Follicular Thyroid Adenoma and Follicular Thyroid Carcinoma—A Common or Distinct Background? Loss of Heterozygosity in Comprehensive Microarray Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hegedüs, L.; Bonnema, S.J.; Bennedbaek, F.N. Management of Simple Nodular Goiter: Current Status and Future Perspectives. Endocr. Rev. 2003, 24, 102–132. [Google Scholar] [CrossRef] [PubMed]
- Haugen, B.R.; Alexander, E.K.; Bible, K.C.; Doherty, G.M.; Mandel, S.J.; Nikiforov, Y.E.; Pacini, F.; Randolph, G.W.; Sawka, A.M.; Schlumberger, M.; et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid 2016, 26, 1–133. [Google Scholar] [CrossRef] [PubMed]
- Hegedüs, L. Clinical Practice. The Thyroid Nodule. N. Engl. J. Med. 2004, 351, 1764–1771. [Google Scholar] [CrossRef] [PubMed]
- Holt, E.H. Current Evaluation of Thyroid Nodules. Med. Clin. N. Am. 2021, 105, 1017–1031. [Google Scholar] [CrossRef]
- Ohori, N.P.; Nishino, M. Follicular Neoplasm of Thyroid Revisited: Current Differential Diagnosis and the Impact of Molecular Testing. Adv. Anat. Pathol. 2022, 30, 11–23. [Google Scholar] [CrossRef]
- Borowczyk, M.; Woliński, K.; Więckowska, B.; Jodłowska-Siewert, E.; Szczepanek-Parulska, E.; Verburg, F.A.; Ruchała, M. Sonographic Features Differentiating Follicular Thyroid Cancer from Follicular Adenoma-A Meta-Analysis. Cancers 2021, 13, 938. [Google Scholar] [CrossRef]
- Filetti, S.; Durante, C.; Hartl, D.; Leboulleux, S.; Locati, L.D.; Newbold, K.; Papotti, M.G.; Berruti, A.; ESMO Guidelines Committee. Thyroid Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2019, 30, 1856–1883. [Google Scholar] [CrossRef]
- McHenry, C.R.; Phitayakorn, R. Follicular Adenoma and Carcinoma of the Thyroid Gland. Oncologist 2011, 16, 585–593. [Google Scholar] [CrossRef]
- Jung, S.-H.; Kim, M.S.; Jung, C.K.; Park, H.-C.; Kim, S.Y.; Liu, J.; Bae, J.-S.; Lee, S.H.; Kim, T.-M.; Lee, S.H.; et al. Mutational Burdens and Evolutionary Ages of Thyroid Follicular Adenoma Are Comparable to Those of Follicular Carcinoma. Oncotarget 2016, 7, 69638–69648. [Google Scholar] [CrossRef]
- Nikiforov, Y.E.; Nikiforova, M.N. Molecular Genetics and Diagnosis of Thyroid Cancer. Nat. Rev. Endocrinol. 2011, 7, 569–580. [Google Scholar] [CrossRef]
- Borowczyk, M.; Szczepanek-Parulska, E.; Olejarz, M.; Więckowska, B.; Verburg, F.A.; Dębicki, S.; Budny, B.; Janicka-Jedyńska, M.; Ziemnicka, K.; Ruchała, M. Evaluation of 167 Gene Expression Classifier (GEC) and ThyroSeq v2 Diagnostic Accuracy in the Preoperative Assessment of Indeterminate Thyroid Nodules: Bivariate/HROC Meta-Analysis. Endocr. Pathol. 2019, 30, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Akbani, R.; Aksoy, B.A.; Ally, A.; Arachchi, H.; Asa, S.L.; Auman, J.T.; Balasundaram, M.; Balu, S.; Baylin, S.B.; et al. Cancer Genome Atlas Research Network Integrated Genomic Characterization of Papillary Thyroid Carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Luthra, R.; Routbort, M.J.; Patel, K.P.; Cabanillas, M.E.; Broaddus, R.R.; Williams, M.D. Molecular Profile of Advanced Thyroid Carcinomas by Next-Generation Sequencing: Characterizing Tumors Beyond Diagnosis for Targeted Therapy. Mol. Cancer Ther. 2018, 17, 1575–1584. [Google Scholar] [CrossRef]
- Prete, A.; Borges de Souza, P.; Censi, S.; Muzza, M.; Nucci, N.; Sponziello, M. Update on Fundamental Mechanisms of Thyroid Cancer. Front. Endocrinol. 2020, 11, 102. [Google Scholar] [CrossRef]
- Borowczyk, M.; Szczepanek-Parulska, E.; Dębicki, S.; Budny, B.; Verburg, F.A.; Filipowicz, D.; Więckowska, B.; Janicka-Jedyńska, M.; Gil, L.; Ziemnicka, K.; et al. Differences in Mutational Profile between Follicular Thyroid Carcinoma and Follicular Thyroid Adenoma Identified Using Next Generation Sequencing. Int. J. Mol. Sci. 2019, 20, 3126. [Google Scholar] [CrossRef] [PubMed]
- Borowczyk, M.; Szczepanek-Parulska, E.; Dębicki, S.; Budny, B.; Verburg, F.A.; Filipowicz, D.; Wrotkowska, E.; Janicka-Jedyńska, M.; Więckowska, B.; Gil, L.; et al. Genetic Heterogeneity of Indeterminate Thyroid Nodules Assessed Preoperatively with Next-Generation Sequencing Reflects the Diversity of the Final Histopathologic Diagnosis. Pol. Arch. Intern. Med. 2019, 129, 761–769. [Google Scholar] [CrossRef]
- Borowczyk, M.; Szczepanek-Parulska, E.; Dębicki, S.; Budny, B.; Janicka-Jedyńska, M.; Gil, L.; Verburg, F.A.; Filipowicz, D.; Wrotkowska, E.; Majchrzycka, B.; et al. High Incidence of FLT3 Mutations in Follicular Thyroid Cancer: Potential Therapeutic Target in Patients with Advanced Disease Stage. Ther. Adv. Med. Oncol. 2020, 12, 1758835920907534. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Gao, J.-B.; Cao, Y.; Bottinger, E.; Zhang, W. Exploiting Noise in Array CGH Data to Improve Detection of DNA Copy Number Change. Nucleic. Acids. Res. 2007, 35, e35. [Google Scholar] [CrossRef]
- Migdalska-Sęk, M.; Czarnecka, K.H.; Kusiński, M.; Pastuszak-Lewandoska, D.; Nawrot, E.; Kuzdak, K.; Brzeziańska-Lasota, E. Clinicopathological Significance of Overall Frequency of Allelic Loss (OFAL) in Lesions Derived from Thyroid Follicular Cell. Mol. Diagn. Ther. 2019, 23, 369–382. [Google Scholar] [CrossRef]
- Migdalska-Sęk, M.; Pastuszak-Lewandoska, D.; Brzeziańska, E. MSI and LOH in the Development and Prognosis of Follicular Cell-Derived Thyroid Tumours. Endokrynol. Pol. 2012, 63, 126–136. [Google Scholar]
- Kim, J.H.; Choi, K.Y.; Lee, D.J.; Rho, Y.-S.; Jo, S.-J. Loss of Heterozygosities in Five Tumor Suppressor Genes (FHIT Gene, P16, PRb, E-Cadherin and P53) in Thyroid Tumors. Clin. Exp. Otorhinolaryngol. 2014, 7, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.M.; Oumie, A.; Togneri, F.S.; Vasques, F.R.; Hau, D.; Taylor, M.; Tinkler-Hundal, E.; Southward, K.; Medlow, P.; McGreeghan-Crosby, K.; et al. Cross-Laboratory Validation of the OncoScan® FFPE Assay, a Multiplex Tool for Whole Genome Tumour Profiling. BMC Med. Genom. 2015, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.R.; Mehrotra, M.; Chen, H.; Almohammedsalim, A.A.; Sahin, A.; Bosamra, A.; Patel, K.P.; Routbort, M.J.; Lu, X.; Ronald, A.; et al. Comprehensive Screening of Gene Copy Number Aberrations in Formalin-Fixed, Paraffin-Embedded Solid Tumors Using Molecular Inversion Probe–Based Single-Nucleotide Polymorphism Array. J. Mol. Diagn. 2016, 18, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Jarząb, B.; Dedecjus, M.; Lewiński, A.; Adamczewski, Z.; Bakuła-Zalewska, E.; Bałdys-Waligórska, A.; Barczyński, M.; Biskup-Frużyńska, M.; Bobek-Billewicz, B.; Bossowski, A.; et al. Diagnosis and Treatment of Thyroid Cancer in Adult Patients—Recommendations of Polish Scientific Societies and the National Oncological Strategy. 2022 Update [Diagnostyka i Leczenie Raka Tarczycy u Chorych Dorosłych—Rekomendacje Polskich Towarzystw Naukowych Oraz Narodowej Strategii Onkologicznej. Aktualizacja Na Rok 2022]. Endokrynol. Pol. 2022, 73, 173–300. [Google Scholar] [CrossRef] [PubMed]
- Sawicka-Gutaj, N.; Gruszczyński, D.; Guzik, P.; Mostowska, A.; Walkowiak, J. Publication Ethics of Human Studies in the Light of the Declaration of Helsinki—A Mini-Review. J.Med. Sci. 2022, 91, e700. [Google Scholar] [CrossRef]
- Weber, F.; Aldred, M.A.; Morrison, C.D.; Plass, C.; Frilling, A.; Broelsch, C.E.; Waite, K.A.; Eng, C. Silencing of the Maternally Imprinted Tumor Suppressor ARHI Contributes to Follicular Thyroid Carcinogenesis. J. Clin. Endocrinol. Metab. 2005, 90, 1149–1155. [Google Scholar] [CrossRef]
- Rodrigues-Serpa, A.; Catarino, A.; Soares, J. Loss of Heterozygosity in Follicular and Papillary Thyroid Carcinomas. Cancer Genet. Cytogenet. 2003, 141, 26–31. [Google Scholar] [CrossRef]
- Trovato, M.; Fraggetta, F.; Villari, D.; Batolo, D.; Mackey, K.; Trimarchi, F.; Benvenga, S. Loss of Heterozygosity of the Long Arm of Chromosome 7 in Follicular and Anaplastic Thyroid Cancer, but Not in Papillary Thyroid Cancer 1. J. Clin. Endocrinol. Metab. 1999, 84, 3235–3240. [Google Scholar] [CrossRef]
- Zhang, J.-S.; Nelson, M.; McIver, B.; Hay, I.D.; Goellner, J.R.; Grant, C.S.; Eberhardt, N.L.; Smith, D.I. Differential Loss of Heterozygosity at 7q31.2 in Follicular and Papillary Thyroid Tumors. Oncogene 1998, 17, 789–793. [Google Scholar] [CrossRef]
- Farrand, K.; Delahunt, B.; Wang, X.-L.; McIver, B.; Hay, I.D.; Goellner, J.R.; Eberhardt, N.L.; Grebe, S.K.G. High Resolution Loss of Heterozygosity Mapping of 17p13 in Thyroid Cancer: Hurthle Cell Carcinomas Exhibit a Small 411-Kilobase Common Region of Allelic Imbalance, Probably Containing a Novel Tumor Suppressor Gene. J. Clin. Endocrinol. Metab. 2002, 87, 4715–4721. [Google Scholar] [CrossRef]
- Grebe, S.K.; McIver, B.; Hay, I.D.; Wu, P.S.; Maciel, L.M.; Drabkin, H.A.; Goellner, J.R.; Grant, C.S.; Jenkins, R.B.; Eberhardt, N.L. Frequent Loss of Heterozygosity on Chromosomes 3p and 17p without VHL or P53 Mutations Suggests Involvement of Unidentified Tumor Suppressor Genes in Follicular Thyroid Carcinoma. J. Clin. Endocrinol. Metab. 1997, 82, 3684–3691. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.-J.; Xu, H.-D.; Zhou, R.; Li, X.-F.; Zhang, H.-Y. Loss of heterozygosity on chromosome 3p in thyroid tumors. Zhonghua Bing Li Xue Za Zhi Chin. J. Pathol. 2008, 37, 305–308. [Google Scholar]
- Nikiforova, M.N.; Lynch, R.A.; Biddinger, P.W.; Alexander, E.K.; Dorn, G.W.; Tallini, G.; Kroll, T.G.; Nikiforov, Y.E. RAS Point Mutations and PAX8-PPAR Gamma Rearrangement in Thyroid Tumors: Evidence for Distinct Molecular Pathways in Thyroid Follicular Carcinoma. J. Clin. Endocrinol. Metab. 2003, 88, 2318–2326. [Google Scholar] [CrossRef] [PubMed]
- Tanikawa, C.; Kamatani, Y.; Toyoshima, O.; Sakamoto, H.; Ito, H.; Takahashi, A.; Momozawa, Y.; Hirata, M.; Fuse, N.; Takai-Igarashi, T.; et al. Genome-Wide Association Study Identifies Gastric Cancer Susceptibility Loci at 12q24.11-12 and 20q11.21. Cancer Sci. 2018, 109, 4015–4024. [Google Scholar] [CrossRef]
- Wolf, M.; Korja, M.; Karhu, R.; Edgren, H.; Kilpinen, S.; Ojala, K.; Mousses, S.; Kallioniemi, A.; Haapasalo, H. Array-Based Gene Expression, CGH and Tissue Data Defines a 12q24 Gain in Neuroblastic Tumors with Prognostic Implication. BMC Cancer 2010, 10, 181. [Google Scholar] [CrossRef]
- Zou, X.; Gao, Y.; Ruvolo, V.R.; Gardner, T.L.; Ruvolo, P.P.; Brown, R.E. Human Glycolipid Transfer Protein Gene (GLTP) Expression Is Regulated by Sp1 and Sp3: Involvement of the Bioactive Sphingolipid Ceramide. J. Biol. Chem. 2011, 286, 1301–1311. [Google Scholar] [CrossRef]
- Hemmer, S.; Wasenius, V.M.; Knuutila, S.; Franssila, K.; Joensuu, H. DNA Copy Number Changes in Thyroid Carcinoma. Am. J. Pathol. 1999, 154, 1539–1547. [Google Scholar] [CrossRef]
- Kitamura, Y.; Shimizu, K.; Ito, K.; Tanaka, S.; Emi, M. Allelotyping of Follicular Thyroid Carcinoma: Frequent Allelic Losses in Chromosome Arms 7q, 11p, and 22q. J. Clin. Endocrinol. Metab. 2001, 86, 4268–4272. [Google Scholar] [CrossRef]
- Ward, L.S.; Brenta, G.; Medvedovic, M.; Fagin, J.A. Studies of Allelic Loss in Thyroid Tumors Reveal Major Differences in Chromosomal Instability between Papillary and Follicular Carcinomas. J. Clin. Endocrinol. Metab. 1998, 83, 525–530. [Google Scholar] [CrossRef]
- Boland, C.R.; Thibodeau, S.N.; Hamilton, S.R.; Sidransky, D.; Eshleman, J.R.; Burt, R.W.; Meltzer, S.J.; Rodriguez-Bigas, M.A.; Fodde, R.; Ranzani, G.N.; et al. A National Cancer Institute Workshop on Microsatellite Instability for Cancer Detection and Familial Predisposition: Development of International Criteria for the Determination of Microsatellite Instability in Colorectal Cancer. Cancer Res. 1998, 58, 5248–5257. [Google Scholar]
- Knudson, A.G. Two Genetic Hits (More or Less) to Cancer. Nat. Rev. Cancer 2001, 1, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Nikitski, A.V.; Nikiforova, M.N.; Yip, L.; Karslioglu-French, E.; Carty, S.E.; Nikiforov, Y.E. Can TP53-Mutant Follicular Adenoma Be a Precursor of Anaplastic Thyroid Carcinoma? Endocr. Relat. Cancer 2021, 28, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.L.; Livolsi, V.A.; Baloch, Z.W.; Swalsky, P.A.; Bakker, A.; Sasatomi, E.; Finkelstein, S.; Barnes, E.L. A Novel Microdissection and Genotyping of Follicular-Derived Thyroid Tumors to Predict Aggressiveness. Hum. Pathol. 2003, 34, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D.J.; Zheng, Z.; Zedenius, J.; Kremer, H.; Padberg, G.W.; Larsson, C.; Longy, M.; Eng, C. Differential Loss of Heterozygosity in the Region of the Cowden Locus within 10q22-23 in Follicular Thyroid Adenomas and Carcinomas. Cancer Res. 1997, 57, 500–503. [Google Scholar] [PubMed]
- Wozniak, A.; Wiench, M.; Olejniczak, A.; Wloch, J.; Lachinski, A.; Lange, D.; Olczyk, T.; Jarzab, B.; Limon, J. Loss of Heterozygosity in 73 Human Thyroid Tumors. Neuroendocrinol. Lett. 2005, 26, 521–525. [Google Scholar]
- Sarquis, M.S.; Weber, F.; Shen, L.; Broelsch, C.E.; Jhiang, S.M.; Zedenius, J.; Frilling, A.; Eng, C. High Frequency of Loss of Heterozygosity in Imprinted, Compared with Nonimprinted, Genomic Regions in Follicular Thyroid Carcinomas and Atypical Adenomas. J. Clin. Endocrinol. Metab. 2006, 91, 262–269. [Google Scholar] [CrossRef]
- Gerashchenko, T.S.; Denisov, E.V.; Litviakov, N.V.; Zavyalova, M.V.; Vtorushin, S.V.; Tsyganov, M.M.; Perelmuter, V.M.; Cherdyntseva, N.V. Intratumor Heterogeneity: Nature and Biological Significance. Biochemistry 2013, 78, 1201–1215. [Google Scholar] [CrossRef]
- Chmielik, E.; Rusinek, D.; Oczko-Wojciechowska, M.; Jarzab, M.; Krajewska, J.; Czarniecka, A.; Jarzab, B. Heterogeneity of Thyroid Cancer. Pathobiology 2018, 85, 117–129. [Google Scholar] [CrossRef]
- Pizzo, L.; Lasser, M.; Yusuff, T.; Jensen, M.; Ingraham, P.; Huber, E.; Singh, M.D.; Monahan, C.; Iyer, J.; Desai, I.; et al. Functional Assessment of the “Two-Hit” Model for Neurodevelopmental Defects in Drosophila and X. Laevis. PLoS Genet. 2021, 17, e1009112. [Google Scholar] [CrossRef]
- Vasko, V.V.; Gaudart, J.; Allasia, C.; Savchenko, V.; Di Cristofaro, J.; Saji, M.; Ringel, M.D.; De Micco, C. Thyroid follicular adenomas may display features of follicular carcinoma and follicular variant of papillary carcinoma. Eur. J. Endocrinol. 2004, 151, 779–786. [Google Scholar] [CrossRef]
- Odermatt, A.; Barton, K.; Khanna, V.K.; Mathieu, J.; Escolar, D.; Kuntzer, T.; Karpati, G.; MacLennan, D.H. The Mutation of Pro789 to Leu Reduces the Activity of the Fast-Twitch Skeletal Muscle Sarco(Endo)Plasmic Reticulum Ca2+ ATPase (SERCA1) and Is Associated with Brody Disease. Hum. Genet. 2000, 106, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Expression of ATP2A1 in Cancer-Summary-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000196296-ATP2A1/pathology (accessed on 31 July 2022).
- Bloise, F.F.; Cordeiro, A.; Ortiga-Carvalho, T.M. Role of Thyroid Hormone in Skeletal Muscle Physiology. J. Endocrinol. 2018, 236, R57–R68. [Google Scholar] [CrossRef] [PubMed]
- GeneCards Database.
- Hartong, R.; Wang, N.; Kurokawa, R.; Lazar, M.A.; Glass, C.K.; Apriletti, J.W.; Dillmann, W.H. Delineation of Three Different Thyroid Hormone-Response Elements in Promoter of Rat Sarcoplasmic Reticulum Ca2+ ATPase Gene. Demonstration That Retinoid X Receptor Binds 5’ to Thyroid Hormone Receptor in Response Element 1. J. Biol. Chem. 1994, 269, 13021–13029. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.; Vanderlinden, G.C.; Zuidwijk, M.J.; Simonides, W.S.; Vanderlaarse, W.J.; Vanhardeveld, C. Differential Effects of Thyroid Hormone on the Expression of Sarcoplasmic Reticulum Ca2+-ATPase Isoforms in Rat Skeletal Muscle Fibers. Biochem. Biophys. Res. Commun. 1994, 203, 1035–1042. [Google Scholar] [CrossRef]
- Simonides, W.S.; Brent, G.A.; Thelen, M.M.; van der Linden, C.G.; Larsen, P.R.; van Hardeveld, C. Characterization of the Promoter of the Rat Sarcoplasmic Endoplasmic Reticulum Ca2+-ATPase 1 Gene and Analysis of Thyroid Hormone Responsiveness. J. Biol. Chem. 1996, 271, 32048–32056. [Google Scholar] [CrossRef]
- Dang, D.; Rao, R. Calcium-ATPases: Gene Disorders and Dysregulation in Cancer. Biochim. Biophys. Acta(BBA)-Mol. Cell Res. 2016, 1863, 1344–1350. [Google Scholar] [CrossRef]
- He, W.; Wang, B.; Mu, K.; Zhang, J.; Yang, Y.; Yao, W.; Li, S.; Zhang, J. Association of Single-Nucleotide Polymorphisms in the IL27 Gene with Autoimmune Thyroid Diseases. Endocr. Connect. 2019, 8, 173–181. [Google Scholar] [CrossRef]
- Saeed, M.-H.; Kurosh, K.; Zahra, A.; Hossein, D.M.; Davood, R.; Ataollahi, M.R. Decreased Serum Levels of IL-27and IL-35 in Patients with Graves Disease. Arch. Endocrinol. Metab. 2020, 64, 521–527. [Google Scholar] [CrossRef]
- Nie, X.; Yuan, F.; Chen, P.; Pu, Y.; Zhu, J.; Wang, Y.; Xiao, X.; Che, G.; Gao, L.; Zhang, L. Association between IL-27 Gene Polymorphisms and Risk of Papillary Thyroid Carcinoma. Biomark. Med. 2017, 11, 141–149. [Google Scholar] [CrossRef]
- Xi, C.; Zhang, G.-Q.; Sun, Z.-K.; Song, H.-J.; Shen, C.-T.; Chen, X.-Y.; Sun, J.-W.; Qiu, Z.-L.; Luo, Q.-Y. Interleukins in Thyroid Cancer: From Basic Researches to Applications in Clinical Practice. Front. Immunol. 2020, 11, 1124. [Google Scholar] [CrossRef]
- Jia, H.; Dilger, P.; Bird, C.; Wadhwa, M. IL-27 Promotes Proliferation of Human Leukemic Cell Lines Through the MAPK/ERK Signaling Pathway and Suppresses Sensitivity to Chemotherapeutic Drugs. J. Interferon Cytokine Res. 2016, 36, 302–316. [Google Scholar] [CrossRef]
- Larousserie, F.; Bardel, E.; Coulomb L’Herminé, A.; Canioni, D.; Brousse, N.; Kastelein, R.; Devergne, O. Variable Expression of Epstein–Barr Virus-Induced Gene 3 during Normal B-Cell Differentiation and among B-Cell Lymphomas. J. Pathol. 2006, 209, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Gonin, J.; Carlotti, A.; Dietrich, C.; Audebourg, A.; Radenen-Bussière, B.; Caignard, A.; Avril, M.-F.; Vacher-Lavenu, M.-C.; Larousserie, F.; Devergne, O. Expression of IL-27 by Tumor Cells in InvasCutaneous and Metastatic Melanomas. PLoS ONE 2013, 8, e75694. [Google Scholar] [CrossRef]
- Kourko, O.; Seaver, K.; Odoardi, N.; Basta, S.; Gee, K. IL-27, IL-30, and IL-35: A Cytokine Triumvirate in Cancer. Front. Oncol. 2019, 9, 969. [Google Scholar] [CrossRef] [PubMed]
- Pisarev, M.A.; Thomasz, L.; Juvenal, G.J. Role of Transforming Growth Factor Beta in the Regulation of Thyroid Function and Growth. Thyroid 2009, 19, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Kardalas, E.; Sakkas, E.; Ruchala, M.; Macut, D.; Mastorakos, G. The Role of Transforming Growth Factor Beta in Thyroid Autoimmunity: Current Knowledge and Future Perspectives. Rev. Endocr. Metab. Disord. 2022, 23, 431–447. [Google Scholar] [CrossRef]
- Mincione, G.; Di Marcantonio, M.C.; Tarantelli, C.; D’Inzeo, S.; Nicolussi, A.; Nardi, F.; Donini, C.F.; Coppa, A. EGF and TGF- <b/> 1 Effects on Thyroid Function. J. Thyroid. Res. 2011, 2011, 1–13. [Google Scholar] [CrossRef]
- Grubeck-Loebenstein, B.; Buchan, G.; Sadeghi, R.; Kissonerghis, M.; Londei, M.; Turner, M.; Pirich, K.; Roka, R.; Niederle, B.; Kassal, H. Transforming Growth Factor Beta Regulates Thyroid Growth. Role in the Pathogenesis of Nontoxic Goiter. J. Clin. Invest. 1989, 83, 764–770. [Google Scholar] [CrossRef]
- Wikipathways MAPK Pathway in Congenital Thyroid Cancer (Homo Sapiens).
- Protein Atlas.
- Knauf, J.A.; Fagin, J.A. Role of MAPK Pathway Oncoproteins in Thyroid Cancer Pathogenesis and as Drug Targets. Curr. Opin. Cell Biol. 2009, 21, 296–303. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Davis, F.B.; Gordinier, J.K.; Martino, L.J.; Davis, P.J. Thyroid Hormone Induces Activation of Mitogen-Activated Protein Kinase in Cultured Cells. Am. J. Physiol.-Cell Physiol. 1999, 276, C1014–C1024. [Google Scholar] [CrossRef]
- Cho, S.Y.; Kim, S.; Kim, G.; Singh, P.; Kim, D.W. Integrative Analysis of KIF4A, 9, 18A, and 23 and Their Clinical Significance in Low-Grade Glioma and Glioblastoma. Sci. Rep. 2019, 9, 4599. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, P.; Chen, J.; He, X. Systemic Characterization of the SLC Family Genes Reveals SLC26A6 as a Novel Oncogene in Hepatocellular Carcinoma. Transl. Cancer Res. 2021, 10, 2882–2894. [Google Scholar] [CrossRef] [PubMed]
- Alper, S.L.; Sharma, A.K. The SLC26 Gene Family of Anion Transporters and Channels. Mol. Asp. Med. 2013, 34, 494–515. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Huang, Y.; Chen, L.; Guo, L.; Wang, L.; Li, M.; Liang, Y. Up-Regulation of SLC26A6 in Hepatocellular Carcinoma and Its Diagnostic and Prognostic Significance. Crit. Rev. Eukaryot.Gene Expr. 2021, 31, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, J.-F.; Mirebeau-Prunier, D.; Franc, B.; Triau, S.; Rodien, P.; Houlgatte, R.; Malthièry, Y.; Savagner, F. Microarray Analysis Refines Classification of Non-Medullary Thyroid Tumours of Uncertain Malignancy. Oncogene 2008, 27, 2228–2236. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Li, Y.; Qin, S.; Jiao, Y.; Hua, F. Ubiquitin-like Modifier-activating Enzyme 7 as a Marker for the Diagnosis and Prognosis of Breast Cancer. Oncol. Lett. 2020, 19, 2773–2784. [Google Scholar] [CrossRef]
- Fan, J.-B.; Miyauchi, S.; Xu, H.-Z.; Liu, D.; Kim, L.J.Y.; Burkart, C.; Cheng, H.; Arimoto, K.; Yan, M.; Zhou, Y.; et al. Type I Interferon Regulates a Coordinated Gene Network to Enhance Cytotoxic T Cell–Mediated Tumor Killing. Cancer Discov. 2020, 10, 382–393. [Google Scholar] [CrossRef]
- Warnier, M.; Roudbaraki, M.; Derouiche, S.; Delcourt, P.; Bokhobza, A.; Prevarskaya, N.; Mariot, P. CACNA2D2 Promotes Tumorigenesis by Stimulating Cell Proliferation and Angiogenesis. Oncogene 2015, 34, 5383–5394. [Google Scholar] [CrossRef]
- Carboni, G.L.; Gao, B.; Nishizaki, M.; Xu, K.; Minna, J.D.; Roth, J.A.; Ji, L. CACNA2D2-Mediated Apoptosis in NSCLC Cells Is Associated with Alterations of the Intracellular Calcium Signaling and Disruption of Mitochondria Membrane Integrity. Oncogene 2003, 22, 615–626. [Google Scholar] [CrossRef]
- Peng, S.; Li, C.; Wang, X.; Liu, X.; Han, C.; Jin, T.; Liu, S.; Zhang, X.; Zhang, H.; He, X.; et al. Increased Toll-Like Receptors Activity and TLR Ligands in Patients with Autoimmune Thyroid Diseases. Front. Immunol. 2016, 7, 578. [Google Scholar] [CrossRef]
- Nihon-Yanagi, Y.; Wakayama, M.; Tochigi, N.; Saito, F.; Ogata, H.; Shibuya, K. Immunohistochemical Analysis of Toll-Like Receptors, MyD88, and TRIF in Human Papillary Thyroid Carcinoma and Anaplastic Thyroid Carcinoma. J. Thyroid. Res. 2021, 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Inoue, N.; Katsumata, Y.; Watanabe, M.; Ishido, N.; Manabe, Y.; Watanabe, A.; Masutani, R.; Hidaka, Y.; Iwatani, Y. Polymorphisms and Expression of Toll-like Receptors in Autoimmune Thyroid Diseases. Autoimmunity 2017, 50, 182–191. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, K.J.; Gallanis, G.T.; Heller, K.A.; Melas, M.; Idos, G.E.; Culver, J.O.; Martin, S.-E.; Peng, D.H.; Gruber, S.B. A Novel BAP1 Mutation Is Associated with Melanocytic Neoplasms and Thyroid Cancer. Cancer Genet. 2016, 209, 75–81. [Google Scholar] [CrossRef]
- Farid, R.M.; Abd El Atti, R.M.; Abd Raboh, N.M. Immunohistochemical Expression of the Cancer Predisposition Gene BRCA1-Associated Protein 1 in Thyroid and Lung Carcinoma. Egypt J. Pathol. 2019, 39, 98. [Google Scholar]
- Haugh, A.M.; Njauw, C.-N.; Bubley, J.A.; Verzì, A.E.; Zhang, B.; Kudalkar, E.; VandenBoom, T.; Walton, K.; Swick, B.L.; Kumar, R.; et al. Genotypic and Phenotypic Features of BAP1 Cancer Syndrome: A Report of 8 New Families and Review of Cases in the Literature. JAMA Dermatol. 2017, 153, 999. [Google Scholar] [CrossRef] [PubMed]
- Gallanis, G.T.; Heller, K.A.; Melas, E.-M.; Gruber, S.B. Abstract 3522: A Novel BAP1 Mutation Is Associated with Melanocytic Neoplasms and Thyroid and Pancreatic Cancers. Cancer Res. 2014, 74, 3522. [Google Scholar] [CrossRef]
- Avilla, E.; Guarino, V.; Visciano, C.; Liotti, F.; Svelto, M.; Krishnamoorthy, G.; Franco, R.; Melillo, R.M. Activation of TYRO3/AXL Tyrosine Kinase Receptors in Thyroid Cancer. Cancer Res. 2011, 71, 1792–1804. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.-L.; Jou, J.; Tsai, S.-J. TYRO3: A Potential Therapeutic Target in Cancer. Exp. Biol. Med. 2019, 244, 83–99. [Google Scholar] [CrossRef]
- Ao, Z.; Chen, Y.; Lu, J.; Shen, J.; Peng, L.; Lin, X.; Peng, C.; Zeng, C.; Wang, X.; Zhou, R.; et al. Identification of Potential Functional Genes in Papillary Thyroid Cancer by Co-expression Network Analysis. Oncol. Lett. 2018, 16, 4871–4878. [Google Scholar] [CrossRef]
- Sato, A.; Matsuda, K.; Motoyama, T.; Mussazhanova, Z.; Otsubo, R.; Kondo, H.; Akazawa, Y.; Higuchi, M.; Suzuki, A.; Hirokawa, M.; et al. 53BP1 Expression as a Biomarker to Differentiate Thyroid Follicular Tumors. Endocr. Connect. 2021, 10, 309–315. [Google Scholar] [CrossRef]
- Xia, Z.; Morales, J.C.; Dunphy, W.G.; Carpenter, P.B. Negative Cell Cycle Regulation and DNA Damage-Inducible Phosphorylation of the BRCT Protein 53BP1. J. Biol. Chem. 2001, 276, 2708–2718. [Google Scholar] [CrossRef] [PubMed]
- Mussazhanova, Z.; Matsuda, K.; Naruke, Y.; Mitsutake, N.; Stanojevic, B.; Rougounovitch, T.; Saenko, V.; Suzuki, K.; Nishihara, E.; Hirokawa, M.; et al. Significance of P53-Binding Protein 1 (53BP1) Expression in Thyroid Papillary Microcarcinoma: Association with BRAF V 600E Mutation Status. Histopathology 2013, 63, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, R.; Matsuda, K.; Mussazhanova, Z.; Sato, A.; Matsumoto, M.; Yano, H.; Oikawa, M.; Kondo, H.; Ito, M.; Miyauchi, A.; et al. A Novel Diagnostic Method for Thyroid Follicular Tumors Based on Immunofluorescence Analysis of P53-Binding Protein 1 Expression: Detection of Genomic Instability. Thyroid 2019, 29, 657–665. [Google Scholar] [CrossRef]
- Zambrano, A.; García-Carpizo, V.; Gallardo, M.E.; Villamuera, R.; Gómez-Ferrería, M.A.; Pascual, A.; Buisine, N.; Sachs, L.M.; Garesse, R.; Aranda, A. The Thyroid Hormone Receptor β Induces DNA Damage and Premature Senescence. J. Cell Biol. 2014, 204, 129–146. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.M.H.; Matsuda, K.; Niino, D.; Kurohama, H.; Ito, M.; Nakashima, M. Significance of Abnormal 53BP1 Expression as a Novel Molecular Pathologic Parameter of Follicular-Shaped B-Cell Lymphoid Lesions in Human Digestive Tract. Sci. Rep. 2021, 11, 3074. [Google Scholar] [CrossRef]
- Lee, A.S.Y.; Kranzusch, P.J.; Cate, J.H.D. EIF3 Targets Cell-Proliferation Messenger RNAs for Translational Activation or Repression. Nature 2015, 522, 111–114. [Google Scholar] [CrossRef]
- Lee, A.S.Y.; Kranzusch, P.J.; Doudna, J.A.; Cate, J.H.D. EIF3d Is an MRNA Cap-Binding Protein That Is Required for Specialized Translation Initiation. Nature 2016, 536, 96–99. [Google Scholar] [CrossRef]
- Chi, N.C.; Shaw, R.M.; De Val, S.; Kang, G.; Jan, L.Y.; Black, B.L.; Stainier, D.Y.R. Foxn4 Directly Regulates Tbx2b Expression and Atrioventricular Canal Formation. Genes Dev. 2008, 22, 734–739. [Google Scholar] [CrossRef]
- Luo, H.; Jin, K.; Xie, Z.; Qiu, F.; Li, S.; Zou, M.; Cai, L.; Hozumi, K.; Shima, D.T.; Xiang, M. Forkhead Box N4 (Foxn4) Activates Dll4-Notch Signaling to Suppress Photoreceptor Cell Fates of Early Retinal Progenitors. Proc. Natl. Acad. Sci. USA 2012, 109. [Google Scholar] [CrossRef]
- MalaCards.
- Weterman, M.A.J.; Barth, P.G.; van Spaendonck-Zwarts, K.Y.; Aronica, E.; Poll-The, B.-T.; Brouwer, O.F.; van Tintelen, J.P.; Qahar, Z.; Bradley, E.J.; de Wissel, M.; et al. Recessive MYL2 Mutations Cause Infantile Type I Muscle Fibre Disease and Cardiomyopathy. Brain 2013, 136, 282–293. [Google Scholar] [CrossRef]
- Manivannan, S.N.; Darouich, S.; Masmoudi, A.; Gordon, D.; Zender, G.; Han, Z.; Fitzgerald-Butt, S.; White, P.; McBride, K.L.; Kharrat, M.; et al. Novel Frameshift Variant in MYL2 Reveals Molecular Differences between Dominant and Recessive Forms of Hypertrophic Cardiomyopathy. PLoS Genet. 2020, 16, e1008639. [Google Scholar] [CrossRef]
- Claes, G.R.F.; van Tienen, F.H.J.; Lindsey, P.; Krapels, I.P.C.; Helderman-van den Enden, A.T.J.M.; Hoos, M.B.; Barrois, Y.E.G.; Janssen, J.W.H.; Paulussen, A.D.C.; Sels, J.-W.E.M.; et al. Hypertrophic Remodelling in Cardiac Regulatory Myosin Light Chain (MYL2) Founder Mutation Carriers. Eur. Heart J. 2016, 37, 1815–1822. [Google Scholar] [CrossRef]
- Lee, E.J.; Shaikh, S.; Choi, D.; Ahmad, K.; Baig, M.H.; Lim, J.H.; Lee, Y.-H.; Park, S.J.; Kim, Y.-W.; Park, S.-Y.; et al. Transthyretin Maintains Muscle Homeostasis through the Novel Shuttle Pathway of Thyroid Hormones during Myoblast Differentiation. Cells 2019, 8, 1565. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Burnett, E.; Kinch, M.; Simon, E.; Wang, B. Activation of EphA2 Kinase Suppresses Integrin Function and Causes Focal-Adhesion-Kinase Dephosphorylation. Nat. Cell Biol. 2000, 2, 62–69. [Google Scholar] [CrossRef]
- Lee, H.-H.; Chang, Z.-F. Regulation of RhoA-Dependent ROCKII Activation by Shp2. J. Cell Biol. 2008, 181, 999–1012. [Google Scholar] [CrossRef]
- Pannone, L.; Bocchinfuso, G.; Flex, E.; Rossi, C.; Baldassarre, G.; Lissewski, C.; Pantaleoni, F.; Consoli, F.; Lepri, F.; Magliozzi, M.; et al. Structural, Functional, and Clinical Characterization of a Novel PTPN11 Mutation Cluster Underlying Noonan Syndrome: HUMAN MUTATION. Hum. Mutat. 2017, 38, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-Q.; Ma, R.; Zhang, C.-M.; Li, J.; Li, L.; Hu, Z.-T.; Gao, Q.; Li, W.-M. Expression and Clinical Significance of Tyrosine Phosphatase SHP2 in Thyroid Carcinoma. Oncol. Lett. 2015, 10, 1507–1512. [Google Scholar] [CrossRef] [PubMed]
- Basel-Vanagaite, L.; Dallapiccola, B.; Ramirez-Solis, R.; Segref, A.; Thiele, H.; Edwards, A.; Arends, M.J.; Miró, X.; White, J.K.; Désir, J.; et al. Deficiency for the Ubiquitin Ligase UBE3B in a Blepharophimosis-Ptosis-Intellectual-Disability Syndrome. Am. J. Hum. Genet. 2012, 91, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Basel-Vanagaite, L.; Yilmaz, R.; Tang, S.; Reuter, M.S.; Rahner, N.; Grange, D.K.; Mortenson, M.; Koty, P.; Feenstra, H.; Farwell Gonzalez, K.D.; et al. Expanding the Clinical and Mutational Spectrum of Kaufman Oculocerebrofacial Syndrome with Biallelic UBE3B Mutations. Hum. Genet. 2014, 133, 939–949. [Google Scholar] [CrossRef]
- Wickenhagen, A.; Sugrue, E.; Lytras, S.; Kuchi, S.; Noerenberg, M.; Turnbull, M.L.; Loney, C.; Herder, V.; Allan, J.; Jarmson, I.; et al. A Prenylated DsRNA Sensor Protects against Severe COVID-19. Science 2021, 374, eabj3624. [Google Scholar] [CrossRef]
- Yamazaki, K.; Suzuki, K.; Yamada, E.; Yamada, T.; Takeshita, F.; Matsumoto, M.; Mitsuhashi, T.; Obara, T.; Takano, K.; Sato, K. Suppression of Iodide Uptake and Thyroid Hormone Synthesis with Stimulation of the Type I Interferon System by Double-Stranded Ribonucleic Acid in Cultured Human Thyroid Follicles. Endocrinology 2007, 148, 3226–3235. [Google Scholar] [CrossRef]
- Stefan, M.; Wei, C.; Lombardi, A.; Li, C.W.; Concepcion, E.S.; Inabnet, W.B.; Owen, R.; Zhang, W.; Tomer, Y. Genetic–Epigenetic Dysregulation of Thymic TSH Receptor Gene Expression Triggers Thyroid Autoimmunity. Proc. Natl. Acad. Sci. USA 2014, 111, 12562–12567. [Google Scholar] [CrossRef]
- Poma, A.M.; Basolo, A.; Bonuccelli, D.; Proietti, A.; Macerola, E.; Ugolini, C.; Torregrossa, L.; Alì, G.; Giannini, R.; Vignali, P.; et al. Activation of Type I and Type II Interferon Signaling in SARS-CoV-2-Positive Thyroid Tissue of Patients Dying from COVID-19. Thyroid 2021, 31, 1766–1775. [Google Scholar] [CrossRef] [PubMed]
- Hébrant, A.; Dom, G.; Dewaele, M.; Andry, G.; Trésallet, C.; Leteurtre, E.; Dumont, J.E.; Maenhaut, C. MRNA Expression in Papillary and Anaplastic Thyroid Carcinoma: Molecular Anatomy of a Killing Switch. PLoS ONE 2012, 7, e37807. [Google Scholar] [CrossRef] [PubMed]
- Zhen, J.; Song, Z.; Su, W.; Zeng, Q.-C.; Li, J.; Sun, Q. Integrated Analysis of RNA-Binding Proteins in Thyroid Cancer. PLoS ONE 2021, 16, e0247836. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, P.; Li, L.-P.; He, Y.-C.; Gao, R.; Gao, Y.-F. Identification of Novel Thyroid Cancer-Related Genes and Chemicals Using Shortest Path Algorithm. BioMed Res. Int. 2015, 2015, 1–8. [Google Scholar] [CrossRef]
- Auslander, N.; Wolf, Y.I.; Koonin, E.V. Interplay between DNA Damage Repair and Apoptosis Shapes Cancer Evolution through Aneuploidy and Microsatellite Instability. Nat. Commun. 2020, 11, 1234. [Google Scholar] [CrossRef]
- Xing, M. RASAL1 in Thyroid Cancer: Promise From a New Friend. J. Clin. Endocrinol. Metab. 2014, 99, 3619–3621. [Google Scholar] [CrossRef]
- Chang, R.-X.; Cui, A.-L.; Dong, L.; Guan, S.-P.; Jiang, L.-Y.; Miao, C.-X. Overexpression of RASAL1 Indicates Poor Prognosis and Promotes Invasion of Ovarian Cancer. Open Life Sci. 2019, 14, 133–140. [Google Scholar] [CrossRef]
- Liu, D.; Yang, C.; Bojdani, E.; Murugan, A.K.; Xing, M. Identification of RASAL1 as a Major Tumor Suppressor Gene in Thyroid Cancer. JNCI J. Natl. Cancer Inst. 2013, 105, 1617–1627. [Google Scholar] [CrossRef]
- Wang, G.; Li, Z.; Li, X.; Zhang, C.; Peng, L. RASAL1 Induces to Downregulate the SCD1, Leading to Suppression of Cell Proliferation in Colon Cancer via LXRα/SREBP1c Pathway. Biol. Res. 2019, 52, 60. [Google Scholar] [CrossRef]
- Hińcza, K.; Kowalik, A.; Kowalska, A. Current Knowledge of Germline Genetic Risk Factors for the Development of Non-Medullary Thyroid Cancer. Genes 2019, 10, 482. [Google Scholar] [CrossRef] [PubMed]
- Lyssikatos, C.; Quezado, M.M.; Faucz, F.R.; Angelousi, A.; Nasiri-Ansari, N.; Stratakis, C.A.; Kassi, E. A Rare Case of Medullary Thyroid Cancer, Mesothelioma and Meningioma, Due to APC and RASAL1 Mutations. In Proceedings of the 19th European Congress of Endocrinology, Lisbon, Portugal, 20–23 May 2017. [Google Scholar] [CrossRef]
- Henderson, Y.C.; Toro-Serra, R.; Chen, Y.; Ryu, J.; Frederick, M.J.; Zhou, G.; Gallick, G.E.; Lai, S.Y.; Clayman, G.L. Src Inhibitors in Suppression of Papillary Thyroid Carcinoma Growth: Effects of SRC Inhibitors in PTC. Head Neck 2014, 36, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Beadnell, T.C.; Nassar, K.W.; Rose, M.M.; Clark, E.G.; Danysh, B.P.; Hofmann, M.-C.; Pozdeyev, N.; Schweppe, R.E. Src-Mediated Regulation of the PI3K Pathway in Advanced Papillary and Anaplastic Thyroid Cancer. Oncogenesis 2018, 7, 23. [Google Scholar] [CrossRef]
- Chan, C.M.; Jing, X.; Pike, L.A.; Zhou, Q.; Lim, D.-J.; Sams, S.B.; Lund, G.S.; Sharma, V.; Haugen, B.R.; Schweppe, R.E. Targeted Inhibition of Src Kinase with Dasatinib Blocks Thyroid Cancer Growth and Metastasis. Clin. Cancer Res. 2012, 18, 3580–3591. [Google Scholar] [CrossRef]
- Lee, W.K.; Kim, W.G.; Fozzatti, L.; Park, S.; Zhao, L.; Willingham, M.C.; Lonard, D.; O’Malley, B.W.; Cheng, S. Steroid Receptor Coactivator-3 as a Target for Anaplastic Thyroid Cancer. Endocr.-Relat. Cancer 2020, 27, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Falola, J.; Zhu, X.; Gu, Y.; Kim, L.T.; Sarosi, G.A.; Anthony, T.; Nwariaku, F.E. Antiproliferative Effects of Src Inhibition on Medullary Thyroid Cancer. J. Clin. Endocrinol. Metab. 2004, 89, 3503–3509. [Google Scholar] [CrossRef]
- Chen, Z. CD82, but Not CD63, Is Linked to Cellular Invasiveness in Human Thyroid Carcinoma. Ph.D. Thesis, Martin-Luther-Universität Halle-Wittenberg, Halle (Saale), Germany, 2007. [Google Scholar] [CrossRef]
- Kim, T.; Kim, Y.; Kwon, H.J. Expression of CD9 and CD82 in Papillary Thyroid Microcarcinoma and Its Prognostic Significance. Endokrynologia Polska 2019, 70, 224–231. [Google Scholar] [CrossRef]
- Qiu, K.; Li, K.; Zeng, T.; Liao, Y.; Min, J.; Zhang, N.; Peng, M.; Kong, W.; Chen, L. Integrative Analyses of Genes Associated with Hashimoto’s Thyroiditis. J. Immunol. Res. 2021, 2021, 1–9. [Google Scholar] [CrossRef]
- Bang, H.S.; Choi, M.H.; Kim, C.S.; Choi, S.J. Gene Expression Profiling in Undifferentiated Thyroid Carcinoma Induced by High-Dose Radiation. J. Radiat. Res. 2016, 57, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, D.; Jovancevic, N.; Zwanziger, D.; Theurer, S.; Hönes, J.; Führer, D.; Hatt, H. Functional Characterization of Olfactory Receptors in the Thyroid Gland. Front. Physiol. 2021, 12, 676907. [Google Scholar] [CrossRef] [PubMed]
- Abaffy, T. Human olfactory receptors expression and their role in non-olfactory tissues-a mini-review. J. Pharm. Pharm. 2015, 6, 1. [Google Scholar] [CrossRef]
- Mitsiades, C.S.; Hayden, P.; Kotoula, V.; McMillin, D.W.; McMullan, C.; Negri, J.; Delmore, J.E.; Poulaki, V.; Mitsiades, N. Bcl-2 Overexpression in Thyroid Carcinoma Cells Increases Sensitivity to Bcl-2 Homology 3 Domain Inhibition. J. Clin. Endocrinol. Metab. 2007, 92, 4845–4852. [Google Scholar] [CrossRef]
- Wang, Q.; Shen, Y.; Ye, B.; Hu, H.; Fan, C.; Wang, T.; Zheng, Y.; Lv, J.; Ma, Y.; Xiang, M. Gene Expression Differences between Thyroid Carcinoma, Thyroid Adenoma and Normal Thyroid Tissue. Oncol. Rep. 2018, 40, 3359–3369. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Qi, B.; Zhou, Q.; Lu, C.; Huang, Q.; Xian, L.; Chen, M. Key Genes and Pathways in Thyroid Cancer Based on Gene Set Enrichment Analysis. Oncol. Rep. 2013, 30, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Rakhsh-Khorshid, H.; Samimi, H.; Torabi, S.; Sajjadi-Jazi, S.M.; Samadi, H.; Ghafouri, F.; Asgari, Y.; Haghpanah, V. Network Analysis Reveals Essential Proteins That Regulate Sodium-Iodide Symporter Expression in Anaplastic Thyroid Carcinoma. Sci. Rep. 2020, 10, 21440. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Zhang, J.; Wang, X.; Song, R.; Qin, Q.; Muhali, F.; Zhou, J.; Xu, J.; Zhang, J. Gene-Gene and Gene-Sex Epistatic Interactions of DNMT1, DNMT3A and DNMT3B in Autoimmune Thyroid Disease. Endocr. J. 2016, 63, 643–653. [Google Scholar] [CrossRef]
- Kyono, Y.; Sachs, L.M.; Bilesimo, P.; Wen, L.; Denver, R.J. Developmental and Thyroid Hormone Regulation of the DNA Methyltransferase 3a Gene in Xenopus Tadpoles. Endocrinology 2016, 157, 4961–4972. [Google Scholar] [CrossRef]
- Coppedè, F. Epigenetics and Autoimmune Thyroid Diseases. Front. Endocrinol. 2017, 8, 149. [Google Scholar] [CrossRef]
- Zafon, C.; Gil, J.; Pérez-González, B.; Jordà, M. DNA Methylation in Thyroid Cancer. Endocr.-Relat. Cancer 2019, 26, R415–R439. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, Y.; Watanabe, M.; Inoue, N.; Sarumaru, M.; Hidaka, Y.; Iwatani, Y. Association of Polymorphisms in DNMT1, DNMT3A, DNMT3B, MTHFR and MTRR Genes with Global DNA Methylation Levels and Prognosis of Autoimmune Thyroid Disease. Clin. Exp. Immunol. 2012, 170, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Muhali, F.; Song, R.; Qin, Q.; Wang, X.; Shi, L.; Jiang, W.; Xiao, L.; Li, D.; Zhang, J. Genome-Wide DNA Methylation Analysis in Graves’ Disease. Genomics 2015, 105, 204–210. [Google Scholar] [CrossRef]
- Wojcicka, A.; Piekielko–Witkowska, A.; Kedzierska, H.; Rybicka, B.; Poplawski, P.; Boguslawska, J.; Master, A.; Nauman, A. Epigenetic Regulation of Thyroid Hormone Receptor Beta in Renal Cancer. PLoS ONE 2014, 9, e97624. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hwang, J.-A.; Lee, E.K. Recent Progress of Genome Study for Anaplastic Thyroid Cancer. Genomics Inform. 2013, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Gobin, E.; Bagwell, K.; Wagner, J.; Mysona, D.; Sandirasegarane, S.; Smith, N.; Bai, S.; Sharma, A.; Schleifer, R.; She, J.-X. A Pan-Cancer Perspective of Matrix Metalloproteases (MMP) Gene Expression Profile and Their Diagnostic/Prognostic Potential. BMC Cancer 2019, 19, 581. [Google Scholar] [CrossRef]
- Rodrigues, R.F.; Roque, L.; Rosa-Santos, J.; Cid, O.; Soares, J. Chromosomal Imbalances Associated with Anaplastic Transformation of Follicular Thyroid Carcinomas. Br. J. Cancer 2004, 90, 492–496. [Google Scholar] [CrossRef]
- Bialek, J.; Kunanuvat, U.; Hombach-Klonisch, S.; Spens, A.; Stetefeld, J.; Sunley, K.; Lippert, D.; Wilkins, J.A.; Hoang-Vu, C.; Klonisch, T. Relaxin Enhances the Collagenolytic Activity and In Vitro Invasiveness by Upregulating Matrix Metalloproteinases in Human Thyroid Carcinoma Cells. Mol. Cancer Res. 2011, 9, 673–687. [Google Scholar] [CrossRef]
| Characteristics | Follicular Thyroid Adenomas n = 16 | Follicular Thyroid Carcinomas n = 16 | p-Value 1 |
|---|---|---|---|
| Male/female, n (%) | 2/14 (12.5%/87.5%) | 2/14 (12.5%/87.5%) | 1.000 |
| Median age at diagnosis, years (range) | 53 (29–81) | 56 (31-82) | 0.649 |
| Age group ≤60 years/>60 years), n (%) | 11/5 (68.8%/31.2%) | 10/6 (62.5%/37.5%) | 1.000 |
| Median length of follow-up, months (range) | 119 (58–162) | 152 (47–174) | 0.587 |
| Multifocality, n (%) | 0 | 2 (12.5%) | 0.4839 |
| Capsule invasion, n (%) | NA | 7 (43.8%) | NA |
| Extracapsular extension, n (%) | NA | 10 (62.5%) | NA |
| Nodal (N) involvement, n (%) | NA | 1 (6.3%) | NA |
| Mean tumor size, mm (range) | 23 (6–50) | 26 (8-50) | 0.112 |
| Tumor diameter ≤10 mm, n (%) | 3 (18.8%) | 1 (6.3%) | 0.5996 |
| Localization in the right/left/both lobes, n (%) | 9/7/0 (56.3%/43.7%) | 8/7/1 (50%/43.8%/6.2%) | 0.7222 |
| Chronic lymphocytic thyroiditis, n (%) | 2 (12.5%) | 3 (18.8%) | 1.000 |
| Radioactive iodine refractoriness n (%) | NA | 1 (6.3%) | NA |
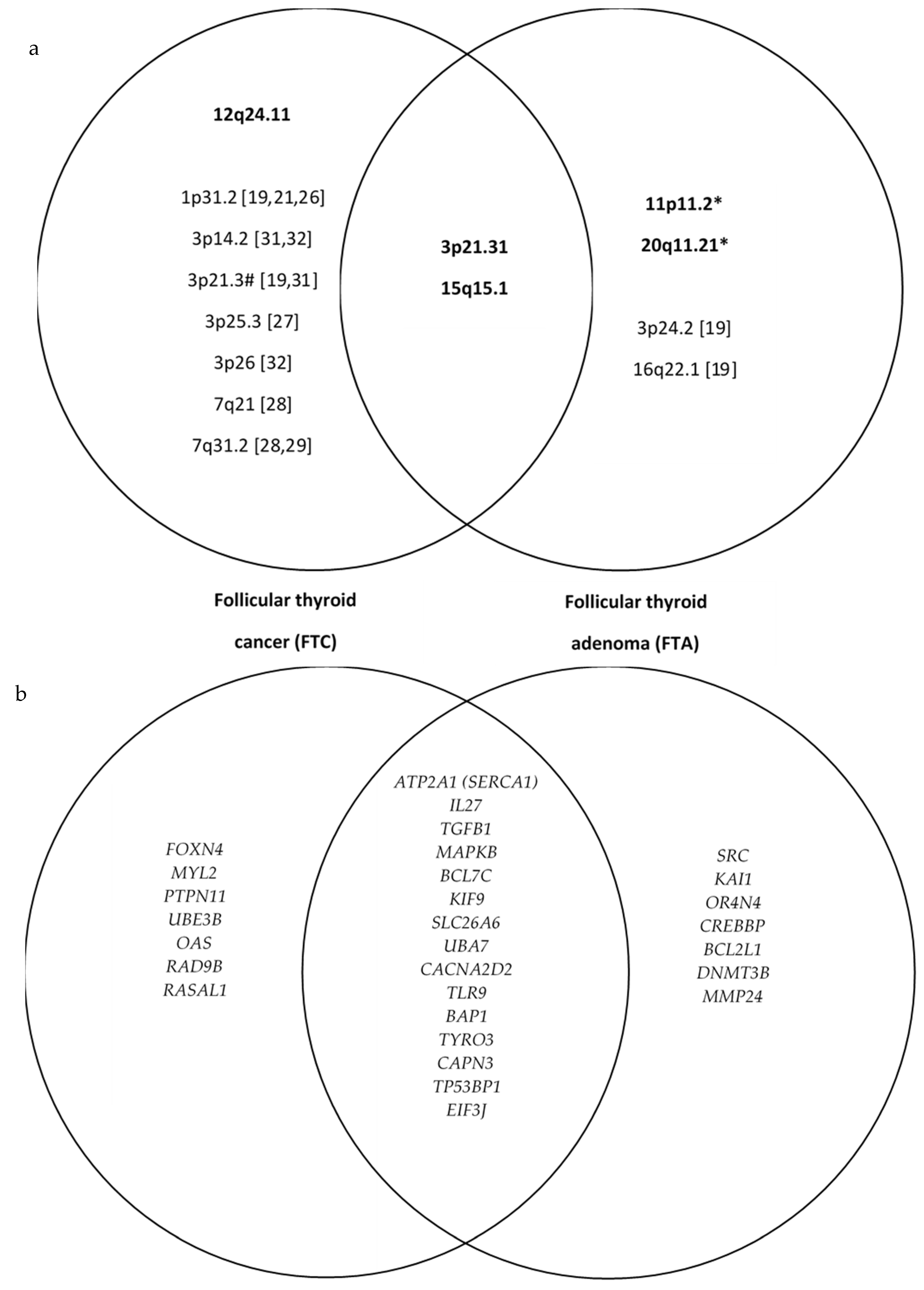
| Chrom. | Cytoband Start | Size (kbp) | Gene Count | Census Genes | Microarray Nomenclature | FTC | FTA | Sum | p-Value | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| LOHs present in both follicular thyroid carcinoma and follicular thyroid adenoma | ||||||||||
| 16 | p12.1 | 7500.913 | 149 | FUS | arr[hg19] 16p12.1-p11.1(27,770,812–35,271,725) hmz | 11 | 10 | 21 | 0.710 | 1.32 (0.31–5.70) |
| 3 | p21.31 | 6391.659 | 172 | SETD2, NCKIPSD, RHOA, BAP1, PBRM1 | arr[hg19] 3p21.31-p21.1(46,778,841–53,170,500) hmz | 9 | 7 | 16 | 0.481 | 1.65 (0.41–6.68) |
| 15 | q15.1 | 3616.641 | 70 | B2M | arr[hg19] 15q15.1-q21.1(41,796,900–45,413,541) hmz | 9 | 5 | 14 | 0.159 | 2.82 (0.67–12.02) |
| LOHs present predominantly in follicular thyroid carcinoma: | ||||||||||
| 12 | q24.11 | 3990.65 | 59 | SH2B3, ALDH2, PTPN11 | arr[hg19] 12q24.11-q24.13(109,669,669–113,660,319) hmz | 6 | 1 | 7 | 0.057 | 9.00 (0.94–86.53) |
| LOHs present predominantly in follicular thyroid adenoma: | ||||||||||
| 11 | p11.2 | 5404.548 | 58 | CREB3L1, DDB2 | arr[hg19] 11p11.2-p11.12(46,171,403–51,575,951) hmz | 0 | 9 | 9 | 0.014 | 41.80 (2.14–816.37) |
| 20 | q11.21 | 6885.552 | 125 | ASXL1, SRC | arr[hg19] 20q11.21-q11.23(29,519,155–36,404,707) hmz | 1 | 5 | 6 | 0.099 | 6.82 (0.69–66.91) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borowczyk, M.; Dobosz, P.; Szczepanek-Parulska, E.; Budny, B.; Dębicki, S.; Filipowicz, D.; Wrotkowska, E.; Oszywa, M.; Verburg, F.A.; Janicka-Jedyńska, M.; et al. Follicular Thyroid Adenoma and Follicular Thyroid Carcinoma—A Common or Distinct Background? Loss of Heterozygosity in Comprehensive Microarray Study. Cancers 2023, 15, 638. https://doi.org/10.3390/cancers15030638
Borowczyk M, Dobosz P, Szczepanek-Parulska E, Budny B, Dębicki S, Filipowicz D, Wrotkowska E, Oszywa M, Verburg FA, Janicka-Jedyńska M, et al. Follicular Thyroid Adenoma and Follicular Thyroid Carcinoma—A Common or Distinct Background? Loss of Heterozygosity in Comprehensive Microarray Study. Cancers. 2023; 15(3):638. https://doi.org/10.3390/cancers15030638
Chicago/Turabian StyleBorowczyk, Martyna, Paula Dobosz, Ewelina Szczepanek-Parulska, Bartłomiej Budny, Szymon Dębicki, Dorota Filipowicz, Elżbieta Wrotkowska, Michalina Oszywa, Frederik A. Verburg, Małgorzata Janicka-Jedyńska, and et al. 2023. "Follicular Thyroid Adenoma and Follicular Thyroid Carcinoma—A Common or Distinct Background? Loss of Heterozygosity in Comprehensive Microarray Study" Cancers 15, no. 3: 638. https://doi.org/10.3390/cancers15030638
APA StyleBorowczyk, M., Dobosz, P., Szczepanek-Parulska, E., Budny, B., Dębicki, S., Filipowicz, D., Wrotkowska, E., Oszywa, M., Verburg, F. A., Janicka-Jedyńska, M., Ziemnicka, K., & Ruchała, M. (2023). Follicular Thyroid Adenoma and Follicular Thyroid Carcinoma—A Common or Distinct Background? Loss of Heterozygosity in Comprehensive Microarray Study. Cancers, 15(3), 638. https://doi.org/10.3390/cancers15030638





