Androgen Receptor/AP-1 Activates UGT2B15 Transcription to Promote Esophageal Squamous Cell Carcinoma Invasion
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmids Construction, Lentivirus Packaging, and Infection
2.3. Chromatin Immunoprecipitation (ChIP) and Re-ChIP Assays and Data Analysis
2.4. Western Blotting
2.5. Invasion Assays
2.6. Cell Proliferation Assays
2.7. Transfection of siRNA
2.8. RNA Extraction and RT-qPCR
2.9. RNA-seq and Data Analysis
2.10. Animal Experiments
2.11. Statistical Analysis
2.12. Survival Analysis
3. Results
3.1. Expression of UGT2B15 Is Upregulated in ESCC and Androgen-Liganded AR Activates UGT2B15 Transcription
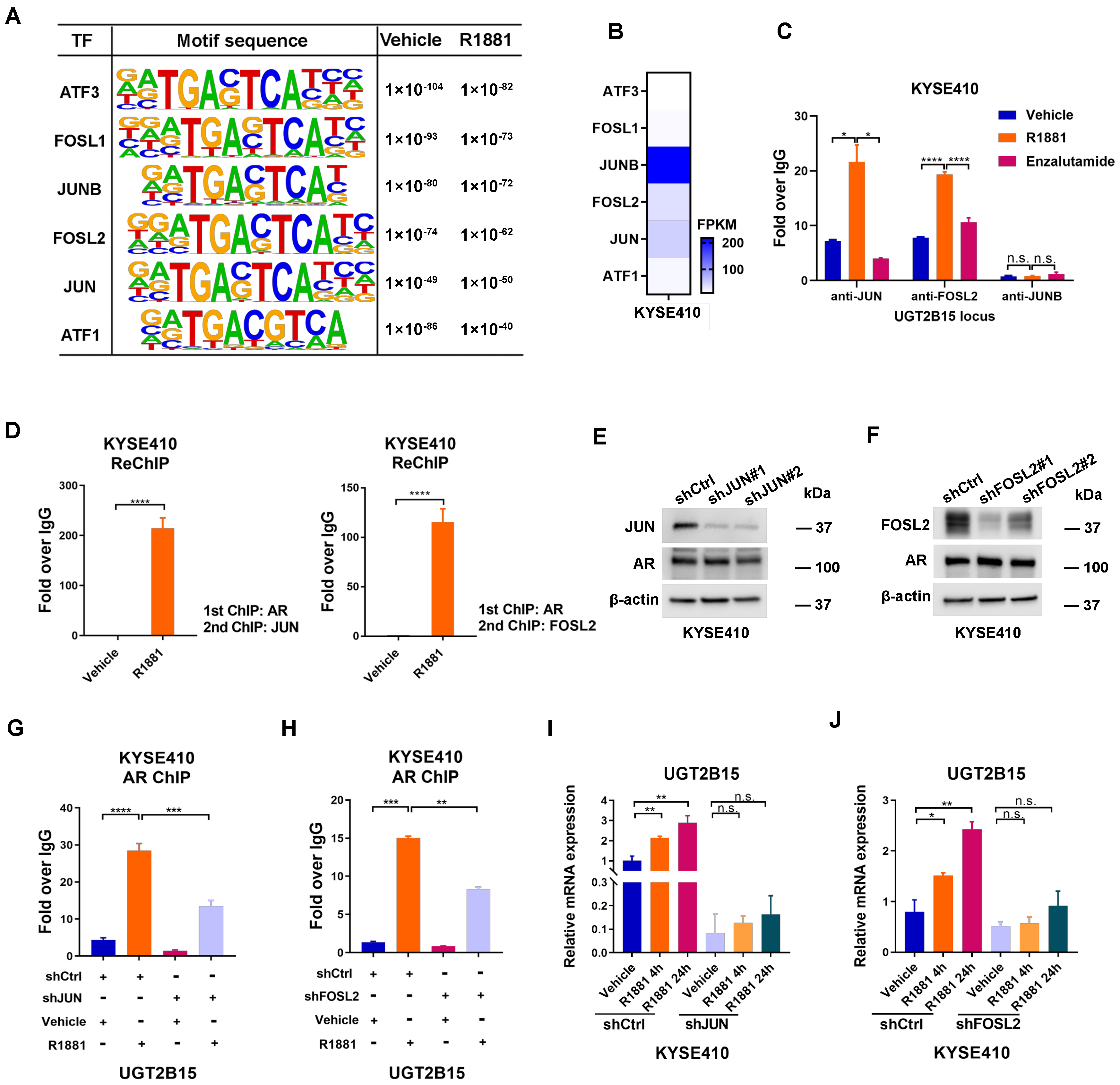
3.2. JUN/FOSL2 Directs Androgen-Induced UGT2B15 Transcription
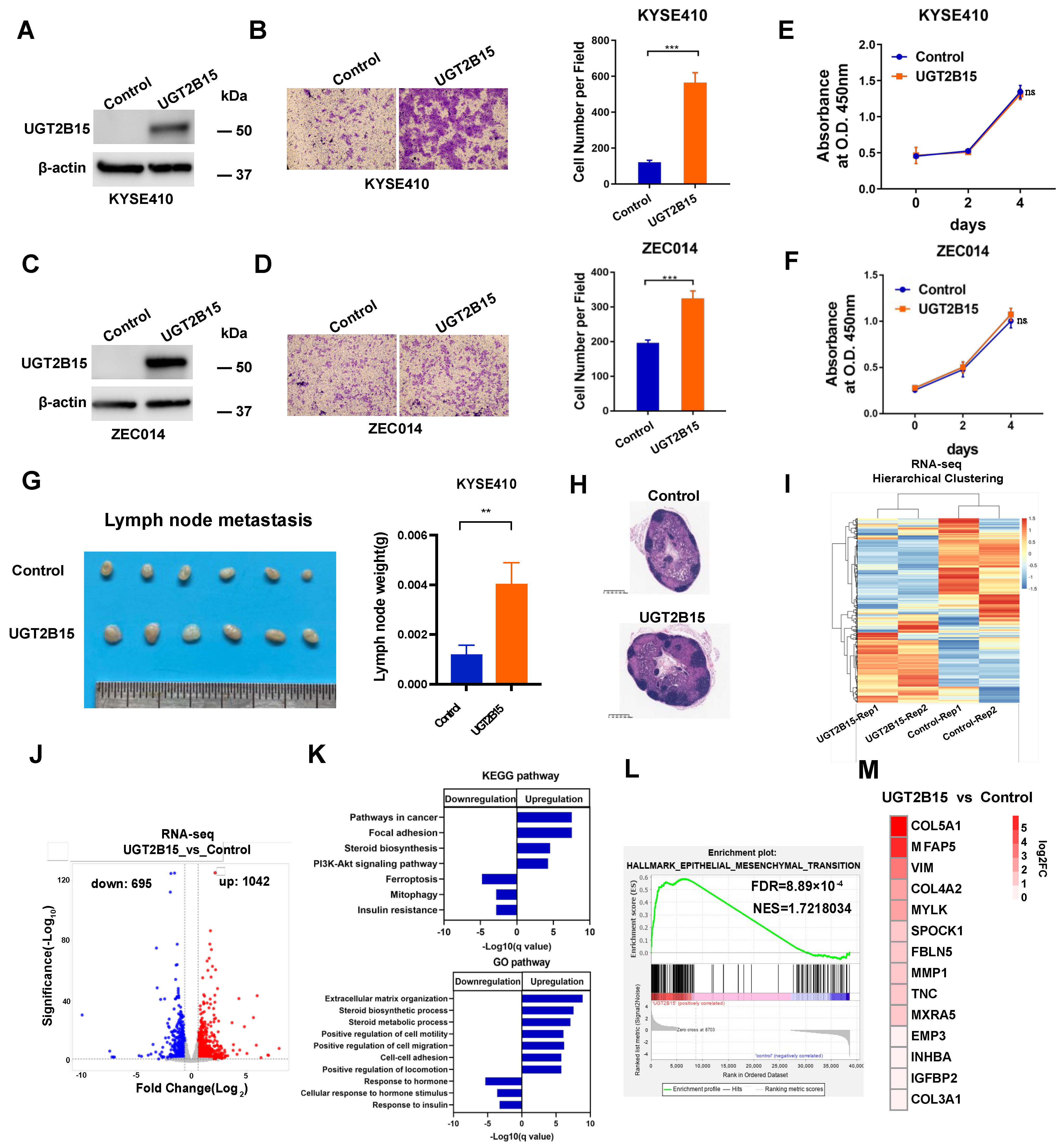
3.3. UGT2B15 Promotes Metastasis in ESCC
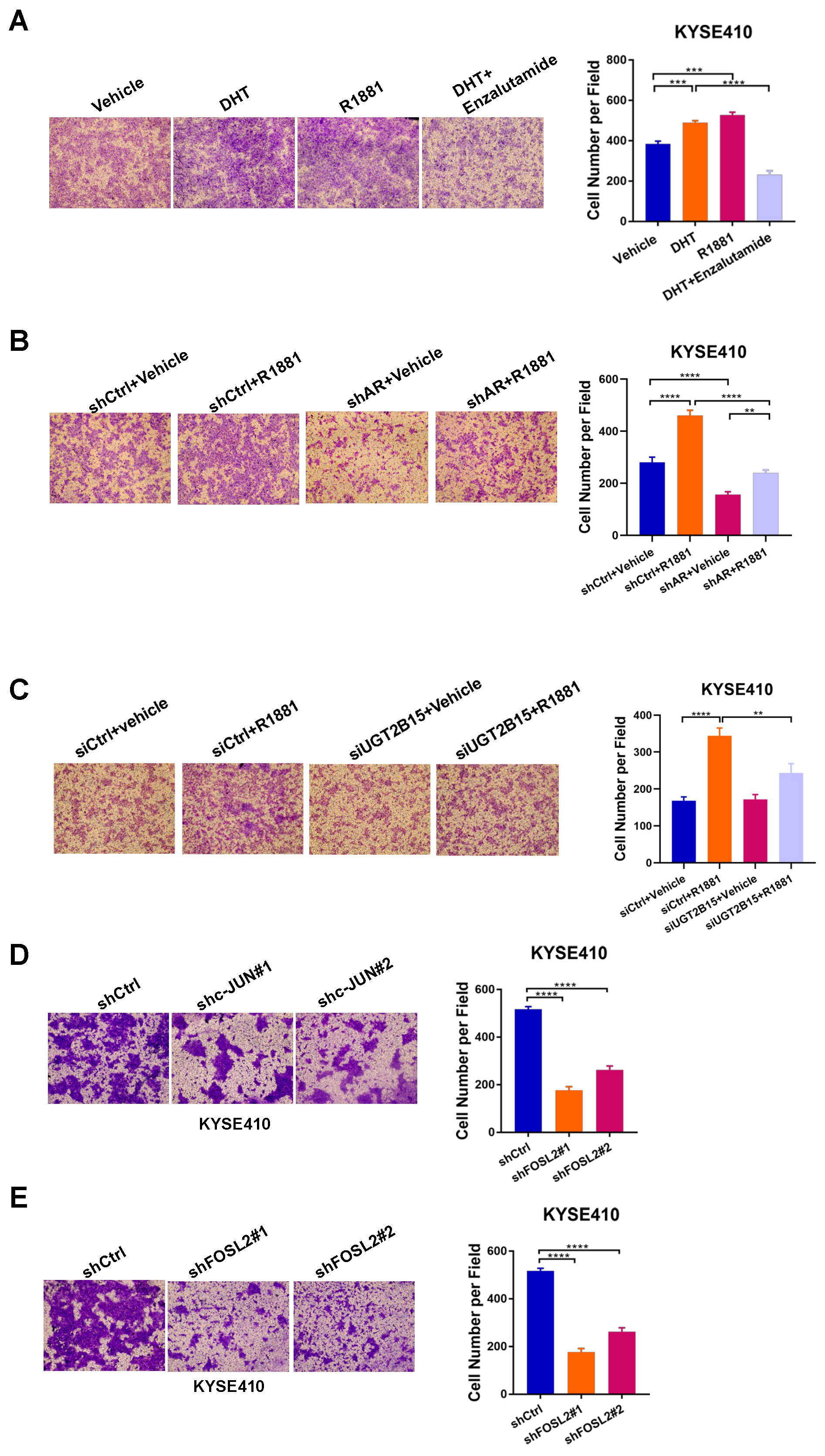
3.4. Androgen-Liganded AR and AP-1 Promote ESCC Cell Invasion
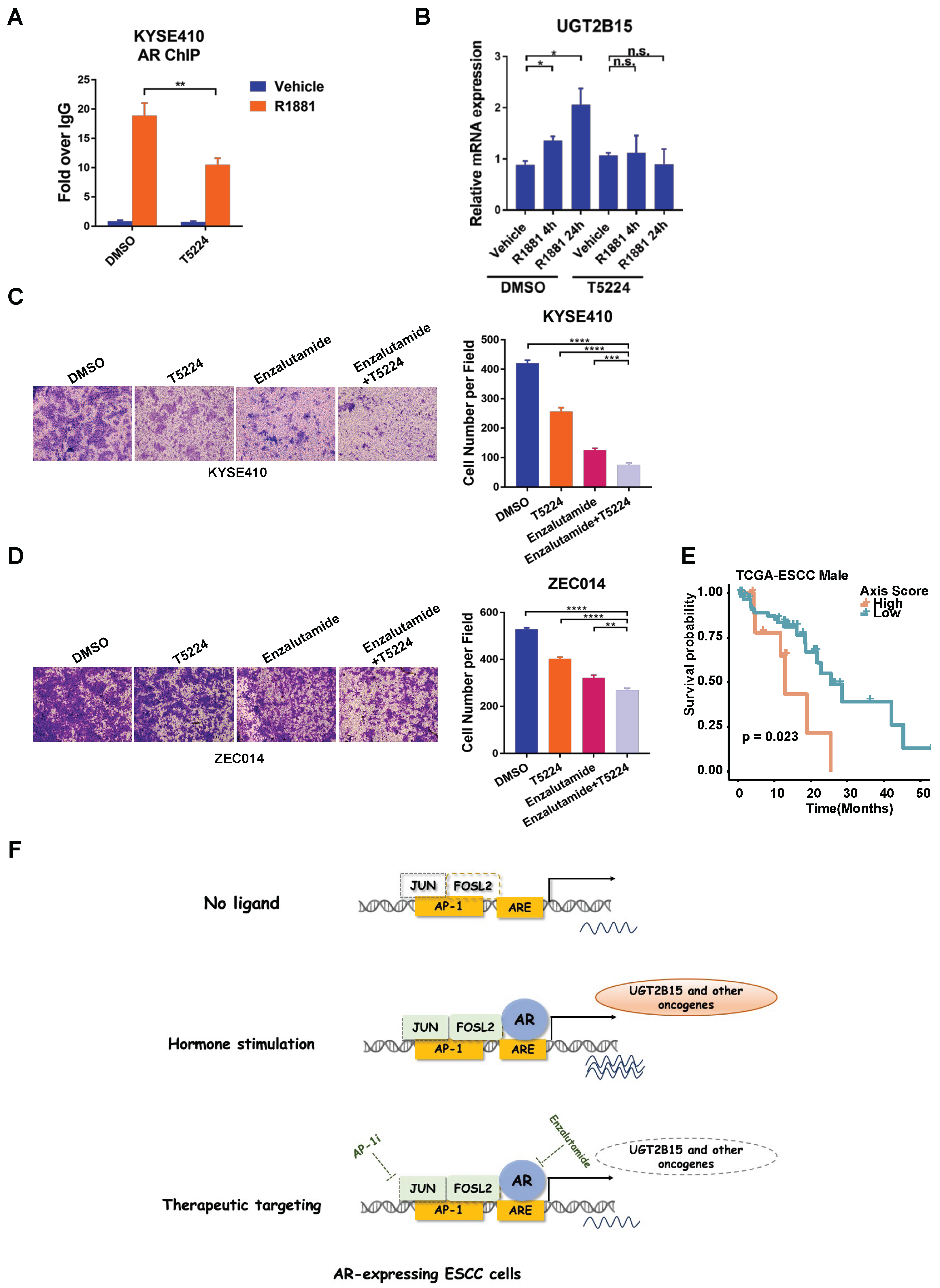
3.5. AP-1 Inhibitor Impairs Androgen-Induced Transcription of UGT2B15 and Invasiveness of ESCC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AR | Androgen receptor |
| ESCC | Esophageal squamous cell carcinoma |
| ChIP | chromatin immunoprecipitation |
| TCGA | The Cancer Genome Atlas |
| AP-1 | Activator protein 1 |
| EMT | Epithelial–mesenchymal transition |
| UGT2B15 | Uridine Diphosphate Glucuronosyltransferase Family 2 Member B15 |
| UGT2B17 | Uridine Diphosphate Glucuronosyltransferase Family 2 Member B17 |
| FOSL2 | FOS-like antigen 2 |
| JUN | Jun proto-oncogene |
| HDAC3 | Histone Deacetylase 3 |
| COL5A1 | Collagen type V alpha 1 chain |
| MFAP5 | Microfibrillar-associated protein 5 |
| ATF | Activating transcription factor |
| MAF | musculoaponeurotic fibrosarcoma |
References
- Abnet, C.C.; Arnold, M.; Wei, W.Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology 2018, 154, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Nozoe, T.; Oyama, T.; Takenoyama, M.; Hanagiri, T.; Sugio, K.; Yasumoto, K. Significance of immunohistochemical expression of estrogen receptors alpha and beta in squamous cell carcinoma of the esophagus. Clin. Cancer Res. 2007, 13, 4046–4050. [Google Scholar] [CrossRef] [PubMed]
- Bohanes, P.; Yang, D.; Chhibar, R.S.; Labonte, M.J.; Winder, T.; Ning, Y.; Gerger, A.; Benhaim, L.; Paez, D.; Wakatsuki, T.; et al. Influence of sex on the survival of patients with esophageal cancer. J. Clin. Oncol. 2012, 30, 2265–2272. [Google Scholar] [CrossRef]
- Matsuoka, H.; Sugimachi, K.; Ueo, H.; Kuwano, H.; Nakano, S.; Nakayama, M. Sex hormone response of a newly established squamous cell line derived from clinical esophageal carcinoma. Cancer Res. 1987, 47, 4134–4140. [Google Scholar]
- Ueo, H.; Matsuoka, H.; Sugimachi, K.; Kuwano, H.; Mori, M.; Akiyoshi, T. Inhibitory effects of estrogen on the growth of a human esophageal carcinoma cell line. Cancer Res. 1990, 50, 7212–7215. [Google Scholar]
- Wu, S.; Zhang, L.; Deng, J.; Guo, B.; Li, F.; Wang, Y.; Wu, R.; Zhang, S.; Lu, J.; Zhou, Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 2020, 80, 2790–2803. [Google Scholar] [CrossRef]
- Dong, H.; Xu, J.; Li, W.; Gan, J.; Lin, W.; Ke, J.; Jiang, J.; Du, L.; Chen, Y.; Zhong, X.; et al. Reciprocal androgen receptor/interleukin-6 crosstalk drives oesophageal carcinoma progression and contributes to patient prognosis. J. Pathol. 2017, 241, 448–462. [Google Scholar] [CrossRef]
- Huang, F.; Chen, H.; Zhu, X.; Gong, T.; Li, X.; Hankey, W.; Wang, H.; Chen, Z.; Wang, Q.; Liu, Z. The oncogenomic function of androgen receptor in esophageal squamous cell carcinoma is directed by GATA3. Cell Res. 2021, 31, 362–365. [Google Scholar] [CrossRef]
- Wang, Q.; Li, W.; Zhang, Y.; Yuan, X.; Xu, K.; Yu, J.; Chen, Z.; Beroukhim, R.; Wang, H.; Lupien, M.; et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 2009, 138, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Sakari, M.; Okada, M.; Yokoyama, A.; Takahashi, S.; Kouzmenko, A.; Kato, S. The androgen receptor in health and disease. Annu. Rev. Physiol. 2013, 75, 201–224. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Liu, B.; Guan, X. The Role of Tumor Microenvironment in Invasion and Metastasis of Esophageal Squamous Cell Carcinoma. Front. Oncol. 2022, 12, 911285. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lin, C.; Liang, W.; Wu, S.; Liu, A.; Wu, J.; Zhang, X.; Ren, P.; Li, M.; Song, L. TBL1XR1 promotes lymphangiogenesis and lymphatic metastasis in esophageal squamous cell carcinoma. Gut 2015, 64, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Chen, X.; Wu, Y.; Li, J.; Zhang, S.; Wang, K.; Guan, X.; Yang, K.; Bai, Y. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018, 25, 1980–1995. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wu, D.; He, X.; Zhao, H.; He, Z.; Lin, J.; Wang, K.; Wang, W.; Pan, Z.; Lin, H.; et al. circGSK3beta promotes metastasis in esophageal squamous cell carcinoma by augmenting beta-catenin signaling. Mol. Cancer 2019, 18, 160. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Liu, Y.; Zhou, H.; Li, L.; Li, Y.; Ding, F.; Cao, X.; Liu, Z. Deubiquitinating enzyme PSMD14 promotes tumor metastasis through stabilizing SNAIL in human esophageal squamous cell carcinoma. Cancer Lett. 2018, 418, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Y.; Zhu, R.; Ding, F.; Cao, X.; Lin, D.; Liu, Z. OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene 2018, 37, 3356–3368. [Google Scholar] [CrossRef]
- Li, L.; Zhou, H.; Zhu, R.; Liu, Z. USP26 promotes esophageal squamous cell carcinoma metastasis through stabilizing Snail. Cancer Lett. 2019, 448, 52–60. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2019, 35, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Gauthier-Landry, L.; Belanger, A.; Barbier, O. Multiple roles for UDP-glucuronosyltransferase (UGT)2B15 and UGT2B17 enzymes in androgen metabolism and prostate cancer evolution. J. Steroid Biochem. Mol. Biol. 2015, 145, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Meech, R.; McKinnon, R.A.; Mackenzie, P.I. Transcriptional regulation of human UDP-glucuronosyltransferase genes. Drug Metab. Rev. 2014, 46, 421–458. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Fan, D.; Xiong, W.; Li, M.; Yuan, J.; Jiang, Q.; Zhao, Y.; Lin, J.; Liu, J.; Lv, Y.; et al. Oncogenic enhancers drive esophageal squamous cell carcinogenesis and metastasis. Nat. Commun. 2021, 12, 4457. [Google Scholar] [CrossRef] [PubMed]
- Westaby, D.; Fenor de La Maza, M.L.D.; Paschalis, A.; Jimenez-Vacas, J.M.; Welti, J.; de Bono, J.; Sharp, A. A New Old Target: Androgen Receptor Signaling and Advanced Prostate Cancer. Annu. Rev. Pharmacol. Toxicol. 2022, 62, 131–153. [Google Scholar] [CrossRef]
- Lin, D.C.; Koeffler, H.P. Genomic lesions drive the metastasis of esophageal squamous cell carcinoma. J. Thorac. Dis. 2017, 9, 3523–3524. [Google Scholar] [CrossRef]
- Grosse, L.; Paquet, S.; Caron, P.; Fazli, L.; Rennie, P.S.; Belanger, A.; Barbier, O. Androgen glucuronidation: An unexpected target for androgen deprivation therapy. with prognosis and diagnostic implications. Cancer Res. 2013, 73, 6963–6971. [Google Scholar] [CrossRef]
- Chouinard, S.; Barbier, O.; Belanger, A. UDP-glucuronosyltransferase 2B15 (UGT2B15) and UGT2B17 enzymes are major determinants of the androgen response in prostate cancer LNCaP cells. J. Biol. Chem. 2007, 282, 33466–33474. [Google Scholar] [CrossRef]
- Bao, B.Y.; Chuang, B.F.; Wang, Q.; Sartor, O.; Balk, S.P.; Brown, M.; Kantoff, P.W.; Lee, G.S. Androgen receptor mediates the expression of UDP-glucuronosyltransferase 2 B15 and B17 genes. Prostate 2008, 68, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Paquet, S.; Fazli, L.; Grosse, L.; Verreault, M.; Tetu, B.; Rennie, P.S.; Belanger, A.; Barbier, O. Differential expression of the androgen-conjugating UGT2B15 and UGT2B17 enzymes in prostate tumor cells during cancer progression. J. Clin. Endocrinol. Metab. 2012, 97, E428–E432. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jing, Y.; Wang, Y.; Tang, J.; Zhu, X.; Jin, W.L.; Wang, Y.; Yuan, W.; Li, X.; Li, X. NAT10 promotes gastric cancer metastasis via N4-acetylated COL5A1. Signal Transduct. Target. Ther. 2021, 6, 173. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.S.; Yeung, T.L.; Yip, K.P.; Pradeep, S.; Balasubramanian, L.; Liu, J.; Wong, K.K.; Mangala, L.S.; Armaiz-Pena, G.N.; Lopez-Berestein, G.; et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat. Commun. 2014, 5, 5092. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wu, P.; Zhang, Q.; Chen, W.; Liu, X.; Zheng, W. MFAP5 promotes basal-like breast cancer progression by activating the EMT program. Cell Biosci. 2019, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Eferl, R.; Wagner, E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 2003, 3, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Shaulian, E.; Karin, M. AP-1 in cell proliferation and survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef]
- Zhao, C.; Qiao, Y.; Jonsson, P.; Wang, J.; Xu, L.; Rouhi, P.; Sinha, I.; Cao, Y.; Williams, C.; Dahlman-Wright, K. Genome-wide profiling of AP-1-regulated transcription provides insights into the invasiveness of triple-negative breast cancer. Cancer Res. 2014, 74, 3983–3994. [Google Scholar] [CrossRef]
- Ding, X.; Pan, H.; Li, J.; Zhong, Q.; Chen, X.; Dry, S.M.; Wang, C.Y. Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Sci. Signal 2013, 6, ra28. [Google Scholar] [CrossRef]
- Aikawa, Y.; Morimoto, K.; Yamamoto, T.; Chaki, H.; Hashiramoto, A.; Narita, H.; Hirono, S.; Shiozawa, S. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat. Biotechnol. 2008, 26, 817–823. [Google Scholar] [CrossRef]
- Bejjani, F.; Evanno, E.; Zibara, K.; Piechaczyk, M.; Jariel-Encontre, I. The AP-1 transcriptional complex: Local switch or remote command? Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Izuta, S.; Ueki, M.; Ueno, M.; Nishina, K.; Shiozawa, S.; Maekawa, N. T-5224. a selective inhibitor of c-Fos/activator protein-1, attenuates lipopolysaccharide-induced liver injury in mice. Biotechnol. Lett. 2012, 34, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, H.; Morishita, J.; Ueki, M.; Nishina, K.; Shiozawa, S.; Maekawa, N. The effects of a selective inhibitor of c-Fos/activator protein-1 on endotoxin-induced acute kidney injury in mice. BMC Nephrol. 2012, 13, 153. [Google Scholar] [CrossRef] [PubMed]
- Makino, H.; Seki, S.; Yahara, Y.; Shiozawa, S.; Aikawa, Y.; Motomura, H.; Nogami, M.; Watanabe, K.; Sainoh, T.; Ito, H.; et al. A selective inhibition of c-Fos/activator protein-1 as a potential therapeutic target for intervertebral disc degeneration and associated pain. Sci. Rep. 2017, 7, 16983. [Google Scholar] [CrossRef] [PubMed]
- Kamide, D.; Yamashita, T.; Araki, K.; Tomifuji, M.; Tanaka, Y.; Tanaka, S.; Shiozawa, S.; Shiotani, A. Selective activator protein-1 inhibitor T-5224 prevents lymph node metastasis in an oral cancer model. Cancer Sci. 2016, 107, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, M.; Li, Y.; Chang, I.; Yuan, Q.; Ekimyan-Salvo, M.; Deng, P.; Yu, B.; Yu, Y.; Dong, J.; et al. Targeting BMI1(+) Cancer Stem Cells Overcomes Chemoresistance and Inhibits Metastases in Squamous Cell Carcinoma. Cell Stem Cell 2017, 20, 621–634. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, J.; Huang, F.; Gao, W.; Gong, T.; Chen, H.; Liu, Z. Androgen Receptor/AP-1 Activates UGT2B15 Transcription to Promote Esophageal Squamous Cell Carcinoma Invasion. Cancers 2023, 15, 5719. https://doi.org/10.3390/cancers15245719
Cai J, Huang F, Gao W, Gong T, Chen H, Liu Z. Androgen Receptor/AP-1 Activates UGT2B15 Transcription to Promote Esophageal Squamous Cell Carcinoma Invasion. Cancers. 2023; 15(24):5719. https://doi.org/10.3390/cancers15245719
Chicago/Turabian StyleCai, Jiahui, Furong Huang, Wenyan Gao, Tongyang Gong, Hongyan Chen, and Zhihua Liu. 2023. "Androgen Receptor/AP-1 Activates UGT2B15 Transcription to Promote Esophageal Squamous Cell Carcinoma Invasion" Cancers 15, no. 24: 5719. https://doi.org/10.3390/cancers15245719
APA StyleCai, J., Huang, F., Gao, W., Gong, T., Chen, H., & Liu, Z. (2023). Androgen Receptor/AP-1 Activates UGT2B15 Transcription to Promote Esophageal Squamous Cell Carcinoma Invasion. Cancers, 15(24), 5719. https://doi.org/10.3390/cancers15245719





