Longitudinal and Multimodal Radiomics Models for Head and Neck Cancer Outcome Prediction
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
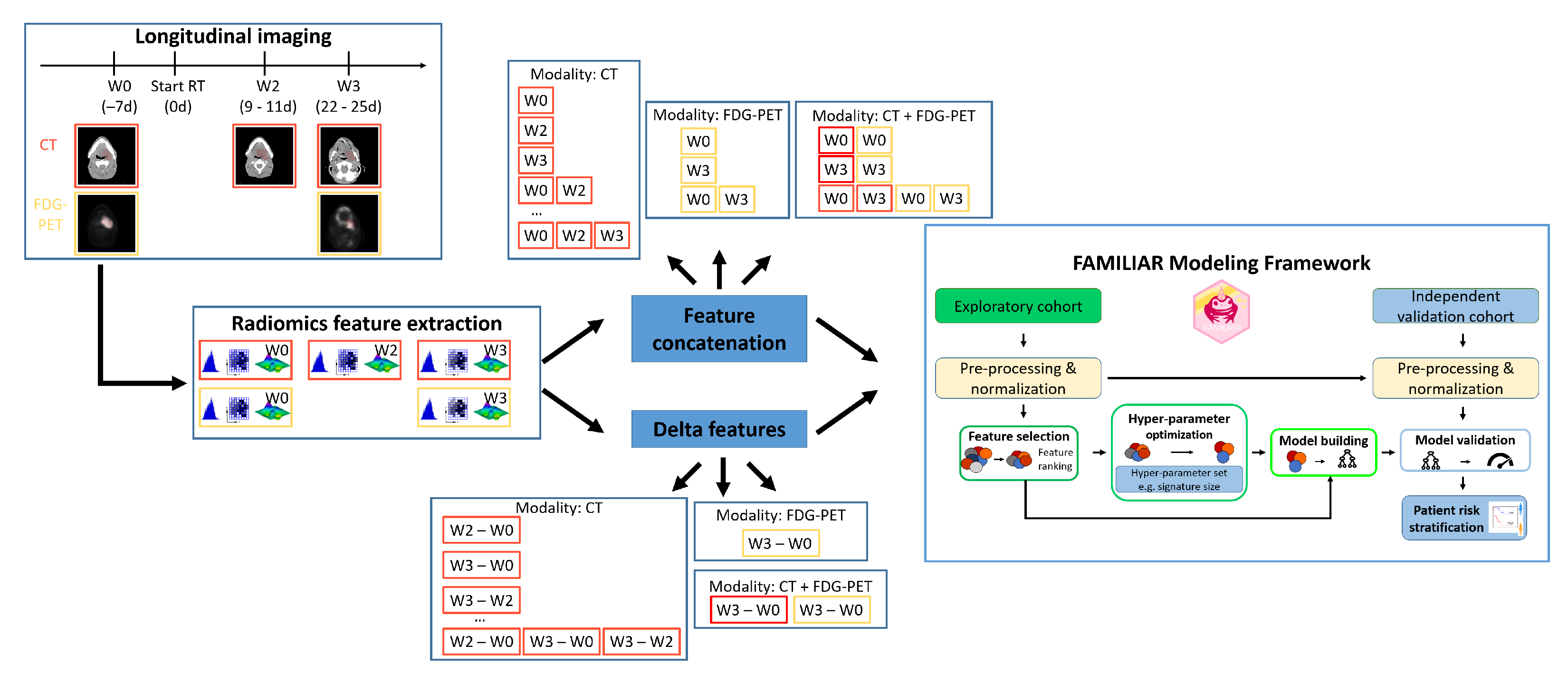
2.2. Image Processing and Feature Extraction
2.3. Analysis Design
2.4. Performance Evaluation
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leemans, C.R.; Braakhuis, B.J.M.; Brakenhoff, R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer 2011, 11, 9–22. [Google Scholar] [CrossRef]
- Baumann, M.; Krause, M.; Overgaard, J.; Debus, J.; Bentzen, S.M.; Daartz, J.; Richter, C.; Zips, D.; Bortfeld, T. Radiation oncology in the era of precision medicine. Nat. Rev. Cancer 2016, 16, 234–249. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Parmar, C.; Grossmann, P.; Rietveld, D.; Rietbergen, M.M.; Lambin, P.; Aerts, H.J. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front. Oncol. 2015, 5, 272. [Google Scholar] [CrossRef] [PubMed]
- Leger, S.; Zwanenburg, A.; Pilz, K.; Lohaus, F.; Linge, A.; Zöphel, K.; Kotzerke, J.; Schreiber, A.; Tinhofer, I.; Budach, V.; et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci. Rep. 2017, 7, 13206. [Google Scholar] [CrossRef]
- Li, Q.; Bai, H.; Chen, Y.; Sun, Q.; Liu, L.; Zhou, S.; Wang, G.; Liang, C.; Li, Z.C. A Fully-Automatic Multiparametric Radiomics Model: Towards Reproducible and Prognostic Imaging Signature for Prediction of Overall Survival in Glioblastoma Multiforme. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ger, R.B.; Zhou, S.; Elgohari, B.; Elhalawani, H.; Mackin, D.M.; Meier, J.G.; Nguyen, C.M.; Anderson, B.M.; Gay, C.; Ning, J.; et al. Radiomics features of the primary tumor fail to improve prediction of overall survival in large cohorts of CT- and PET-imaged head and neck cancer patients. PLoS ONE 2019, 14, e0222509. [Google Scholar] [CrossRef]
- Vallières, M.; Kay-Rivest, E.; Perrin, L.J.; Liem, X.; Furstoss, C.; Aerts, H.J.W.L.; Khaouam, N.; Nguyen-Tan, P.F.; Wang, C.S.; Sultanem, K.; et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci. Rep. 2017, 7, 10117. [Google Scholar] [CrossRef] [PubMed]
- Bogowicz, M.; Leijenaar, R.T.H.; Tanadini-Lang, S.; Riesterer, O.; Pruschy, M.; Studer, G.; Unkelbach, J.; Guckenberger, M.; Konukoglu, E.; Lambin, P. Post-radiochemotherapy PET radiomics in head and neck cancer – The influence of radiomics implementation on the reproducibility of local control tumor models. Radiother. Oncol. 2017, 125, 385–391. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Qian, Z.; Sun, Z.; Xu, K.; Wang, K.; Liu, S.; Fan, X.; Li, S.; Zhang, Z.; et al. A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. NeuroImage: Clin. 2018, 20, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, Y.; Liu, D.; Lv, R.; Huang, Y.; Peng, C.; Jiang, S.; Wang, Y.; He, Y.; Lan, X.; et al. Predicting Progression-Free Survival Using MRI-Based Radiomics for Patients With Nonmetastatic Nasopharyngeal Carcinoma. Front. Oncol. 2020, 10, 618. [Google Scholar] [CrossRef]
- Starke, S.; Thalmeier, D.; Steinbach, P.; Piraud, M. A Hybrid Radiomics Approach to Modeling Progression-Free Survival in Head and Neck Cancers. In Proceedings of the Head and Neck Tumor Segmentation and Outcome Prediction, Strasbourg, France, 27 September 2021; Andrearczyk, V., Oreiller, V., Hatt, M., Depeursinge, A., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 266–277. [Google Scholar] [CrossRef]
- Kwan, J.Y.Y.; Su, J.; Huang, S.H.; Ghoraie, L.S.; Xu, W.; Chan, B.; Yip, K.W.; Giuliani, M.; Bayley, A.; Kim, J.; et al. Radiomic Biomarkers to Refine Risk Models for Distant Metastasis in HPV-related Oropharyngeal Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gensheimer, M.F.; Zhang, N.; Han, F.; Liang, R.; Qian, Y.; Zhang, C.; Fischbein, N.; Pollom, E.L.; Beadle, B.; et al. Integrating Tumor and Nodal Imaging Characteristics at Baseline and Mid-Treatment Computed Tomography Scans to Predict Distant Metastasis in Oropharyngeal Cancer Treated With Concurrent Chemoradiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 942–952. [Google Scholar] [CrossRef]
- Rabasco Meneghetti, A.; Zwanenburg, A.; Leger, S.; Leger, K.; Troost, E.G.; Linge, A.; Lohaus, F.; Schreiber, A.; Kalinauskaite, G.; Tinhofer, I.; et al. Definition and validation of a radiomics signature for loco-regional tumour control in patients with locally advanced head and neck squamous cell carcinoma. Clin. Transl. Radiat. Oncol. 2021, 26, 62–70. [Google Scholar] [CrossRef]
- Mes, S.W.; van Velden, F.H.P.; Peltenburg, B.; Peeters, C.F.W.; Beest, D.E.T.; van de Wiel, A.A.; Mekke, J.; Mulder, D.C.; Martens, R.M.; Castelijns, J.A.; et al. Outcome prediction of head and neck squamous cell carcinoma by MRI radiomic signatures. Eur. Radiol. 2020, 30, 6311–6321. [Google Scholar] [CrossRef]
- Alfieri, S.; Romanò, R.; Bologna, M.; Calareso, G.; Corino, V.; Mirabile, A.; Ferri, A.; Bellanti, L.; Poli, T.; Marcantoni, A.; et al. Prognostic role of pre-treatment magnetic resonance imaging (MRI)-based radiomic analysis in effectively cured head and neck squamous cell carcinoma (HNSCC) patients. Acta Oncol. 2021, 60, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ling, H.; Chen, J.; Tan, L.; Gao, Y.; Li, H.; Tan, P.; Huang, D.; Zhang, X.; Liu, Y.; et al. MRI-based radiomics analysis for preoperative evaluation of lymph node metastasis in hypopharyngeal squamous cell carcinoma. Front. Oncol. 2022, 12, 936040. [Google Scholar] [CrossRef] [PubMed]
- Zips, D.; Zöphel, K.; Abolmaali, N.; Perrin, R.; Abramyuk, A.; Haase, R.; Appold, S.; Steinbach, J.; Kotzerke, J.; Baumann, M. Exploratory prospective trial of hypoxia-specific PET imaging during radiochemotherapy in patients with locally advanced head-and-neck cancer. Radiother. Oncol. 2012, 105, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.; Leijenaar, R.T.H.; Troost, E.G.C.; van Elmpt, W.; Muratet, J.P.; Denis, F.; Ruysscher, D.D.; Aerts, H.J.; Lambin, P. Early variation of FDG-PET radiomics features in NSCLC is related to overall survival—The “delta radiomics” concept. Radiother. Oncol. 2016, 118, S20–S21. [Google Scholar] [CrossRef]
- Löck, S.; Perrin, R.; Seidlitz, A.; Bandurska-Luque, A.; Zschaeck, S.; Zöphel, K.; Krause, M.; Steinbach, J.; Kotzerke, J.; Zips, D.; et al. Residual tumour hypoxia in head-and-neck cancer patients undergoing primary radiochemotherapy, final results of a prospective trial on repeat FMISO-PET imaging. Radiother. Oncol. 2017, 124, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Starke, S.; Leger, S. and Zwanenburg, A.; Leger, K.; Lohaus, F.; Linge, A.; Schreiber, A.; Kalinauskaite, G.; Tinhofer, I.; Guberina, N.; Guberina, M.; et al. 2D and 3D convolutional neural networks for outcome modelling of locally advanced head and neck squamous cell carcinoma. Sci. Rep. 2020, 10, 15625. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, E.; Kurz, C.; Marschner, S.; Avanzo, M.; Gagliardi, V.; Fanetti, G.; Franchin, G.; Stancanello, J.; Corradini, S.; Niyazi, M.; et al. Distant metastasis time to event analysis with CNNs in independent head and neck cancer cohorts. Sci. Rep. 2021, 11, 6418. [Google Scholar] [CrossRef] [PubMed]
- Saeed, N.; Al Majzoub, R.; Sobirov, I.; Yaqub, M. An Ensemble Approach for Patient Prognosis of Head and Neck Tumor Using Multimodal Data. In Proceedings of the Head and Neck Tumor Segmentation and Outcome Prediction, Strasbourg, France, 27 September 2021; Springer International Publishing: Cham, Switzerland, 2022; pp. 278–286. [Google Scholar] [CrossRef]
- Naser, M.A.; Wahid, K.A.; Mohamed, A.S.R.; Abdelaal, M.A.; He, R.; Dede, C.; van Dijk, L.V.; Fuller, C.D. Progression Free Survival Prediction for Head and Neck Cancer Using Deep Learning Based on Clinical and PET/CT Imaging Data. In Proceedings of the Head and Neck Tumor Segmentation and Outcome Prediction, Strasbourg, France, 27 September 2021; Springer International Publishing: Cham, Switzerland, 2022; pp. 287–299. [Google Scholar] [CrossRef]
- Wang, Y.; Lombardo, E.; Avanzo, M.; Zschaek, S.; Weingärtner, J.; Holzgreve, A.; Albert, N.L.; Marschner, S.; Fanetti, G.; Franchin, G.; et al. Deep learning based time-to-event analysis with PET, CT and joint PET/CT for head and neck cancer prognosis. Comput. Methods Progr. Biomed. 2022, 222, 106948. [Google Scholar] [CrossRef]
- Shahin, A.H.; Jacob, J.; Alexander, D.C.; Barber, D. Survival Analysis for Idiopathic Pulmonary Fibrosis using CT Images and Incomplete Clinical Data. In Proceedings of the Medical Imaging with Deep Learning, Zurich, Switzerland, 6–8 July 2022. [Google Scholar]
- Sollini, M.; Antunovic, L.; Chiti, A.; Kirienko, M. Towards clinical application of image mining: A systematic review on artificial intelligence and radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2656–2672. [Google Scholar] [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Leger, S.; Zwanenburg, A.; Pilz, K.; Zschaeck, S.; Zöphel, K.; Kotzerke, J.; Schreiber, A.; Zips, D.; Krause, M.; Baumann, M.; et al. CT imaging during treatment improves radiomic models for patients with locally advanced head and neck cancer. Radiother. Oncol. 2019, 130, 10–17. [Google Scholar] [CrossRef]
- Nasief, H.; Zheng, C.; Schott, D.; Hall, W.; Tsai, S.; Erickson, B.; Allen Li, X. A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. NPJ Precis. Oncol. 2019, 3, 1–10. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; van Elmpt, W.; Leijenaar, R.; Reymen, B.; Monshouwer, R.; Bussink, J.; Paelinck, L.; Bogaert, E.; De Wagter, C.; Elhaseen, E.; et al. Longitudinal radiomics of cone-beam CT images from non-small cell lung cancer patients: Evaluation of the added prognostic value for overall survival and locoregional recurrence. Radiother. Oncol. 2019, 136, 78–85. [Google Scholar] [CrossRef]
- Sellami, S.; Bourbonne, V.; Hatt, M.; Tixier, F.; Bouzid, D.; Lucia, F.; Pradier, O.; Goasduff, G.; Visvikis, D.; Schick, U. Predicting response to radiotherapy of head and neck squamous cell carcinoma using radiomics from cone-beam CT images. Acta Oncol. 2021, 61, 73–80. [Google Scholar] [CrossRef]
- Forouzannezhad, P.; Maes, D.; Hippe, D.S.; Thammasorn, P.; Iranzad, R.; Han, J.; Duan, C.; Liu, X.; Wang, S.; Chaovalitwongse, W.A.; et al. Multitask Learning Radiomics on Longitudinal Imaging to Predict Survival Outcomes following Risk-Adaptive Chemoradiation for Non-Small Cell Lung Cancer. Cancers 2022, 14, 1228. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Leger, S.; Starke, S. Medical Image Radiomics Processor (MIRP). Available online: https://github.com/oncoray/mirp (accessed on 16 September 2022).
- Zwanenburg, A.; Leger, S.; Agolli, L.; Pilz, K.; Troost, E.G.; Richter, C.; Löck, S. Assessing robustness of radiomic features by image perturbation. Sci. Rep. 2019, 9, 614. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Löck, S. familiar: End-to-End Automated Machine Learning and Model Evaluation. 2021. Available online: https://github.com/alexzwanenburg/familiar (accessed on 29 June 2022).
- Peng, H.; Long, F.; Ding, C. Feature Selection Based on Mutual Information: Criteria of Max-Dependency, Max-Relevance, and Min-Redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Cox, D.R. Regression Models and Life-Tables. J. R. Stat. Society. Ser. B (Methodol.) 1972, 34, 187–220. [Google Scholar] [CrossRef]
- Meinshausen, N.; Bühlmann, P. Stability selection. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2010, 72, 417–473. [Google Scholar] [CrossRef]
- Harrell, F.E.; Lee, K.L.; Mark, D.B. Tutorial in biostatistics multivariable prognostic models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 1996, 15, 361–387. [Google Scholar] [CrossRef]
- Linge, A.; Lohaus, F.; Löck, S.; Nowak, A.; Gudziol, V.; Valentini, C.; von Neubeck, C.; Jütz, M.; Tinhofer, I.; Budach, V.; et al. HPV status, cancer stem cell marker expression, hypoxia gene signatures and tumour volume identify good prognosis subgroups in patients with HNSCC after primary radiochemotherapy: A multicentre retrospective study of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG). Radiother. Oncol. 2016, 121, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2006, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Ligero, M.; Jordi-Ollero, O.; Bernatowicz, K.; Garcia-Ruiz, A.; Delgado-Muñoz, E.; Leiva, D.; Mast, R.; Suarez, C.; Sala-Llonch, R.; Calvo, N.; et al. Minimizing acquisition-related radiomics variability by image resampling and batch effect correction to allow for large-scale data analysis. Eur. Radiol. 2021, 31, 1460–1470. [Google Scholar] [CrossRef]
- Fave, X.; Zhang, L.; Yang, J.; Mackin, D.; Balter, P.; Gomez, D.; Followill, D.; Jones, A.K.; Stingo, F.; Liao, Z.; et al. Delta-radiomics features for the prediction of patient outcomes in non–small cell lung cancer. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Eicheler, W.; Krause, M.; Hessel, F.; Zips, D.; Baumann, M. Kinetics of EGFR expression during fractionated irradiation varies between different human squamous cell carcinoma lines in nude mice. Radiother. Oncol. 2005, 76, 151–156. [Google Scholar] [CrossRef]
- Yaromina, A.; Kroeber, T.; Meinzer, A.; Boeke, S.; Thames, H.; Baumann, M.; Zips, D. Exploratory study of the prognostic value of microenvironmental parameters during fractionated irradiation in human squamous cell carcinoma xenografts. Int. J. Radiat. Oncol. Biol. Phys. 2011, 80, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.J.; Mao, W.; Liu, C.; Aref, I.; Elshaikh, M.; Lee, J.K.; Pradhan, D.; Movsas, B.; Chetty, I.J.; Siddiqui, F. Improvements in CBCT Image Quality Using a Novel Iterative Reconstruction Algorithm: A Clinical Evaluation. Adv. Radiat. Oncol. 2019, 4, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, T.; Nishio, T.; Moriya, S.; Tsuneda, M.; Karasawa, K. Feasibility of image quality improvement for high-speed CBCT imaging using deep convolutional neural network for image-guided radiotherapy in prostate cancer. Eur. J. Med. Phys. 2020, 80, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Bertasius, G.; Wang, H.; Torresani, L. Is Space-Time Attention All You Need for Video Understanding? arXiv 2021, arXiv:2102.05095. [Google Scholar] [CrossRef]
| Modality | Imaging Timepoint | Exploration | Independent Validation |
|---|---|---|---|
| CT | |||
| Week 0 | −7 | −6 | |
| Week 2 | 11 | 8.5 | |
| Week 3 | 25 | 22 | |
| FDG | |||
| Week 0 | −7 | −6 | |
| Week 3 | 25 | 22 |
| Variable | Exploratory Cohort (n = 37) | Independent Validation Cohort (n = 18) | p-Value | ||
|---|---|---|---|---|---|
| Median | (Range) | Median | (Range) | ||
| Age (years) | 54 | (42–76) | 54 | (43–67) | 0.67 |
| Primary tumor volume at treatment planning (cm) | 31.20 | (5.06–141.41) | 27.53 | (7.02–183.56) | 0.087 |
| Follow up time of patients alive (months) | 37 | (24–70) | 62 | (8–63) | 0.34 |
| Observed loco-regional recurrence time (months) | 9 | (4–20) | 10 | (3–23) | 1.00 |
| Number of Patients | (%) | Number of Patients | (%) | ||
| Observed loco-regional recurrence | 12 | (32) | 7 | (39) | 0.86 |
| Gender | |||||
| male/female | 32 / 5 | (86 / 14) | 17 / 1 | (94/6) | 0.67 |
| cT-stage | |||||
| T1/T2/T3/T4 | 0/1/15/21 | (0/3/40/57) | 0/8/3/7 | (0/44/17/39) | <0.001 |
| cN-stage | |||||
| N0/N1/N2/N3/unknown | 3/6/27/1/0 | (8/16/73/3/0) | 3/0/13/1/1 | (17/0/71/6/6) | 0.26 |
| UICC-stage | |||||
| I/II/III/IV | 0/0/6/31 | (0/0/16/84) | 0/1/1/16 | (0/6/6/88) | 0.20 |
| Tumor site | |||||
| oropharynx/oral cavity/hypopharynx/larynx | 10/12/11/4 | (27/32/30/11) | 6/6/5/1 | (33/33/28/6) | 0.91 |
| p16 status | |||||
| negative/positive/unknown | 29/2/6 | (78/6/16) | 6/2/10 | (33/11/56) | 0.37 |
| Pathological grading | |||||
| 0/1/2/3/unknown | 0/0/21/16/0 | (0/0/57/43/0) | 0/0/6/9/3 | (0/0/33/50/17) | 0.37 |
| Smoking status | |||||
| no/yes/unknown | 7/30/0 | (19/81/0) | 1/7/10 | (6/39/55) | 1.00 |
| Alcohol consumption | |||||
| no/yes/unknown | 18/17/2 | (49/46/5) | 1/7/10 | (6/39/55) | 0.11 |
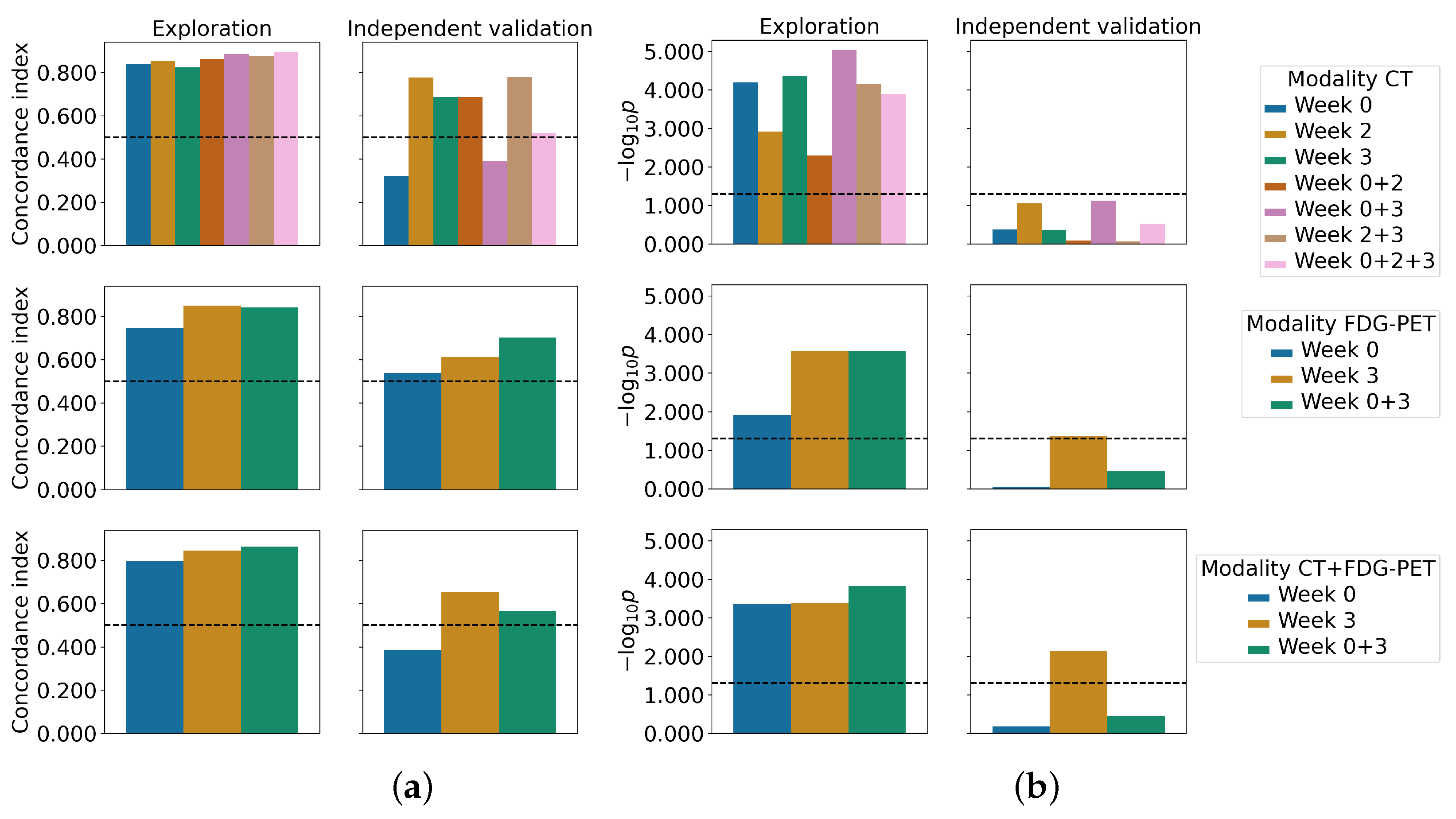
| Experiment | Feature Selection | C-Index | Log-Rank p-Value | Patients in Low-Risk Group | ||
|---|---|---|---|---|---|---|
| Exploration | Ind. Validation | Exploration | Ind. Validation | Ind. Validation | ||
| Week 0 | MRMR | 0.84 (0.70–0.95 ) | 0.32 (0.05–0.64) | <0.001 | 0.42 | 10/18 |
| univariate | 0.76 (0.62 - 0.89) | 0.49 (0.18–0.80) | 0.006 | 0.17 | 9/18 | |
| – (Volume) | 0.62 (0.42–0.78) | 0.45 (0.14–0.75) | 0.062 | 0.074 | 7/18 | |
| Week 2 | MRMR | 0.85 (0.74–0.95) | 0.78 (0.54–0.94) | 0.001 | 0.087 | 10/18 |
| univariate | 0.85 (0.74–0.95) | 0.78 (0.54–0.94) | 0.001 | 0.087 | 10/18 | |
| – (Volume) | 0.72 (0.52–0.87) | 0.78 (0.49–1.00) | 0.020 | 0.61 | 9/18 | |
| Week 3 | MRMR | 0.82 (0.69–0.92) | 0.69 (0.32–0.95) | <0.001 | 0.43 | 11/18 |
| univariate | 0.82 (0.69–0.92) | 0.69 (0.32–0.95) | <0.001 | 0.39 | 14/18 | |
| – (Volume) | 0.74 (0.59–0.86) | 0.59 (0.27–0.85) | 0.003 | 0.47 | 1/18 | |
| Week 0 + 2 | MRMR | 0.86 (0.75–0.96) | 0.69 (0.39–0.95) | 0.005 | 0.80 | 6/18 |
| univariate | 0.86 (0.73–0.96) | 0.76 (0.45–0.94) | 0.002 | 0.017 | 11/18 | |
| – (Volume) | 0.71 (0.56–0.86) | 0.78 (0.49–1.00) | 0.020 | 0.61 | 9/18 | |
| Week 0 + 3 | MRMR | 0.89 (0.76–0.97) | 0.39 (0.13–0.69) | <0.001 | 0.075 | 5/18 |
| univariate | 0.86 (0.71–0.94) | 0.40 (0.10–0.66) | 0.001 | 0.075 | 5/18 | |
| – (Volume) | 0.77 (0.61–0.88) | 0.62 (0.27–0.88) | <0.001 | 0.67 | 1/18 | |
| Week 2 + 3 | MRMR | 0.87 (0.75–0.95) | 0.78 (0.48–0.94) | <0.001 | 0.85 | 6/18 |
| univariate | 0.87 (0.75–0.95) | 0.78 (0.48–0.94) | <0.001 | 0.85 | 6/18 | |
| – (Volume) | 0.75 (0.59–0.86) | 0.56 (0.21–0.81) | 0.003 | 0.47 | 1/18 | |
| Week 0 + 2 + 3 | MRMR | 0.89 (0.80–0.97) | 0.52 (0.20–0.80) | <0.001 | 0.30 | 4/18 |
| univariate | 0.86 (0.73–0.96) | 0.53 (0.16–0.87) | <0.001 | 0.075 | 4/18 | |
| – (Volume) | 0.77 (0.62–0.89) | 0.66 (0.38–0.88) | <0.001 | 0.40 | 2/18 | |
| Experiment | Feature Selection | C-Index | Log-Rank p-Value | Patients in Low-Risk Group | ||
|---|---|---|---|---|---|---|
| Exploration | Ind. Validation | Exploration | Ind. Validation | Ind. Validation | ||
| Week 0 | MRMR | 0.75 (0.59–0.86) | 0.54 (0.24–0.85) | 0.012 | 0.90 | 10/18 |
| univariate | 0.72 (0.53–0.85) | 0.51 (0.19–0.81) | 0.72 | 0.78 | 11/18 | |
| – (Volume) | 0.60 (0.41–0.79) | 0.49 (0.18–0.78) | 0.062 | 0.17 | 5/18 | |
| Week 3 | MRMR | 0.85 (0.73–0.94) | 0.61 (0.05–0.97) | <0.001 | 0.044 * | 17/18 |
| univariate | 0.88 (0.80–0.95) | 0.64 (0.13–1.0) | <0.001 | <0.001* | 16/18 | |
| – (Volume) | 0.74 (0.58–0.87) | 0.72 (0.48–0.92) | 0.003 | 0.90 | 3/18 | |
| Week 0 + 3 | MRMR | 0.84 (0.71–0.93) | 0.70 (0.23–1.0) | <0.001 | 0.36 | 16/18 |
| univariate | 0.89 (0.79–0.96) | 0.42 (0.0–0.87) | <0.001 | 0.45 | 12/18 | |
| – (Volume) | 0.76 (0.61–0.88) | 0.66 (0.37–0.88) | <0.001 | 0.90 | 3/18 | |
| Experiment | Feature Selection | C-Index | Log-Rank p-Value | Patients in Low-Risk Group | ||
|---|---|---|---|---|---|---|
| Exploration | Ind. Validation | Exploration | Ind. Validation | Ind. Validation | ||
| Week 0 | MRMR | 0.80 (0.64–0.94) | 0.39 (0.10–0.73) | <0.001 | 0.66 | 13/18 |
| univariate | 0.76 (0.60–0.92) | 0.56 (0.24–0.85) | 0.007 | 0.89 | 9/18 | |
| Week 3 | MRMR | 0.84 (0.74–0.93) | 0.65 (0.31–0.94) | <0.001 | 0.007 * | 15/18 |
| univariate | 0.84 (0.72–0.93) | 0.56 (0.07–0.89) | <0.001 | 0.044 * | 17/18 | |
| Week 0 + 3 | MRMR | 0.86 (0.74–0.94) | 0.57 (0.11–0.88) | <0.001 | 0.36 | 16/18 |
| univariate | 0.89 (0.82–0.96) | 0.38 (0.0–0.84) | <0.001 | 0.55 | 11/18 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Starke, S.; Zwanenburg, A.; Leger, K.; Zöphel, K.; Kotzerke, J.; Krause, M.; Baumann, M.; Troost, E.G.C.; Löck, S. Longitudinal and Multimodal Radiomics Models for Head and Neck Cancer Outcome Prediction. Cancers 2023, 15, 673. https://doi.org/10.3390/cancers15030673
Starke S, Zwanenburg A, Leger K, Zöphel K, Kotzerke J, Krause M, Baumann M, Troost EGC, Löck S. Longitudinal and Multimodal Radiomics Models for Head and Neck Cancer Outcome Prediction. Cancers. 2023; 15(3):673. https://doi.org/10.3390/cancers15030673
Chicago/Turabian StyleStarke, Sebastian, Alexander Zwanenburg, Karoline Leger, Klaus Zöphel, Jörg Kotzerke, Mechthild Krause, Michael Baumann, Esther G. C. Troost, and Steffen Löck. 2023. "Longitudinal and Multimodal Radiomics Models for Head and Neck Cancer Outcome Prediction" Cancers 15, no. 3: 673. https://doi.org/10.3390/cancers15030673
APA StyleStarke, S., Zwanenburg, A., Leger, K., Zöphel, K., Kotzerke, J., Krause, M., Baumann, M., Troost, E. G. C., & Löck, S. (2023). Longitudinal and Multimodal Radiomics Models for Head and Neck Cancer Outcome Prediction. Cancers, 15(3), 673. https://doi.org/10.3390/cancers15030673







