A High-Throughput Sequencing Data-Based Classifier Reveals the Metabolic Heterogeneity of Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Preparation
2.2. Classification of Liver Cancer Metabolic Subtypes
2.3. Prognostic Analysis of Metabolic Subtypes
2.4. Estimation of Immune Cell Infiltration
2.5. Differential Gene Expression Analysis and Pathway Enrichment Analysis
2.6. Comparison of Oncogenic Pathway Activity
2.7. Somatic Mutation Patterns
2.8. Cancer Stemness Index (CSI)
2.9. Human Liver Cancer Cell Lines and Cell Proliferation Assay
2.10. Lactate Production Measurement
2.11. Cell Cycle Analysis
2.12. Statistical Analysis
3. Results
3.1. Metabolic Pattern Classifies HCC into Two Subtypes with Clinical Significance
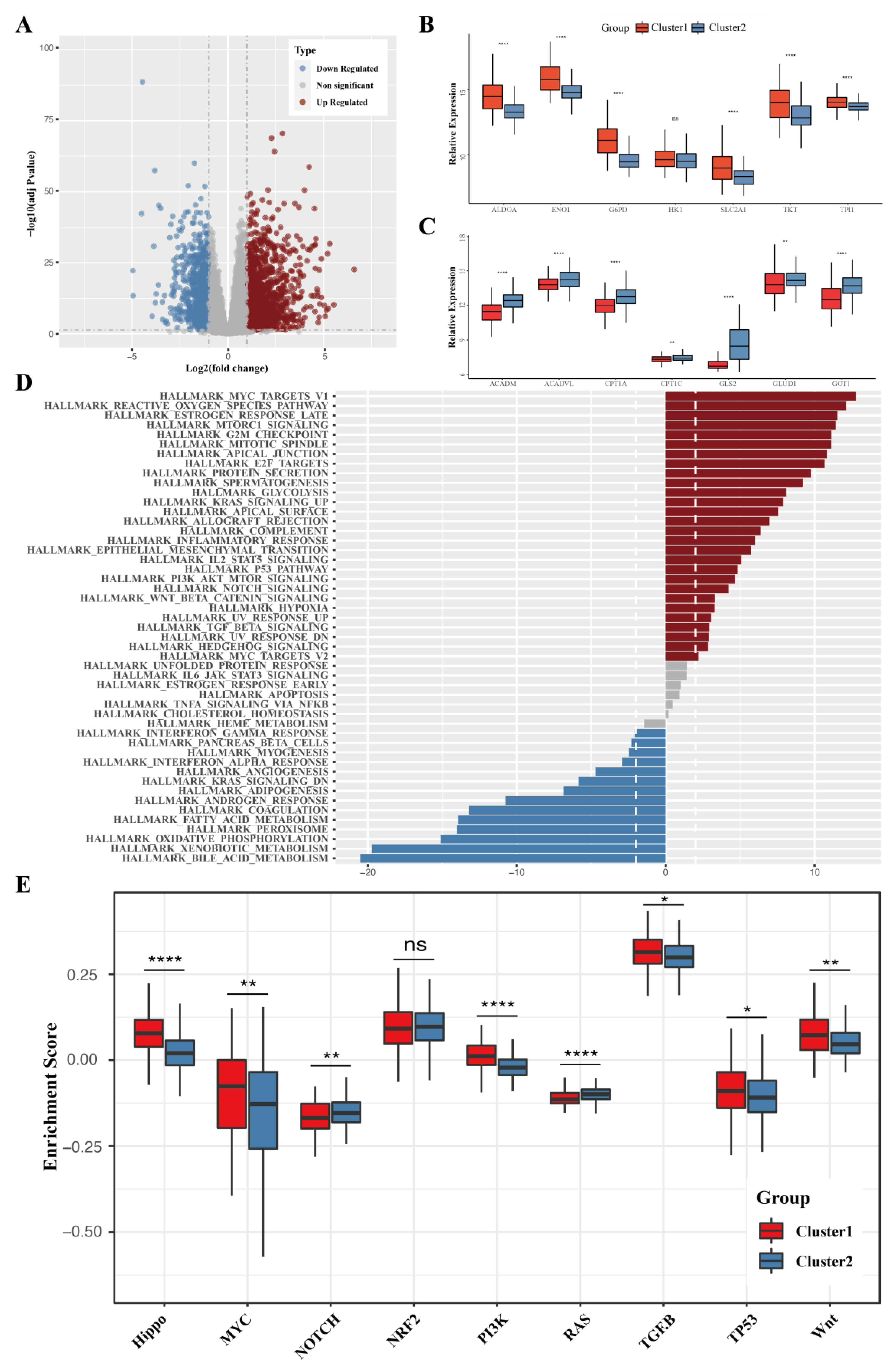
3.2. The Differentially Expressed Genes and the Difference in Pathway Enrichment
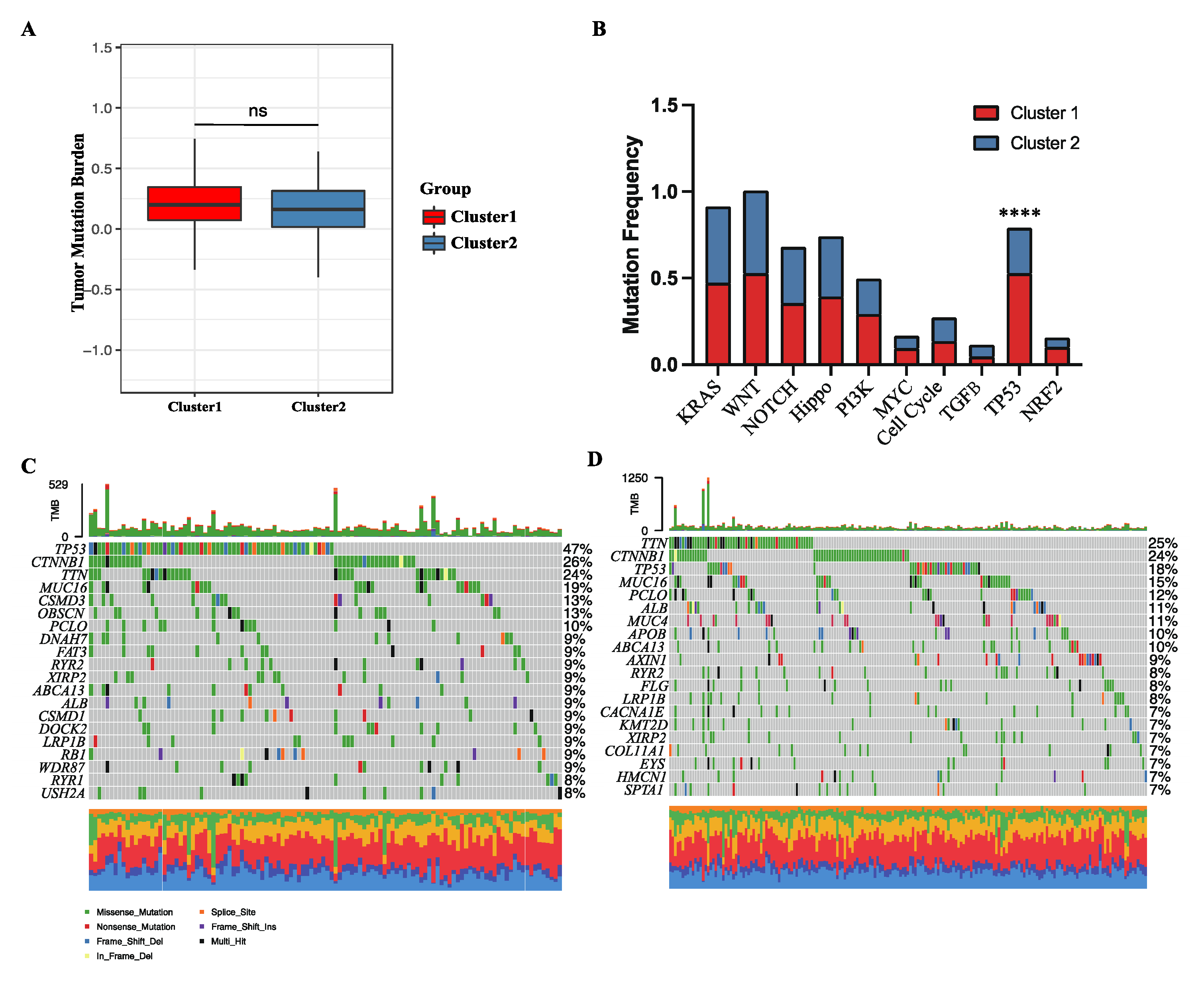
3.3. Metabolic Status-Specific Somatic Mutation Pattern of HCC
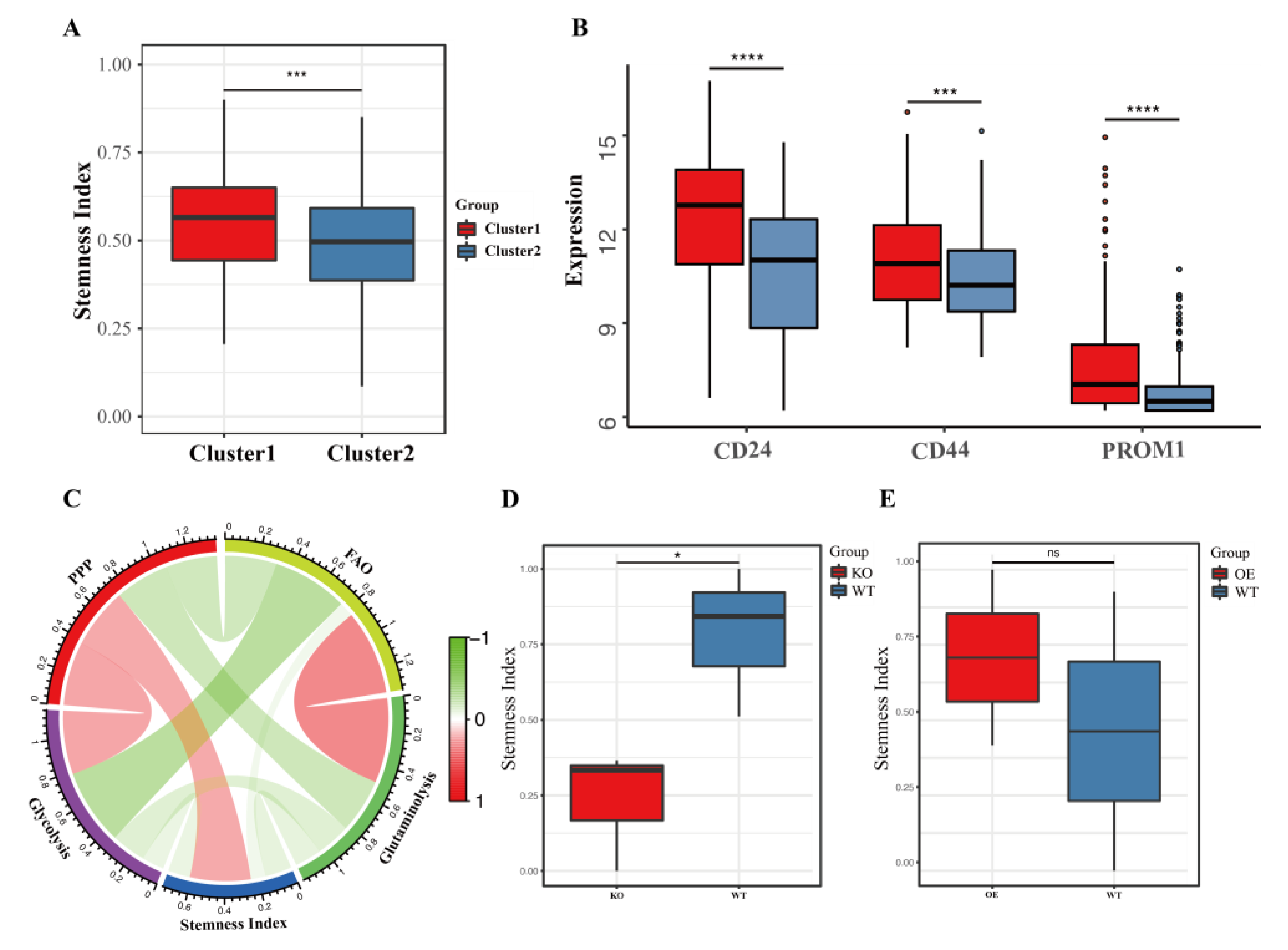
3.4. Cancer Stem Cell Index and Its Correlation with Energy Metabolic Pathways
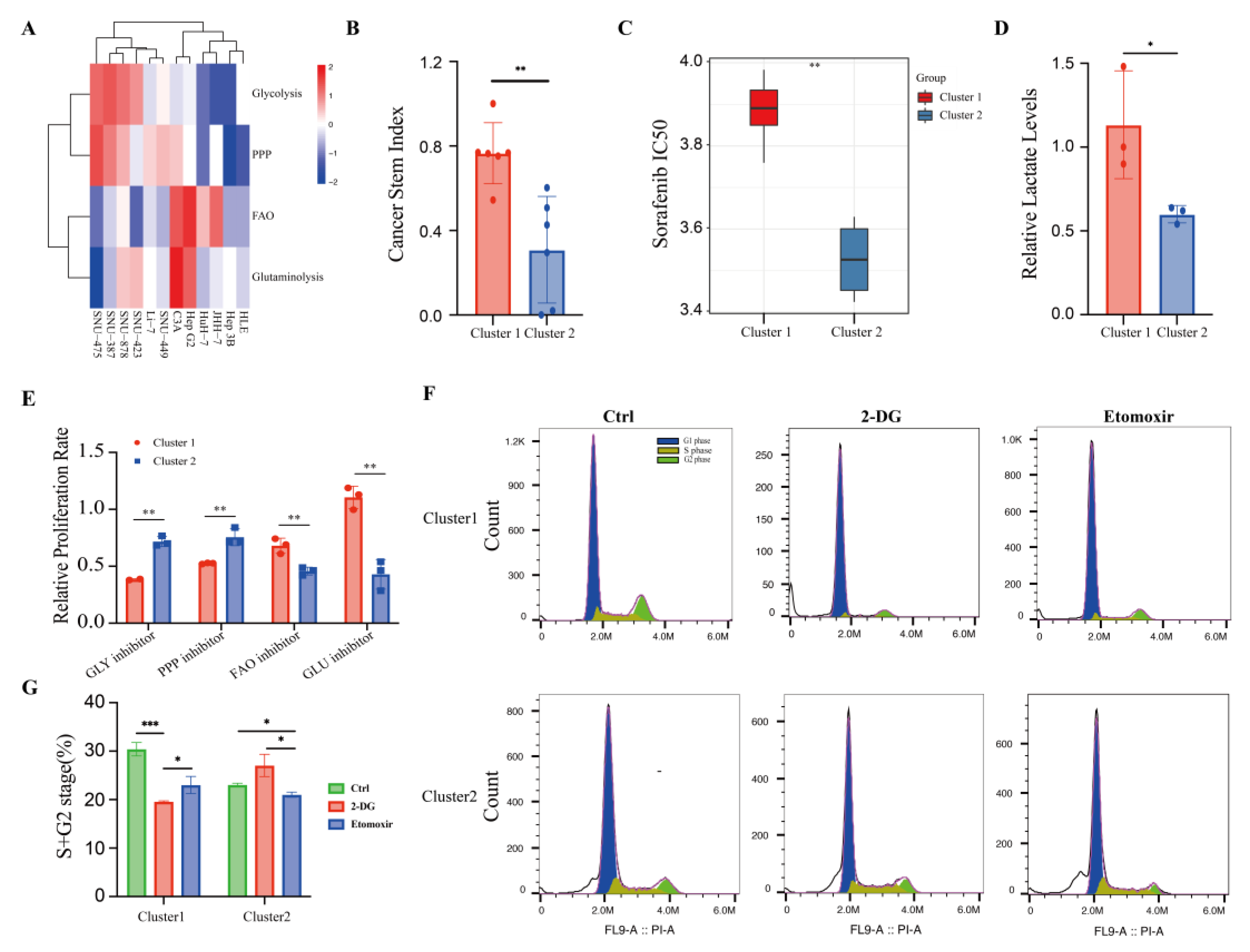
3.5. HCC Cell Lines Mimic the Different Metabolic Patterns of Corresponding Human Tumors
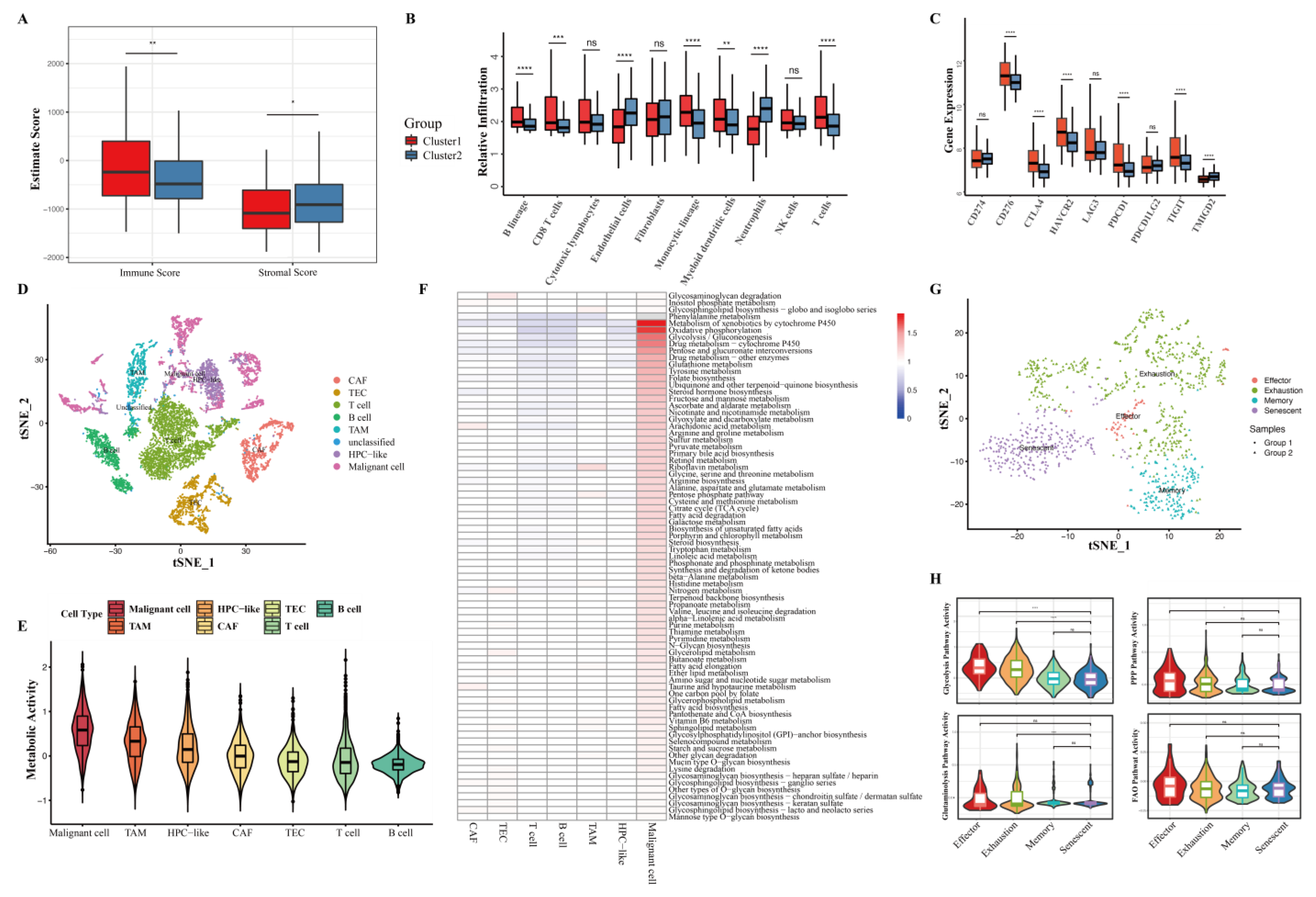
3.6. Distinct Immune Microenvironments between HCC Clusters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Heimbach, J.K. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ 2020, 371, m3544. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef]
- Jin, H.; Wang, S.; Zaal, E.A.; Wang, C.; Wu, H.; Bosma, A.; Jochems, F.; Isima, N.; Jin, G.; Lieftink, C.; et al. A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer. eLife 2020, 9, e56749. [Google Scholar] [CrossRef]
- Gao, Q.; Zhu, H.; Dong, L.; Shi, W.; Chen, R.; Song, Z.; Huang, C.; Li, J.; Dong, X.; Zhou, Y.; et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 2019, 179, 561–577.e522. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Huang, X.; Liu, Z.; Qin, W.; Wang, C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol. Oncol. 2020, 14, 896–913. [Google Scholar] [CrossRef]
- Bidkhori, G.; Benfeitas, R.; Klevstig, M.; Zhang, C.; Nielsen, J.; Uhlen, M.; Boren, J.; Mardinoglu, A. Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc. Natl. Acad. Sci. USA 2018, 115, e11874–e11883. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; Ghoshal, K.; Fernandez, S.; Li, L. Identification of a Subtype of Hepatocellular Carcinoma with Poor Prognosis Based on Expression of Genes within the Glucose Metabolic Pathway. Cancers 2019, 11, 2023. [Google Scholar] [CrossRef]
- Zhu, P.; Zhang, F.; Deng, W.; Chen, W. Integrative analysis of the characteristic of lipid metabolism-related genes for the prognostic prediction of hepatocellular carcinoma. Medicine 2022, 101, e30695. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Tang, R.; Zhou, K.; Chang, J.; Wang, H.; Zhang, Q.; Shi, J.; Sun, C. An asparagine metabolism-based classification reveals the metabolic and immune heterogeneity of hepatocellular carcinoma. BMC Med. Genom. 2022, 15, 222. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ramadori, P.; Pfister, D.; Seehawer, M.; Zender, L.; Heikenwalder, M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer 2021, 21, 541–557. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 2016, 73, 377–392. [Google Scholar] [CrossRef]
- Yu, T.J.; Ma, D.; Liu, Y.Y.; Xiao, Y.; Gong, Y.; Jiang, Y.Z.; Shao, Z.M.; Hu, X.; Di, G.H. Bulk and single-cell transcriptome profiling reveal the metabolic heterogeneity in human breast cancers. Mol. Ther. 2021, 29, 2350–2365. [Google Scholar] [CrossRef]
- Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e1323. [CrossRef]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Whitburn, J.; Rao, S.R.; Morris, E.V.; Tabata, S.; Hirayama, A.; Soga, T.; Edwards, J.R.; Kaya, Z.; Palmer, C.; Hamdy, F.C.; et al. Metabolic profiling of prostate cancer in skeletal microenvironments identifies G6PD as a key mediator of growth and survival. Sci. Adv. 2022, 8, eabf9096. [Google Scholar] [CrossRef]
- Li, R.; Ke, M.; Qi, M.; Han, Z.; Cao, Y.; Deng, Z.; Qian, J.; Yang, Y.; Gu, C. G6PD promotes cell proliferation and dexamethasone resistance in multiple myeloma via increasing anti-oxidant production and activating Wnt/β-catenin pathway. Exp. Hematol. Oncol. 2022, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Hernandez, M.O.; Zhao, Y.; Mehta, M.; Tran, B.; Kelly, M.; Rae, Z.; Hernandez, J.M.; Davis, J.L.; Martin, S.P.; et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell 2019, 36, 418–430.e416. [Google Scholar] [CrossRef]
- van der Leun, A.M.; Thommen, D.S.; Schumacher, T.N. CD8(+) T cell states in human cancer: Insights from single-cell analysis. Nat. Rev. Cancer 2020, 20, 218–232. [Google Scholar] [CrossRef]
- Mogilenko, D.A.; Shpynov, O.; Andhey, P.S.; Arthur, L.; Swain, A.; Esaulova, E.; Brioschi, S.; Shchukina, I.; Kerndl, M.; Bambouskova, M.; et al. Comprehensive Profiling of an Aging Immune System Reveals Clonal GZMK(+) CD8(+) T Cells as Conserved Hallmark of Inflammaging. Immunity 2021, 54, 99–115.e112. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e310. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kamińska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354.e315. [Google Scholar] [CrossRef]
- Calderaro, J.; Ziol, M.; Paradis, V.; Zucman-Rossi, J. Molecular and histological correlations in liver cancer. J. Hepatol. 2019, 71, 616–630. [Google Scholar] [CrossRef]
- Merkle, F.T.; Ghosh, S.; Kamitaki, N.; Mitchell, J.; Avior, Y.; Mello, C.; Kashin, S.; Mekhoubad, S.; Ilic, D.; Charlton, M.; et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 2017, 545, 229–233. [Google Scholar] [CrossRef]
- Nio, K.; Yamashita, T.; Kaneko, S. The evolving concept of liver cancer stem cells. Mol. Cancer 2017, 16, 4. [Google Scholar] [CrossRef]
- Luengo, A.; Gui, D.Y.; Vander Heiden, M.G. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017, 24, 1161–1180. [Google Scholar] [CrossRef]
- Tu, D.; Gao, Y.; Yang, R.; Guan, T.; Hong, J.S.; Gao, H.M. The pentose phosphate pathway regulates chronic neuroinflammation and dopaminergic neurodegeneration. J. Neuroinflamm. 2019, 16, 255. [Google Scholar] [CrossRef]
- Wu, T.; Luo, G.; Lian, Q.; Sui, C.; Tang, J.; Zhu, Y.; Zheng, B.; Li, Z.; Zhang, Y.; Zhang, Y.; et al. Discovery of a Carbamoyl Phosphate Synthetase 1-Deficient HCC Subtype with Therapeutic Potential Through Integrative Genomic and Experimental Analysis. Hepatology 2021, 74, 3249–3268. [Google Scholar] [CrossRef]
- Grinde, M.T.; Hilmarsdottir, B.; Tunset, H.M.; Henriksen, I.M.; Kim, J.; Haugen, M.H.; Rye, M.B.; Mælandsmo, G.M.; Moestue, S.A. Glutamine to proline conversion is associated with response to glutaminase inhibition in breast cancer. Breast Cancer Res. 2019, 21, 61. [Google Scholar] [CrossRef] [PubMed]
- Callao, V.; Montoya, E. Toxohormone-like factor from microorganisms with impaired respiration. Science 1961, 134, 2041–2042. [Google Scholar] [CrossRef]
- Hirschey, M.D.; DeBerardinis, R.J.; Diehl, A.M.E.; Drew, J.E.; Frezza, C.; Green, M.F.; Jones, L.W.; Ko, Y.H.; Le, A.; Lea, M.A.; et al. Dysregulated metabolism contributes to oncogenesis. Semin. Cancer Biol. 2015, 35, S129–S150. [Google Scholar] [CrossRef] [PubMed]
- La Vecchia, S.; Sebastián, C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin. Cell Dev. Biol. 2020, 98, 63–70. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Liang, Y.; Wu, R.; Zhao, Y.; Hong, X.; Lin, M.; Yu, H.; Liu, L.; Levine, A.J.; et al. Tumour-associated mutant p53 drives the Warburg effect. Nat. Commun. 2013, 4, 2935. [Google Scholar] [CrossRef]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef]
- Jiang, P.; Du, W.; Wang, X.; Mancuso, A.; Gao, X.; Wu, M.; Yang, X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011, 13, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Fersht, A.R. Structural biology of the tumor suppressor p53 and cancer-associated mutants. Adv. Cancer Res. 2007, 97, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.; Terzic, A. Energy metabolism in the acquisition and maintenance of stemness. Semin. Cell Dev. Biol. 2016, 52, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Tsogtbaatar, E.; Landin, C.; Minter-Dykhouse, K.; Folmes, C.D.L. Energy Metabolism Regulates Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 87. [Google Scholar] [CrossRef]
- Li, H.; Feng, Z.; He, M.L. Lipid metabolism alteration contributes to and maintains the properties of cancer stem cells. Theranostics 2020, 10, 7053–7069. [Google Scholar] [CrossRef]
- Yang, T.; Shu, X.; Zhang, H.W.; Sun, L.X.; Yu, L.; Liu, J.; Sun, L.C.; Yang, Z.H.; Ran, Y.L. Enolase 1 regulates stem cell-like properties in gastric cancer cells by stimulating glycolysis. Cell Death Dis. 2020, 11, 870. [Google Scholar] [CrossRef]
- Li, M.; He, X.; Guo, W.; Yu, H.; Zhang, S.; Wang, N.; Liu, G.; Sa, R.; Shen, X.; Jiang, Y.; et al. Aldolase B suppresses hepatocellular carcinogenesis by inhibiting G6PD and pentose phosphate pathways. Nat. Cancer 2020, 1, 735–747. [Google Scholar] [CrossRef]
- Jhunjhunwala, S.; Hammer, C.; Delamarre, L. Antigen presentation in cancer: Insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer 2021, 21, 298–312. [Google Scholar] [CrossRef]
- Feng, J.; Li, J.; Wu, L.; Yu, Q.; Ji, J.; Wu, J.; Dai, W.; Guo, C. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 126. [Google Scholar] [CrossRef]
- Xu, S.; Herschman, H.R. A Tumor Agnostic Therapeutic Strategy for Hexokinase 1-Null/Hexokinase 2-Positive Cancers. Cancer Res. 2019, 79, 5907–5914. [Google Scholar] [CrossRef]
- Chang, C.H.; Curtis, J.D.; Maggi, L.B., Jr.; Faubert, B.; Villarino, A.V.; O′Sullivan, D.; Huang, S.C.; van der Windt, G.J.; Blagih, J.; Qiu, J.; et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 2013, 153, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Ghesquière, B.; Wong, B.W.; Kuchnio, A.; Carmeliet, P. Metabolism of stromal and immune cells in health and disease. Nature 2014, 511, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Han, Y.; Wu, N.; Yi, P.; Tang, L.; Pan, Q.; Rao, S.; et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 2021, 20, 28. [Google Scholar] [CrossRef] [PubMed]
- Leone, R.D.; Powell, J.D. Metabolism of immune cells in cancer. Nat. Rev. Cancer 2020, 20, 516–531. [Google Scholar] [CrossRef] [PubMed]

| Overall | Cluster 1 | Cluster 2 | p | |
|---|---|---|---|---|
| N | 363 | 133 | 230 | |
| Gender = female/male (%) | 118/245 (32.5/67.5) | 48/85 (36.1/63.9) | 70/160 (30.4/69.6) | 0.321 |
| Race (%) | 0.062 | |||
| American Indian or Alaska Native | 1 (0.3) | 0 (0.0) | 1 (0.4) | |
| Asian | 155 (42.7) | 65 (48.9) | 90 (39.1) | |
| Black or African American | 17 (4.7) | 9 (6.8) | 8 (3.5) | |
| Not Reported | 10 (2.8) | 1 (0.8) | 9 (3.9) | |
| White | 180 (49.6) | 58 (43.6) | 122 (53.0) | |
| Age (median [IQR]) | 61.53 [51.97, 69.04] | 61.09 [51.46, 68.29] | 62.05 [52.78, 69.53] | 0.446 |
| AFP (median [IQR]) | 15.00 [4.00, 264.75] | 79.00 [9.50, 3088.50] | 8.00 [4.00, 47.00] | <0.001 |
| Stage (%) | <0.001 | |||
| Not reported | 24 (6.6) | 8 (6.0) | 16 (7.0) | |
| Stage i | 170 (46.8) | 42 (31.6) | 128 (55.7) | |
| Stage ii | 84 (23.1) | 38 (28.6) | 46 (20.0) | |
| Stage iii | 3 (0.8) | 0 (0.0) | 3 (1.3) | |
| Stage iiia | 61 (16.8) | 33 (24.8) | 28 (12.2) | |
| Stage iiib | 8 (2.2) | 5 (3.8) | 3 (1.3) | |
| Stage iiic | 9 (2.5) | 6 (4.5) | 3 (1.3) | |
| Stage iv | 1 (0.3) | 0 (0.0) | 1 (0.4) | |
| Stage iva | 1 (0.3) | 0 (0.0) | 1 (0.4) | |
| Stage ivb | 2 (0.6) | 1 (0.8) | 1 (0.4) | |
| T Stage (%) | <0.001 | |||
| T1 | 180 (49.9) | 43 (32.3) | 137 (60.1) | |
| T2 | 89 (24.7) | 42 (31.6) | 47 (20.6) | |
| T2a | 1 (0.3) | 1 (0.8) | 0 (0.0) | |
| T2b | 1 (0.3) | 1 (0.8) | 0 (0.0) | |
| T3 | 42 (11.6) | 22 (16.5) | 20 (8.8) | |
| T3a | 28 (7.8) | 14 (10.5) | 14 (6.1) | |
| T3b | 6 (1.7) | 2 (1.5) | 4 (1.8) | |
| T4 | 13 (3.6) | 8 (6.0) | 5 (2.2) | |
| TX | 1 (0.3) | 0 (0.0) | 1 (0.4) | |
| N Stage (%) | 0.346 | |||
| N0 | 246 (68.0) | 95 (72.0) | 151 (65.7) | |
| N1 | 4 (1.1) | 2 (1.5) | 2 (0.9) | |
| NX | 112 (30.9) | 35 (26.5) | 77 (33.5) | |
| M Stage (%) | 0.148 | |||
| M0 | 262 (72.2) | 104 (78.2) | 158 (68.7) | |
| M1 | 3 (0.8) | 1 (0.8) | 2 (0.9) | |
| MX | 98 (27.0) | 28 (21.1) | 70 (30.4) | |
| Tumor Weight (median [IQR]) | 150.00 [70.00, 315.00] | 220.00 [110.00, 460.00] | 110.00 [50.00, 250.00] | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, M.; Li, X.; Chen, L.; Mo, S.; Liu, J.; Huang, T.; Luo, F.; Zhang, J. A High-Throughput Sequencing Data-Based Classifier Reveals the Metabolic Heterogeneity of Hepatocellular Carcinoma. Cancers 2023, 15, 592. https://doi.org/10.3390/cancers15030592
Ye M, Li X, Chen L, Mo S, Liu J, Huang T, Luo F, Zhang J. A High-Throughput Sequencing Data-Based Classifier Reveals the Metabolic Heterogeneity of Hepatocellular Carcinoma. Cancers. 2023; 15(3):592. https://doi.org/10.3390/cancers15030592
Chicago/Turabian StyleYe, Maolin, Xuewei Li, Lirong Chen, Shaocong Mo, Jie Liu, Tiansheng Huang, Feifei Luo, and Jun Zhang. 2023. "A High-Throughput Sequencing Data-Based Classifier Reveals the Metabolic Heterogeneity of Hepatocellular Carcinoma" Cancers 15, no. 3: 592. https://doi.org/10.3390/cancers15030592
APA StyleYe, M., Li, X., Chen, L., Mo, S., Liu, J., Huang, T., Luo, F., & Zhang, J. (2023). A High-Throughput Sequencing Data-Based Classifier Reveals the Metabolic Heterogeneity of Hepatocellular Carcinoma. Cancers, 15(3), 592. https://doi.org/10.3390/cancers15030592







