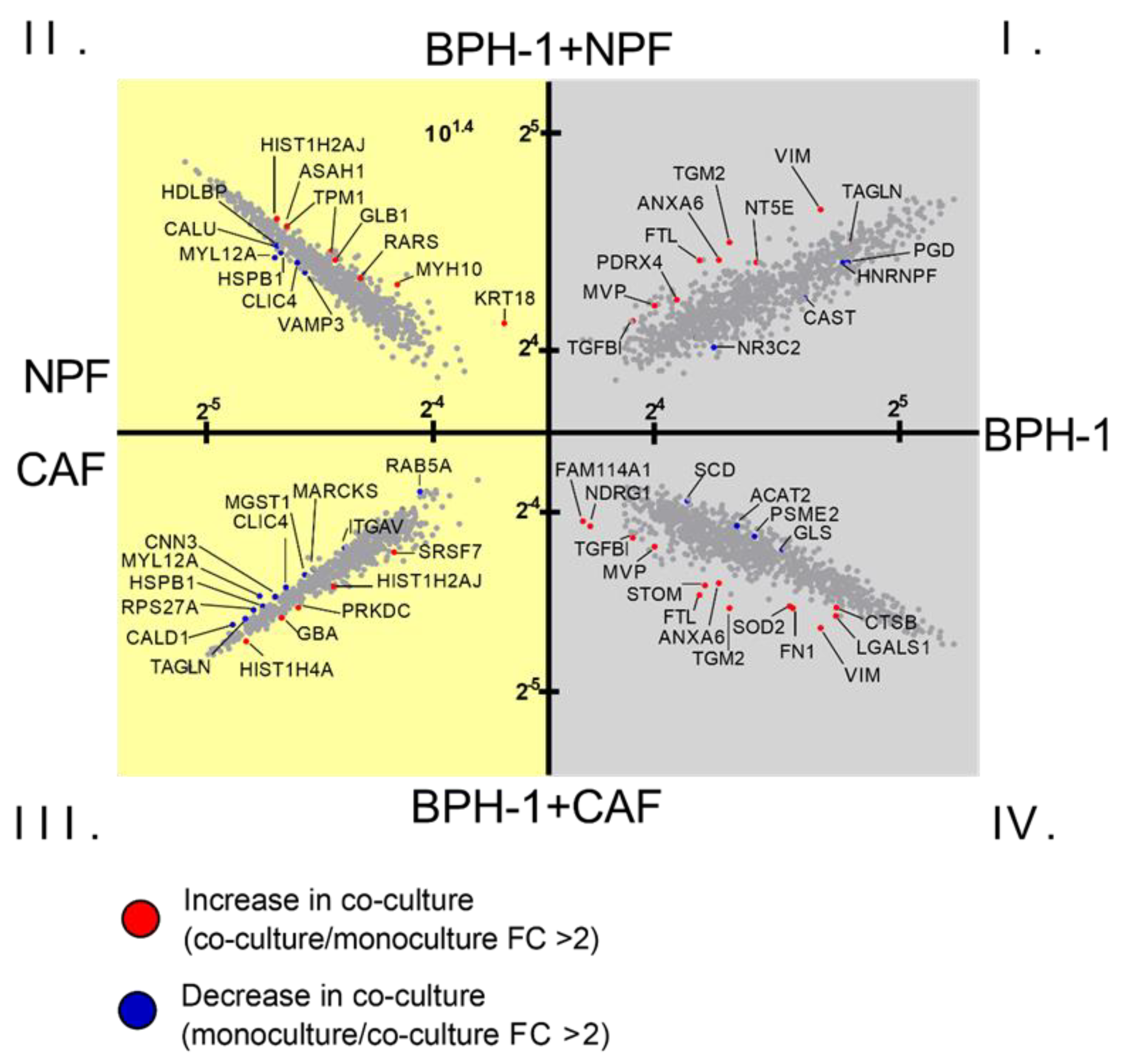
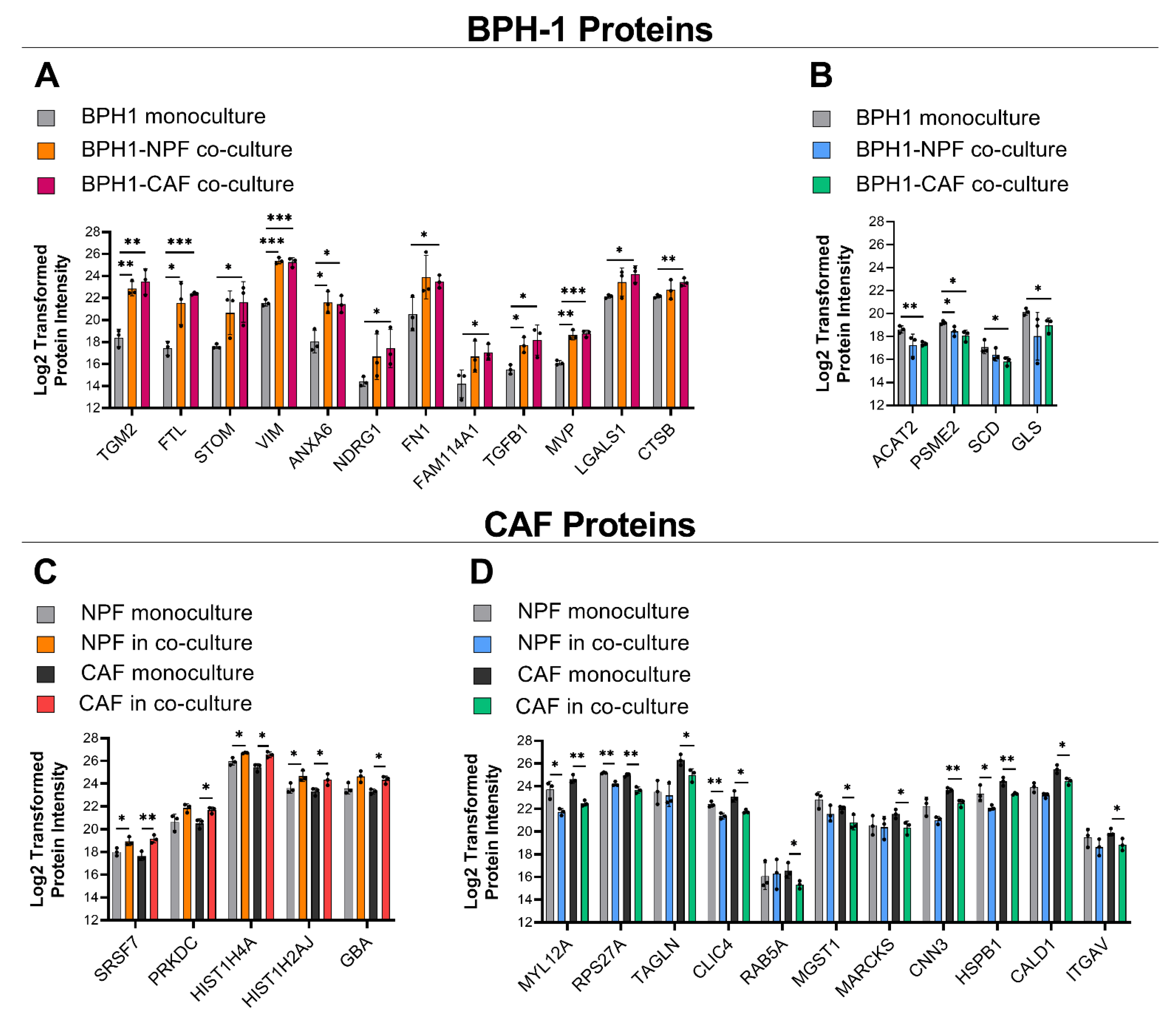
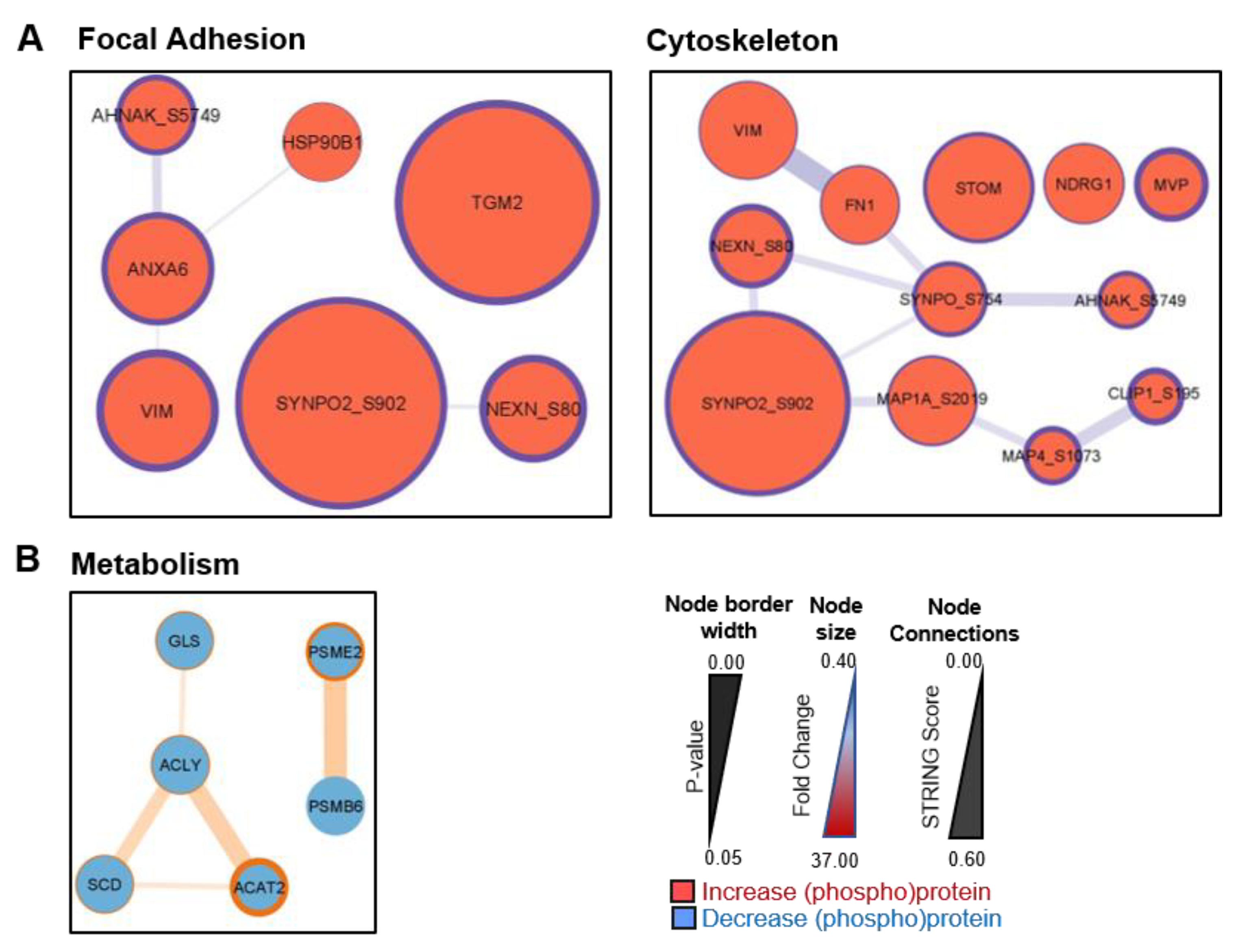
| BPH-1 Protein Fold-Change | ||||
|---|---|---|---|---|
| Protein | BPH-NPF/Monoculture | BPH-CAF/Monoculture | ||
| Fold-Change ± SD | p-Value | Fold-Change ± SD | p-Value | |
| TGM2 | 23.51 ± 9.10 | ** 0.002 | 35.72 ± 10.27 | ** 0.003 |
| FTL | 22.60 ± 17.46 | * 0.026 | 32.90 ± 12.73 | *** 0.000 |
| STOM | 13.78 ± 10.71 | 0.057 | 26.10 ± 20.71 | * 0.020 |
| VIM | 14.85 ± 5.32 | *** 0.000 | 13.21 ± 3.16 | *** 0.000 |
| ANXA6 | 12.26 ± 4.57 | * 0.012 | 15.64 ± 14.00 | * 0.010 |
| NDRG1 | 11.40 ± 16.33 | 0.135 | 15.69 ± 20.80 | * 0.042 |
| FN1 | 11.24 ± 5.28 | 0.081 | 10.06 ± 6.67 | * 0.036 |
| FAM114A1 | 6.19 ± 3.31 | 0.087 | 8.34 ± 5.01 | * 0.027 |
| TGFB1 | 4.89 ± 1.66 | * 0.010 | 8.97 ± 6.43 | * 0.033 |
| MVP | 5.99 ± 2.22 | ** 0.001 | 6.52 ± 2.00 | *** 0.000 |
| LGALS1 | 2.95 ± 1.78 | 0.154 | 4.34 ± 1.74 | * 0.010 |
| CTSB | 1.77 ± 1.23 | 0.318 | 2.53 ± 0.91 | ** 0.005 |
| ACAT2 | 0.47 ± 0.30 | 0.079 | 0.43 ± 0.12 | ** 0.004 |
| PSME2 | 0.60 ± 0.11 | * 0.045 | 0.47 ± 0.10 | * 0.017 |
| SCD | 0.64 ± 0.04 | 0.216 | 0.48 ± 0.25 | * 0.035 |
| GLS | 0.41 ± 0.37 | 0.153 | 0.50 ± 0.26 | * 0.045 |
| Fibroblast Protein Fold-Change | ||||
|---|---|---|---|---|
| Protein | NPF-BPH/Monoculture | CAF-BPH/Monoculture | ||
| Fold-Change ± SD | p-Value | Fold-Change ± SD | p-Value | |
| SRSF7 | 1.92 ± 0.30 | * 0.034 | 2.96 ± 1.08 | ** 0.008 |
| PRKDC | 2.79 ± 2.05 | 0.058 | 2.39 ± 0.96 | * 0.015 |
| HIST1H4A | 1.72 ± 0.41 | * 0.016 | 2.18 ± 0.29 | * 0.010 |
| HIST1H2AJ | 2.34 ± 1.34 | * 0.044 | 2.09 ± 0.35 | * 0.038 |
| GBA | 2.22 ± 0.94 | 0.056 | 2.09 ± 0.53 | * 0.011 |
| MYL12A | 0.27 ± 0.09 | * 0.013 | 0.24 ± 0.08 | ** 0.001 |
| RPS27A | 0.52 ± 0.07 | ** 0.001 | 0.40 ± 0.05 | ** 0.002 |
| TAGLN | 0.85 ± 0.28 | 0.749 | 0.40 ± 0.06 | * 0.037 |
| CLIC4 | 0.50 ± 0.11 | ** 0.007 | 0.42 ± 0.08 | * 0.011 |
| RAB5A | 2.00 ± 2.47 | 0.840 | 0.45 ± 0.15 | * 0.046 |
| MGST1 | 0.43 ± 0.12 | 0.095 | 0.46 ± 0.19 | * 0.040 |
| MARCKS | 1.08 ± 0.79 | 0.841 | 0.48 ± 0.25 | * 0.041 |
| CNN3 | 0.44 ± 0.15 | 0.068 | 0.45 ± 0.05 | ** 0.007 |
| HSPB1 | 0.44 ± 0.13 | * 0.044 | 0.47 ± 0.09 | ** 0.007 |
| CALD1 | 0.61 ± 0.11 | 0.058 | 0.48 ± 0.03 | * 0.017 |
| ITGAV | 0.59 ± 0.23 | 0.241 | 0.51 ± 0.25 | * 0.039 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clark, K.C.; Nguyen, E.V.; Niranjan, B.; Wu, Y.; Lim Kam Sian, T.C.C.; Horvath, L.G.; Taylor, R.A.; Daly, R.J. Cell-Type-Specific Signalling Networks Impacted by Prostate Epithelial-Stromal Intercellular Communication. Cancers 2023, 15, 699. https://doi.org/10.3390/cancers15030699
Clark KC, Nguyen EV, Niranjan B, Wu Y, Lim Kam Sian TCC, Horvath LG, Taylor RA, Daly RJ. Cell-Type-Specific Signalling Networks Impacted by Prostate Epithelial-Stromal Intercellular Communication. Cancers. 2023; 15(3):699. https://doi.org/10.3390/cancers15030699
Chicago/Turabian StyleClark, Kimberley C., Elizabeth V. Nguyen, Birunthi Niranjan, Yunjian Wu, Terry C. C. Lim Kam Sian, Lisa G. Horvath, Renea A. Taylor, and Roger J. Daly. 2023. "Cell-Type-Specific Signalling Networks Impacted by Prostate Epithelial-Stromal Intercellular Communication" Cancers 15, no. 3: 699. https://doi.org/10.3390/cancers15030699
APA StyleClark, K. C., Nguyen, E. V., Niranjan, B., Wu, Y., Lim Kam Sian, T. C. C., Horvath, L. G., Taylor, R. A., & Daly, R. J. (2023). Cell-Type-Specific Signalling Networks Impacted by Prostate Epithelial-Stromal Intercellular Communication. Cancers, 15(3), 699. https://doi.org/10.3390/cancers15030699