DDX5 Functions as a Tumor Suppressor in Tongue Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmid and Lentivirus Preparation
2.3. Establishment of Stable Tongue Cancer Cell Lines
2.4. Cell Proliferation and Colony Formation Analysis
2.5. Wound Healing Assay
2.6. Cisplatin Treatment
2.7. Real-Time PCR
2.8. Western Blot Analysis
2.9. Xenograft Tumor Mouse Models
2.10. Human Tongue Cancer Samples
2.11. Immunohistochemical and Immunofluorescence Staining
2.12. RNA Sequencing and Analysis
2.13. Data Collection and Procession
2.14. Differential Expression Analysis, Gene Set Enrichment Analysis (GSEA), and Gene Set Variation Analysis (GSVA)
2.15. Identification of Malignant Cells via Copy Number Variation (CNV)
2.16. Monocle 2
2.17. Drug Sensitivity Analysis
2.18. Analysis of the Link between DDX5 Expression in Tumor Cells and Infiltrated Immune Cells in the Tumor Microenvironment
2.19. Statistical Analysis
3. Results
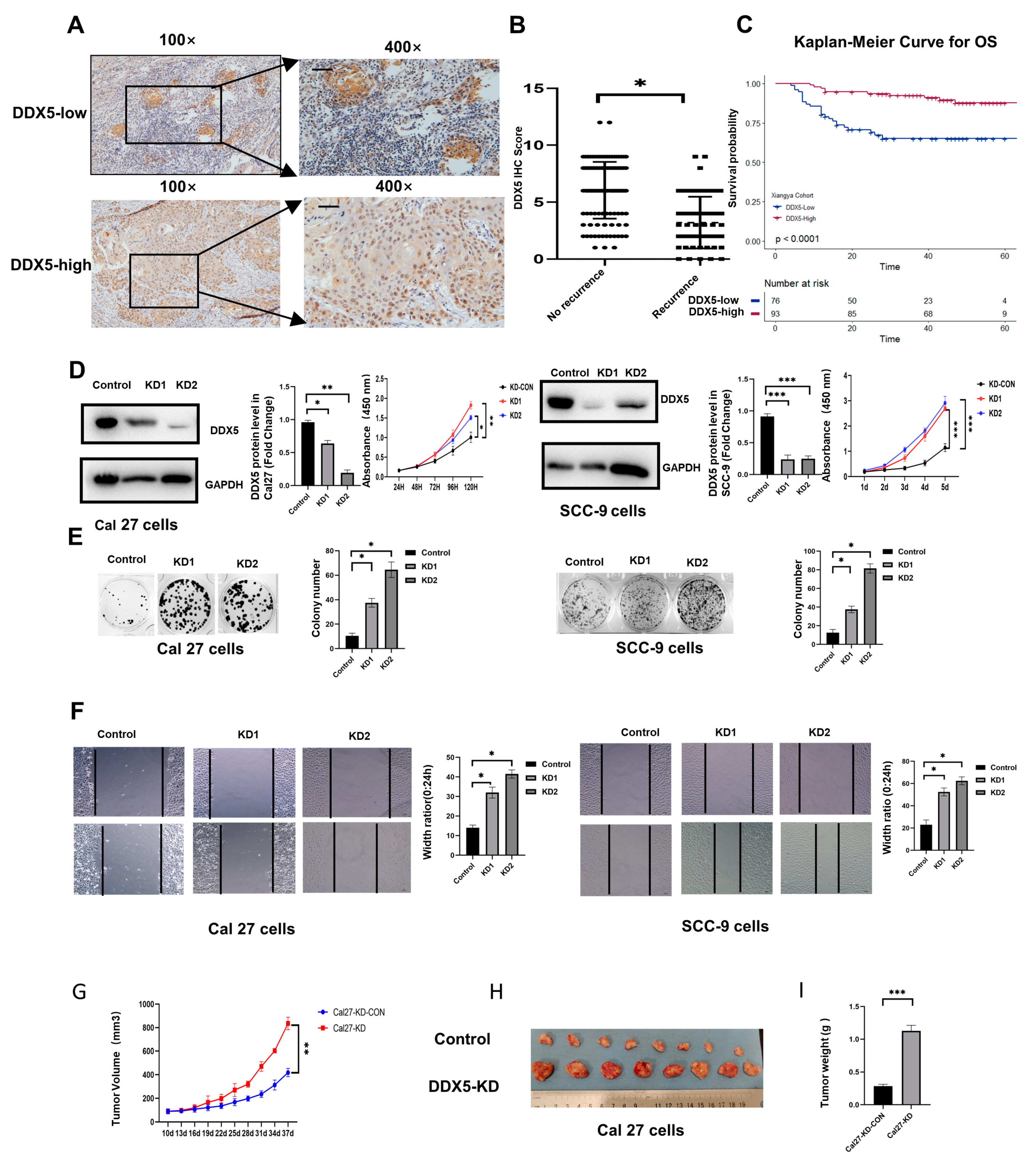
3.1. High Expression of DDX5 in Tongue Cancer Is Associated with Better Clinical Prognosis
3.2. DDX5 Knockdown Promotes Tongue Cancer Cell Proliferation and Mobility
3.3. DDX5 Knockdown Promotes Tongue Cancer Xenograft Tumor Development
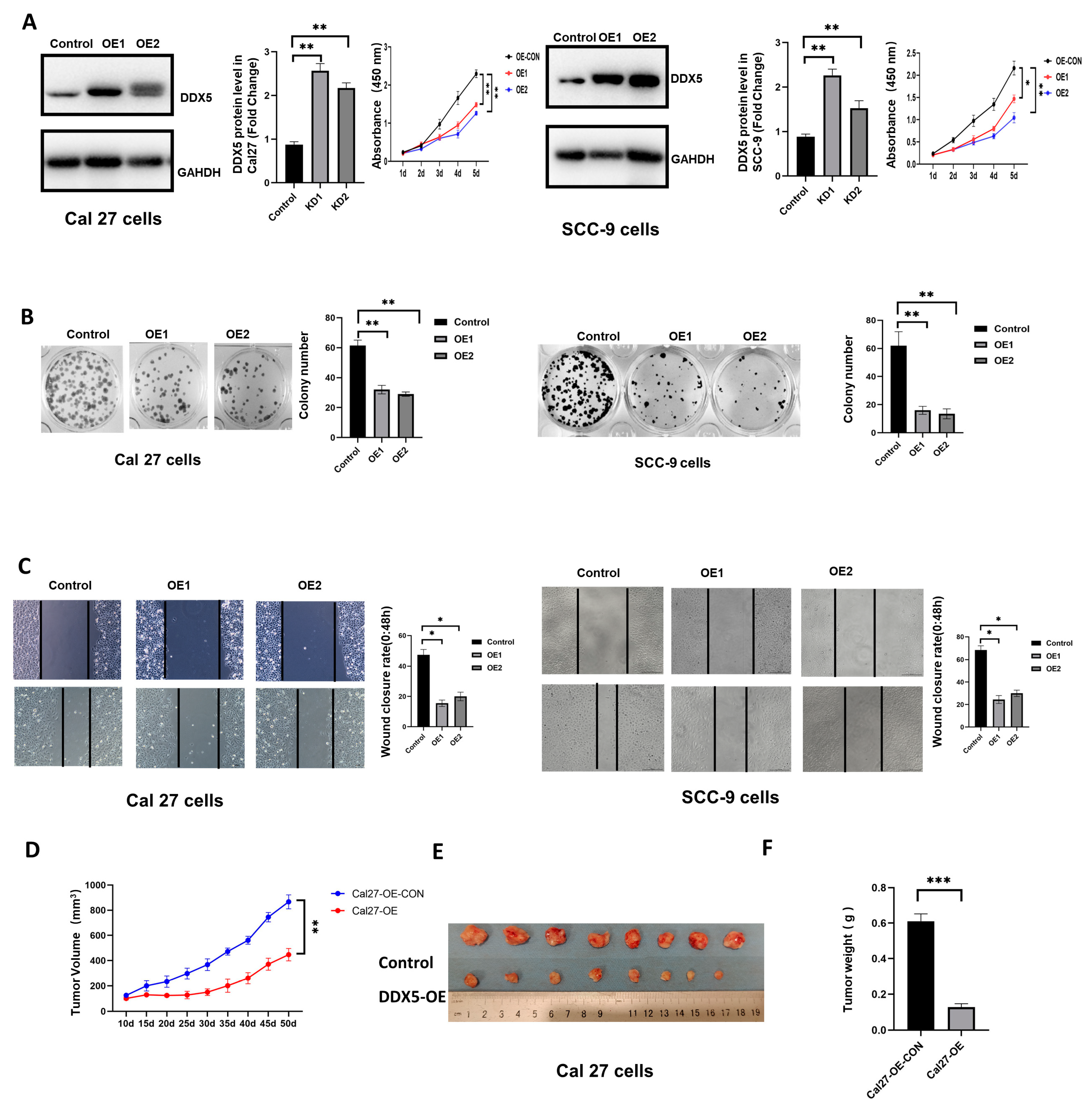
3.4. Overexpression of DDX5 Inhibits Tongue Cancer Cell Proliferation and Mobility
3.5. DDX5 Overexpression Suppresses Tongue Cancer Xenograft Tumor Development
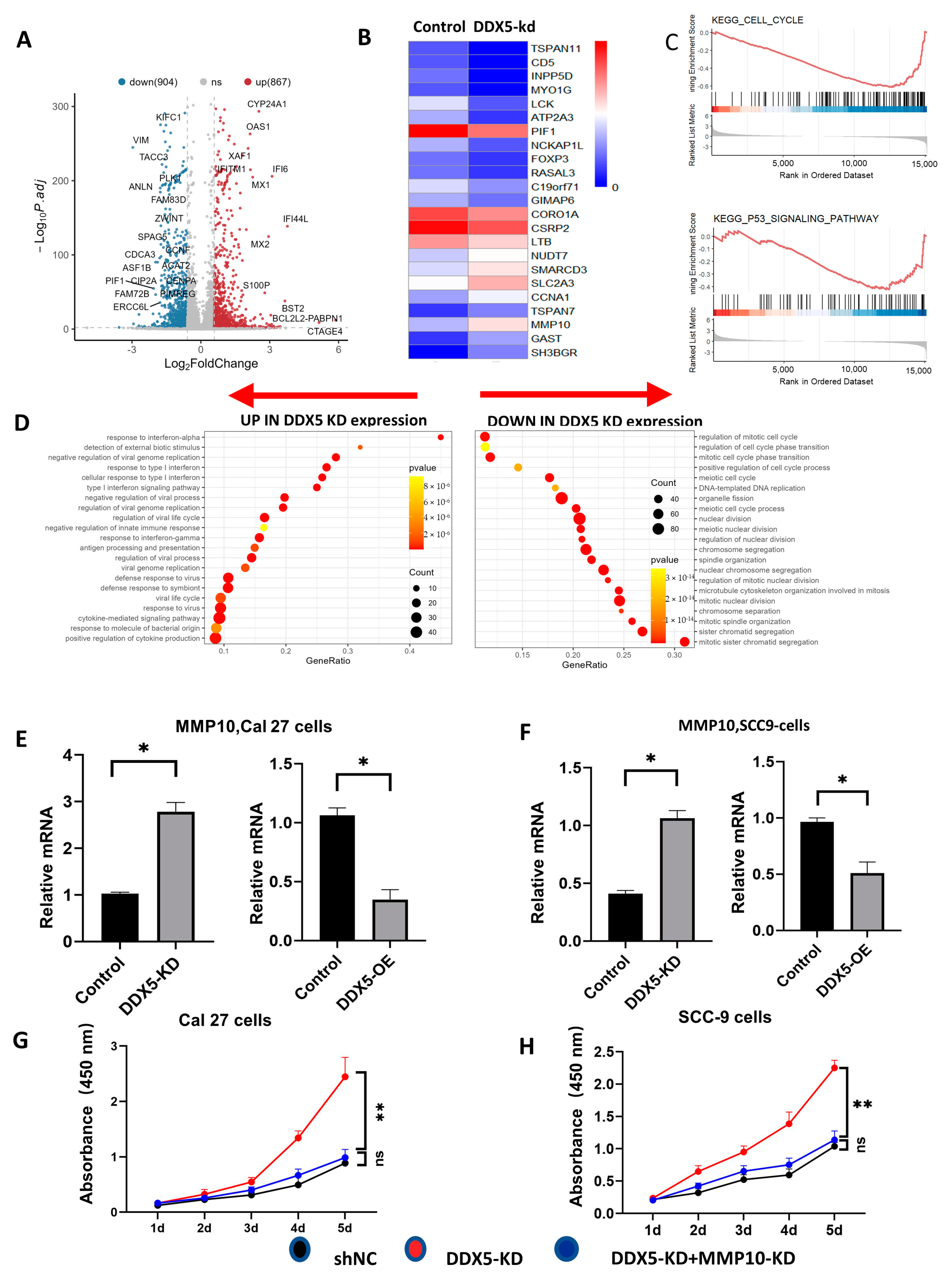
3.6. Knockdown of DDX5 Upregulates the DDX5s Associated with Tongue Cancer Progression
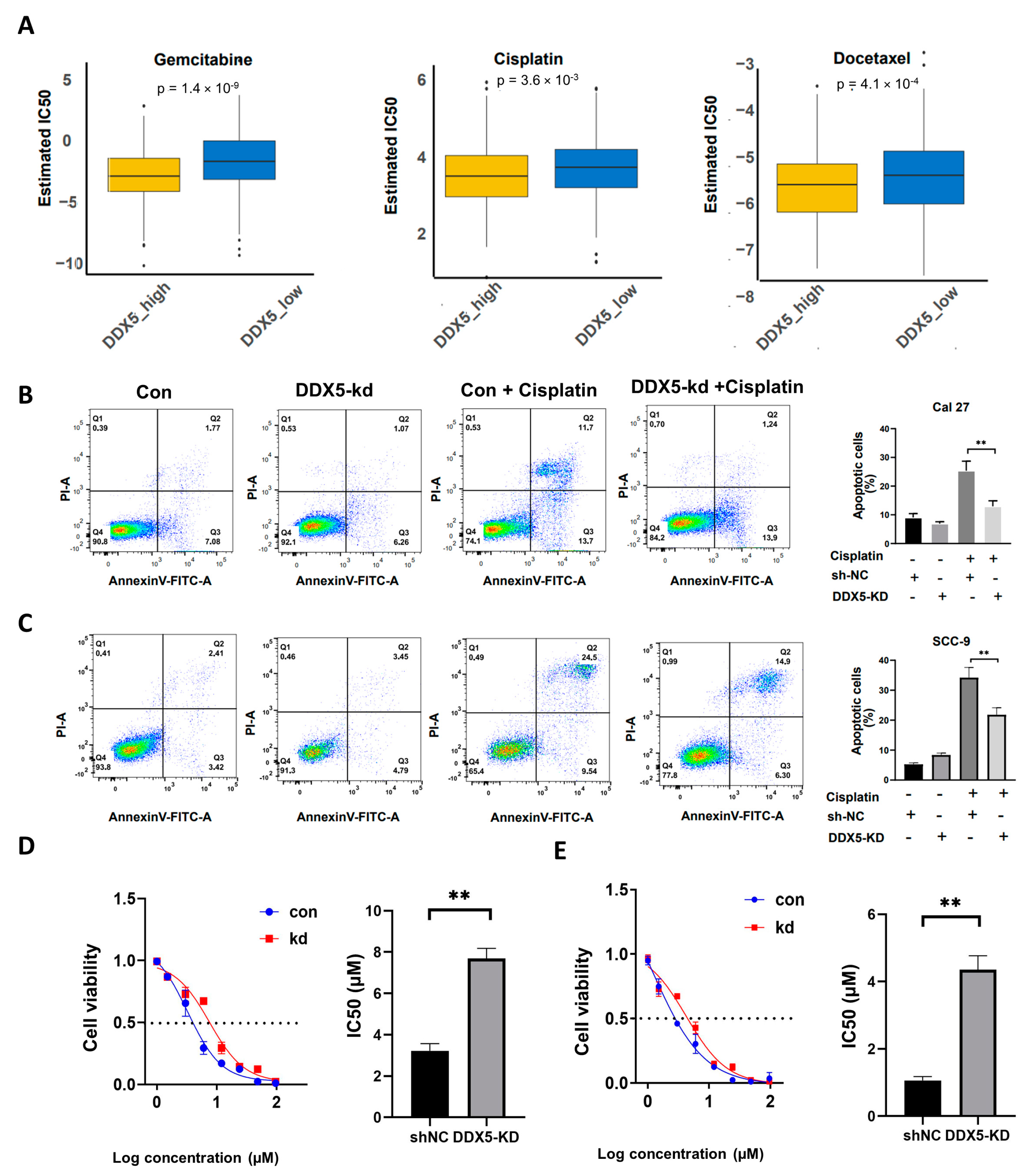
3.7. The Suppression of DDX5 Enhances Chemoresistance in Tongue Cancer Cells
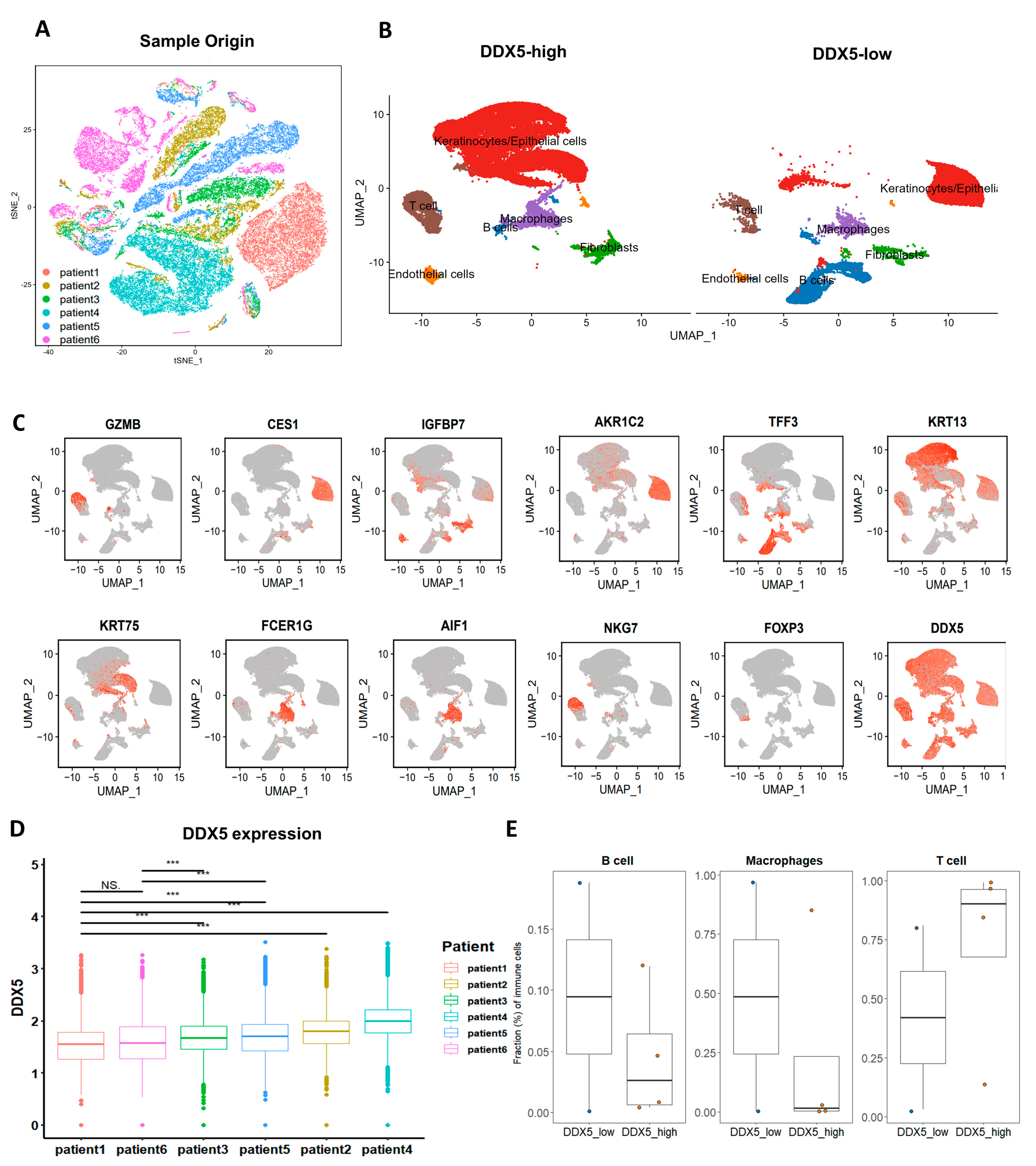
3.8. DDX5 Expression in Tongue Cancer Cells Potentially Modulates Immune Cell Infiltration in the Tumor Microenvironment
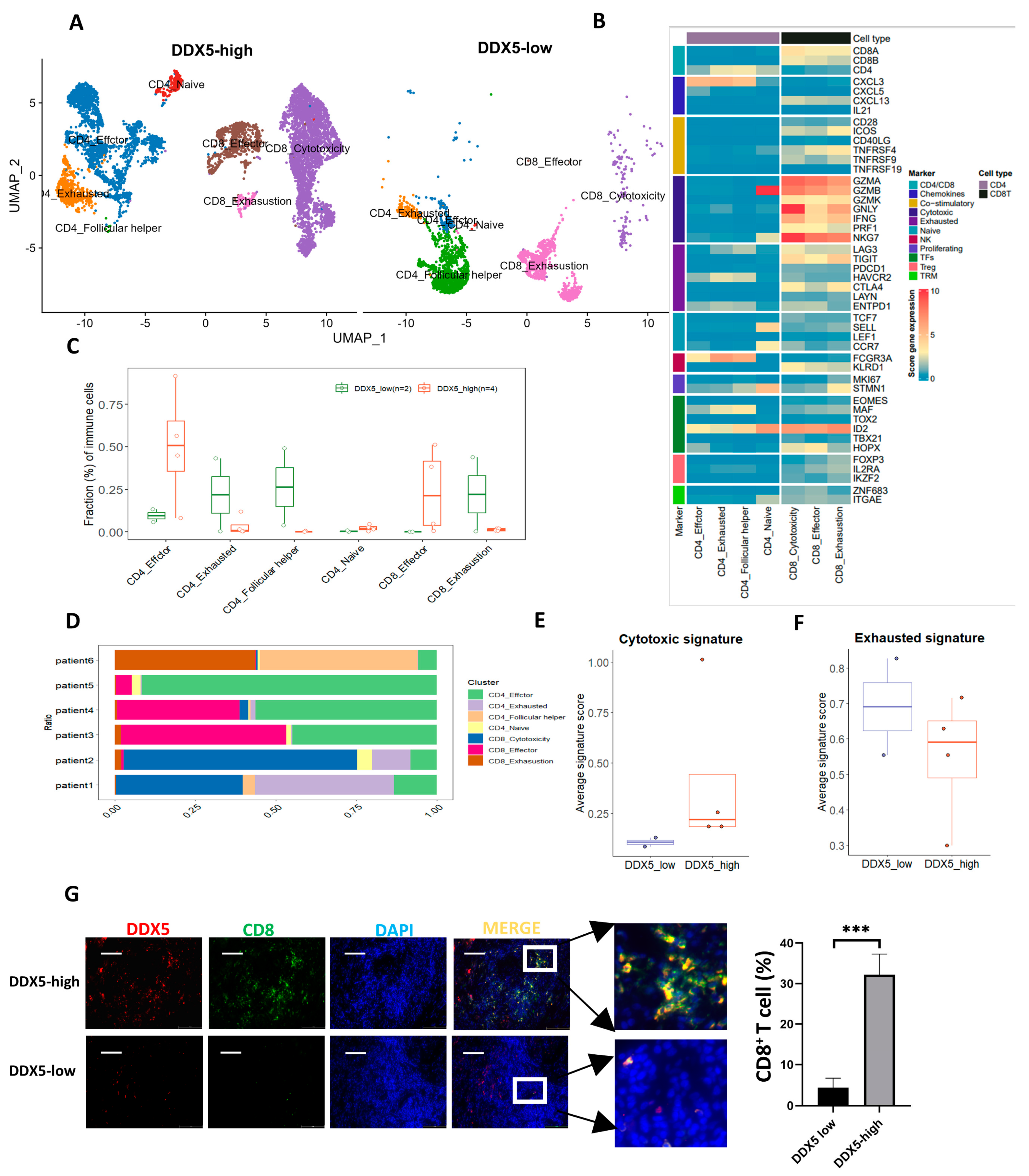
3.9. DDX5 Expression in Tongue Cancer Cells Favors the Infiltration of T Cell Clusters Exhibiting Anticancer Activities
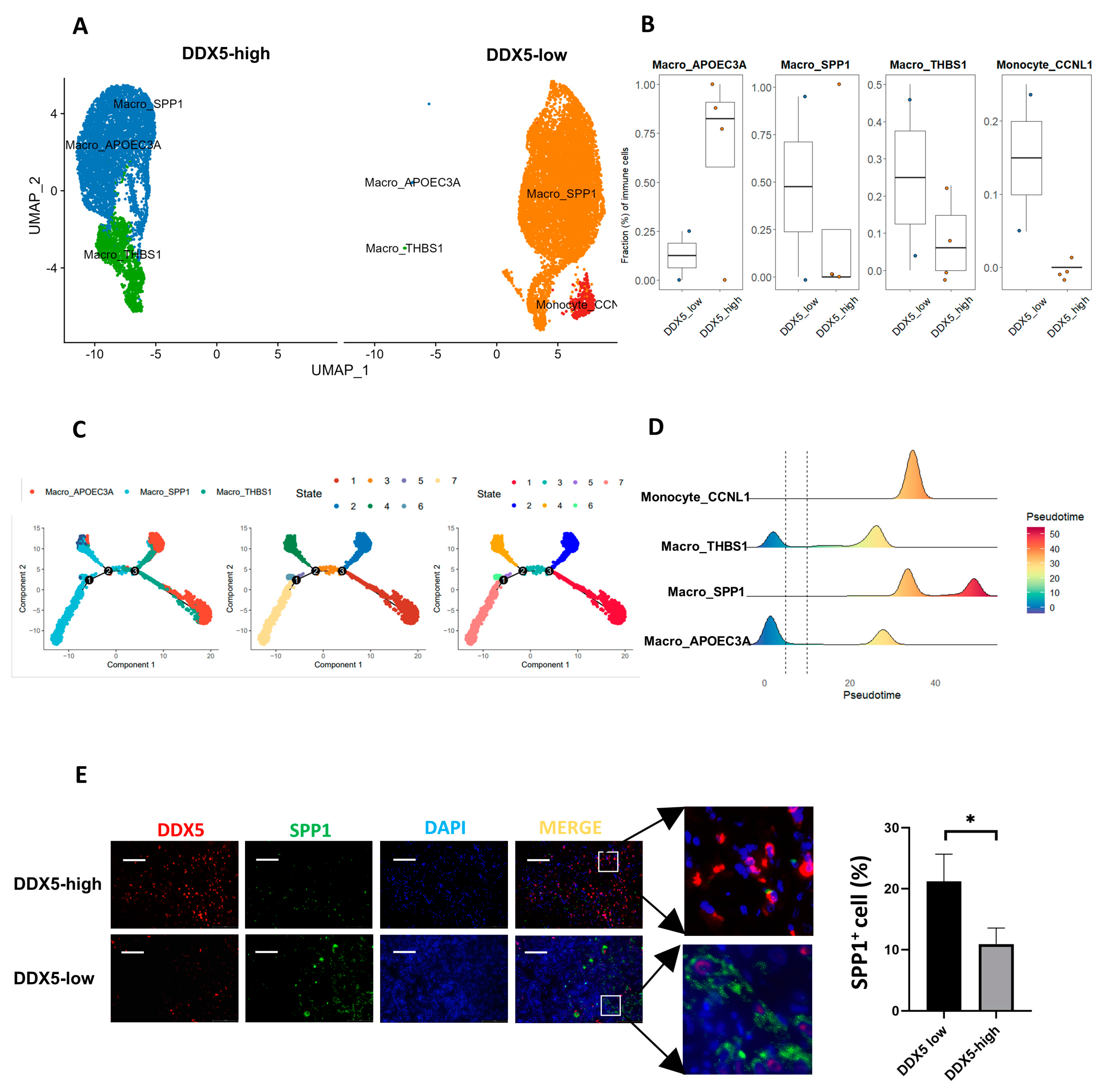
3.10. DDX5 Expression in Tongue Cancer Cells Is Negatively Correlated with the M2 Macrophage Infiltration in the TME
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Cramer, J.D.; Burtness, B.; Le, Q.T.; Ferris, R.L. The changing therapeutic landscape of head and neck cancer. Nat. Rev. Clin. Oncol. 2019, 16, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Cao, C.; Kumar, M.; Sinha, S.; Chanda, A.; McNeil, R.; Samuel, D.; Arora, R.K.; Matthews, T.W.; Chandarana, S.; et al. Spatial transcriptomics reveals distinct and conserved tumor core and edge architectures that predict survival and targeted therapy response. Nat. Commun. 2023, 14, 5029. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Zhang, X.; Song, X.; Zhang, B.; Zhang, Y. Pan-cancer analysis of the prognostic and immunological roles of DEAD-box helicase 5 (DDX5) in human tumors. Front. Genet. 2022, 13, 1039440. [Google Scholar] [CrossRef] [PubMed]
- Terrone, S.; Valat, J.; Fontrodona, N.; Giraud, G.; Claude, J.B.; Combe, E.; Lapendry, A.; Polvèche, H.; Ameur, L.B.; Duvermy, A.; et al. RNA helicase-dependent gene looping impacts messenger RNA processing. Nucleic Acids Res. 2022, 50, 9226–9246. [Google Scholar] [CrossRef]
- Secchi, M.; Lodola, C.; Garbelli, A.; Bione, S.; Maga, G. DEAD-Box RNA Helicases DDX3X and DDX5 as Oncogenes or Oncosuppressors: A Network Perspective. Cancers 2022, 14, 3820. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fountzilas, C.; Puzanov, I.; Attwood, K.M.; Morrison, C.; Ling, X. Multiple functions of the DEAD-box RNA helicase, DDX5 (p68), make DDX5 a superior oncogenic biomarker and target for targeted cancer therapy. Am. J. Cancer Res. 2021, 11, 5190–5213. [Google Scholar] [PubMed]
- Nyamao, R.M.; Wu, J.; Yu, L.; Xiao, X.; Zhang, F.M. Roles of DDX5 in the tumorigenesis, proliferation, differentiation, metastasis and pathway regulation of human malignancies. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 85–98. [Google Scholar] [CrossRef]
- Qiu, P.; Jie, Y.; Ma, C.; Chen, H.; Qin, Y.; Tu, K.; Wang, L.; Zhang, Z. Paired box 8 facilitates the c-MYC related cell cycle progress in TP53-mutation uterine corpus endometrial carcinoma through interaction with DDX5. Cell Death Discov. 2022, 8, 276. [Google Scholar] [CrossRef]
- Xu, J.; Cai, Y.; Ma, Z.; Jiang, B.; Liu, W.; Cheng, J.; Guo, N.; Wang, Z.; Sealy, J.E.; Song, C.; et al. The RNA helicase DDX5 promotes viral infection via regulating N6-methyladenosine levels on the DHX58 and NFκB transcripts to dampen antiviral innate immunity. PLoS Pathog. 2021, 17, e1009530. [Google Scholar] [CrossRef]
- Che, Y.; Bai, M.; Lu, K.; Fu, L. Splicing factor SRSF3 promotes the progression of cervical cancer through regulating DDX5. Mol. Carcinog. 2023, 62, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Xiao, L.; Rong, H.; Ou, Z.; Cai, T.; Liu, N.; Li, B.; Zhang, L.; Wu, F.; Lan, T.; et al. Single-cell profiling of tumor-infiltrating TCF1/TCF7(+) T cells reveals a T lymphocyte subset associated with tertiary lymphoid structures/organs and a superior prognosis in oral cancer. Oral Oncol. 2021, 119, 105348. [Google Scholar] [CrossRef] [PubMed]
- Doescher, J.; von Witzleben, A.; Boukas, K.; Weissinger, S.E.; Thomas, G.J.; Laban, S.; Thomas, J.; Hoffmann, T.K.; Ottensmeier, C.H. Changes in Gene Expression Patterns in the Tumor Microenvironment of Head and Neck Squamous Cell Carcinoma Under Chemoradiotherapy Depend on Response. Front. Oncol. 2022, 12, 862694. [Google Scholar] [CrossRef]
- Bouaoud, J.; Foy, J.P.; Tortereau, A.; Michon, L.; Lavergne, V.; Gadot, N.; Boyault, S.; Valantin, J.; De Souza, G.; Zrounba, P.; et al. Early changes in the immune microenvironment of oral potentially malignant disorders reveal an unexpected association of M2 macrophages with oral cancer free survival. Oncoimmunology 2021, 10, 1944554. [Google Scholar] [CrossRef]
- Chen, C.; Méndez, E.; Houck, J.; Fan, W.; Lohavanichbutr, P.; Doody, D.; Yueh, B.; Futran, N.D.; Upton, M.; Farwell, D.G.; et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2152–2162. [Google Scholar] [CrossRef]
- Gao, R.; Bai, S.; Henderson, Y.C.; Lin, Y.; Schalck, A.; Yan, Y.; Kumar, T.; Hu, M.; Sei, E.; Davis, A.; et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat. Biotechnol. 2021, 39, 599–608. [Google Scholar] [CrossRef]
- Liang, X.; Tang, Z.; Zhang, Y.; Sun, Y.; Wang, J. NAP1L1 promotes the growth of colon cancer by activating HDGF/DDX5. Acta Biochim. Biophys. Sin. 2022, 54, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Song, D.; Sinha, I.; Hessling, B.; Li, X.; Haldosen, L.A.; Zhao, C. Endogenous interaction profiling identifies DDX5 as an oncogenic coactivator of transcription factor Fra-1. Oncogene 2019, 38, 5725–5738. [Google Scholar] [CrossRef]
- Le, T.K.; Cherif, C.; Omabe, K.; Paris, C.; Lannes, F.; Audebert, S.; Baudelet, E.; Hamimed, M.; Barbolosi, D.; Finetti, P.; et al. DDX5 mRNA-targeting antisense oligonucleotide as a new promising therapeutic in combating castration-resistant prostate cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2023, 31, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Beier, U.H.; Maune, S.; Meyer, J.E.; Görögh, T. Overexpression of p68 mRNA in head and neck squamous cell carcinoma cells. Anticancer. Res. 2006, 26, 1941–1946. [Google Scholar]
- Hashemi, V.; Masjedi, A.; Hazhir-Karzar, B.; Tanomand, A.; Shotorbani, S.S.; Hojjat-Farsangi, M.; Ghalamfarsa, G.; Azizi, G.; Anvari, E.; Baradaran, B.; et al. The role of DEAD-box RNA helicase p68 (DDX5) in the development and treatment of breast cancer. J. Cell Physiol. 2019, 234, 5478–5487. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Xu, C.; Yu, D. Mechanisms correlated with chemotherapy resistance in tongue cancers. J. Cancer Res. Ther. 2018, 14, 1–5. [Google Scholar] [CrossRef]
- Louis, T.L.; Wong, W.H.; Yao, P.; Kurd, N.S.; Tysl, T.; Indralingam, C.S.; Ma, S.; Huang, W.J.M.; Chang, J.T. Regulation of CD8 T Cell Differentiation by the RNA-Binding Protein DDX5. J. Immunol. 2023, 211, 241–251. [Google Scholar] [CrossRef]
- Xiao, G.; Li, L.; Tanzhu, G.; Liu, Z.; Gao, X.; Wan, X.; Xiao, D.; Chen, L.; Xia, X.; Zhou, R. Heterogeneity of tumor immune microenvironment of EGFR/ALK-positive tumors versus EGFR/ALK-negative tumors in resected brain metastases from lung adenocarcinoma. J. Immunother. Cancer 2023, 11, e006243. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Ma, S.; Sun, B.; Duan, S.; Han, J.; Barr, T.; Zhang, J.; Bissonnette, M.B.; Kortylewski, M.; He, C.; Chen, J.; et al. YTHDF2 orchestrates tumor-associated macrophage reprogramming and controls antitumor immunity through CD8(+) T cells. Nat. Immunol. 2023, 24, 255–266. [Google Scholar] [CrossRef]
- Wang, X.; Tokheim, C.; Gu, S.S.; Wang, B.; Tang, Q.; Li, Y.; Traugh, N.; Zeng, Z.; Zhang, Y.; Li, Z.; et al. In vivo CRISPR screens identify the E3 ligase Cop1 as a modulator of macrophage infiltration and cancer immunotherapy target. Cell 2021, 184, 5357–5374.e5322. [Google Scholar] [CrossRef]
| Characteristics | DDX5 Expression | |||
|---|---|---|---|---|
| Overall | Low (n = 76) | High (n = 93) | p Value | |
| Age (n, %) | ||||
| <50 | 71(42.0) | 31(40.8) | 40(43.0) | |
| ≥50 | 98(58) | 45(59.2) | 53(57) | 0.893 |
| Sex (n, %) | ||||
| Male | 151(89.3) | 63(82.9) | 88(94.6) | |
| Female | 18(10.7) | 13(17.1) | 5(5.4) | 0.027 |
| T Stage (n, %) | ||||
| I | 62(36.7) | 26(34.2) | 36(38.7) | |
| II | 72(42.6) | 32(42.1) | 40(43) | |
| III | 24(14.2) | 11(14.5) | 13(14) | |
| IV | 11(6.5) | 7(9.2) | 4(4.3) | 0.616 |
| N Stage (n, %) | ||||
| I | 106(62.7) | 40(52.6) | 66(71.0) | |
| II | 36(21.3) | 22(28.9) | 14(15.1) | |
| III | 27(16.0) | 14(18.4) | 13(14.0) | 0.038 |
| Metastasis (n, %) | ||||
| Absence | 153(90.5) | 63(82.9) | 90(96.8) | |
| Presence | 16(9.5) | 13(17.1) | 3(3.2) | 0.005 |
| AJCC Stage (n, %) | ||||
| I | 50(29.6) | 16(21.1) | 34(36.6) | |
| II | 46(27.2) | 19(25) | 27(29) | |
| III | 41(24.3) | 24(31.6) | 17(18.3) | |
| IV | 32(18.9) | 17(22.4) | 15(16.1) | 0.056 |
| Pathology Grade (n, %) | ||||
| Grade 1 | 110(65.1) | 49(64.5) | 61(65.6) | |
| Grade 2 | 50(29.6) | 21(27.6) | 29(31.2) | |
| Grade 3 | 9(5.3) | 6(7.9) | 3(3.2) | 0.387 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Sun, Y.; Long, M.; Chen, X.; Zhong, S.; Huang, C.; Wei, R.; Luo, J.-L. DDX5 Functions as a Tumor Suppressor in Tongue Cancer. Cancers 2023, 15, 5882. https://doi.org/10.3390/cancers15245882
Liu Q, Sun Y, Long M, Chen X, Zhong S, Huang C, Wei R, Luo J-L. DDX5 Functions as a Tumor Suppressor in Tongue Cancer. Cancers. 2023; 15(24):5882. https://doi.org/10.3390/cancers15245882
Chicago/Turabian StyleLiu, Qingqing, Yangqing Sun, Min Long, Xueyan Chen, Shangwei Zhong, Changhao Huang, Rui Wei, and Jun-Li Luo. 2023. "DDX5 Functions as a Tumor Suppressor in Tongue Cancer" Cancers 15, no. 24: 5882. https://doi.org/10.3390/cancers15245882
APA StyleLiu, Q., Sun, Y., Long, M., Chen, X., Zhong, S., Huang, C., Wei, R., & Luo, J.-L. (2023). DDX5 Functions as a Tumor Suppressor in Tongue Cancer. Cancers, 15(24), 5882. https://doi.org/10.3390/cancers15245882






