HSP110 Inhibition in Primary Effusion Lymphoma Cells: One Molecule, Many Pro-Survival Targets
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Treatments
2.2. HSP110 Silencing
2.3. Trypan Blue Exclusion Assay
2.4. Cell Cycle Analysis
2.5. Western Blot Analysis
2.6. Antibodies
2.7. Acridine Orange Staining
2.8. Immunofluorescence
2.9. Densitometric and Statistical Analysis
3. Results
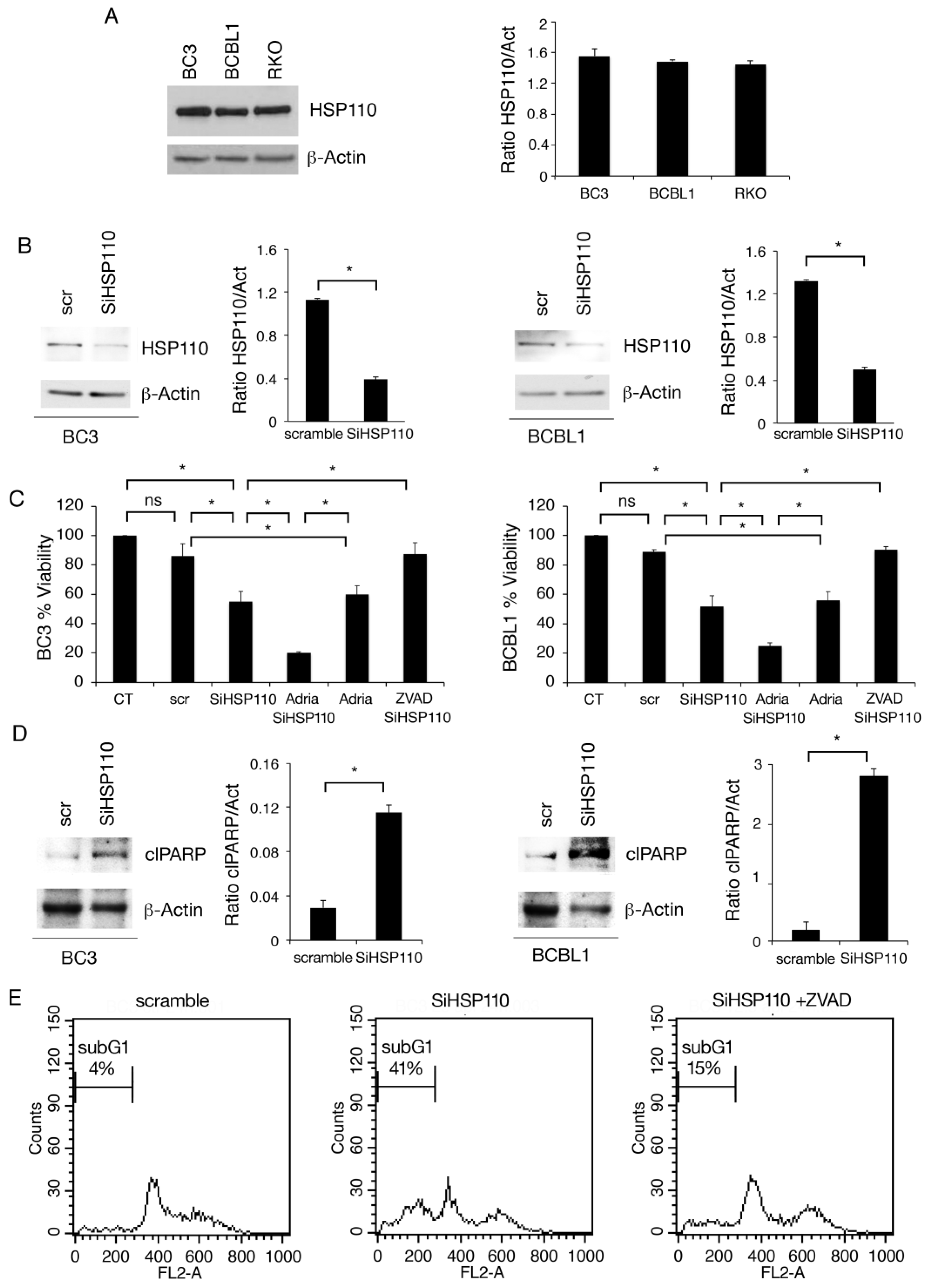
3.1. HSP110 Is Highly Expressed in PEL Cells and Sustains Cell Survival
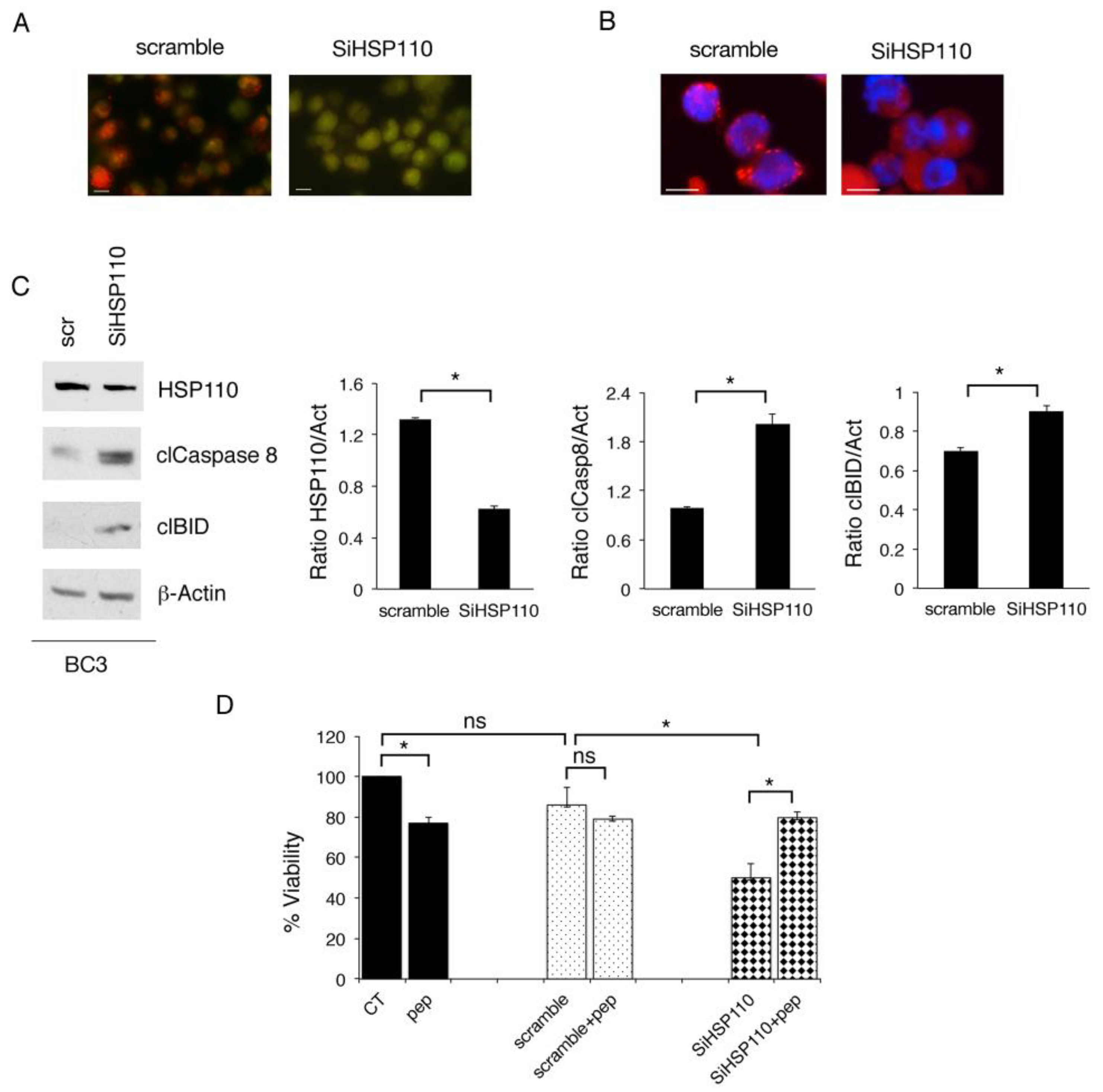
3.2. Lysosomal Permeabilization Contributes to the Cytotoxic Effect of HSP110 Inhibition
3.3. HSP110 Knockdown Induces DNA Damage by Impairing HR and NHEJ DNA Repair, an Effect That Is Also Mediated by the Release of Lysosomal Cathepsins
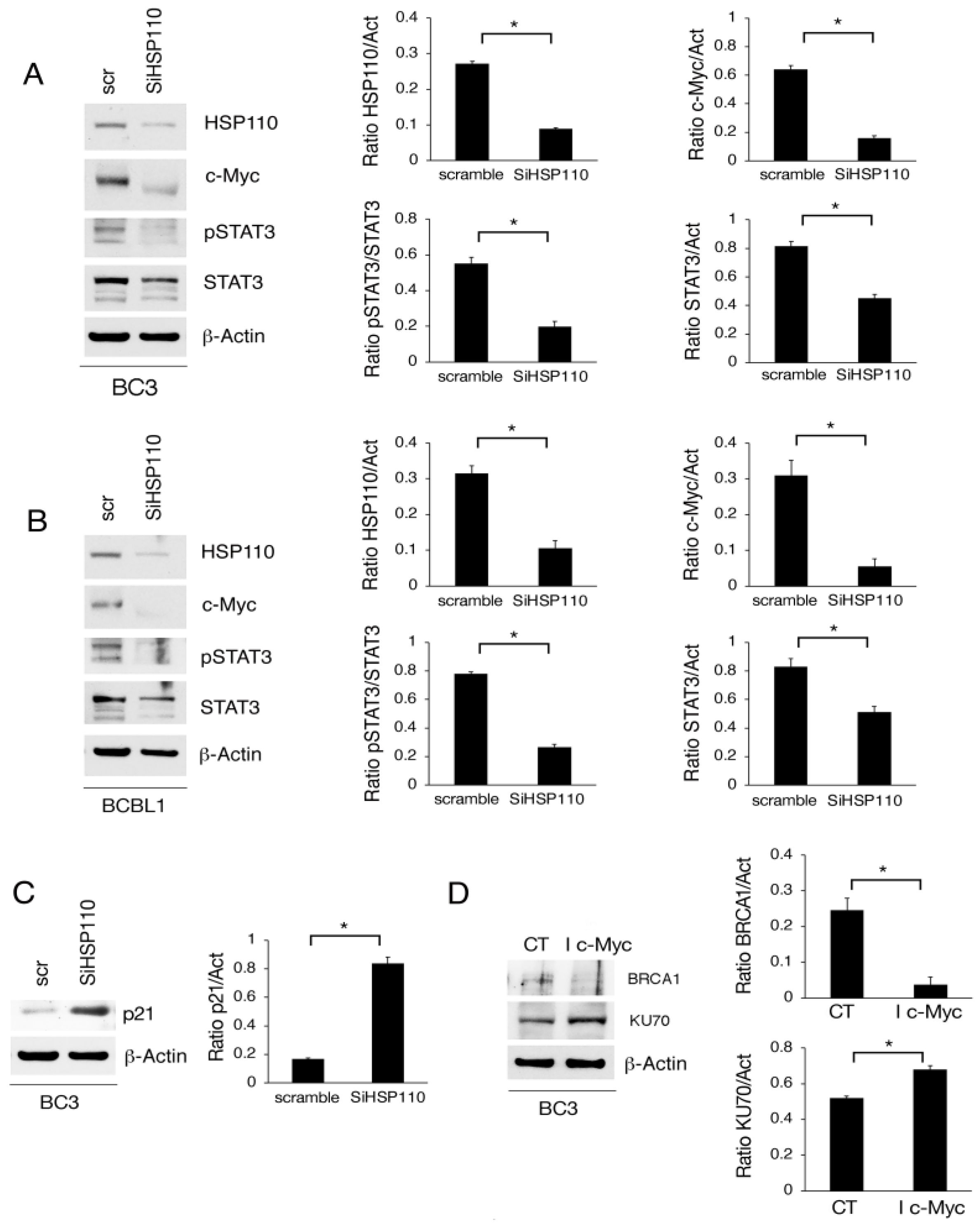
3.4. Myc Downregulation via HSP110 Silencing Contributes to wtp53 Activation and DNA Damage Induction in PEL Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, S.; Xiao, H.; Cao, L. Recent advances in heat shock proteins in cancer diagnosis, prognosis, metabolism and treatment. Biomed. Pharmacother. 2021, 142, 112074. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xie, S. Therapeutic targeting of cellular stress responses in cancer. Thorac. Cancer 2018, 9, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Briones, J. Heat-shock proteins: A c-Myc lymphoma target? Blood 2015, 125, 1685–1686. [Google Scholar] [CrossRef][Green Version]
- Alexandrova, E.M.; Marchenko, N.D. Mutant p53—Heat Shock Response Oncogenic Cooperation: A New Mechanism of Cancer Cell Survival. Front. Endocrinol. 2015, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Gilardini Montani, M.S.; Cecere, N.; Granato, M.; Romeo, M.A.; Falcinelli, L.; Ciciarelli, U.; D’Orazi, G.; Faggioni, A.; Cirone, M. Mutant p53, Stabilized by Its Interplay with HSP90, Activates a Positive Feed-Back Loop Between NRF2 and p62 that Induces Chemo-Resistance to Apigenin in Pancreatic Cancer Cells. Cancers 2019, 11, 703. [Google Scholar] [CrossRef] [PubMed]
- Boudesco, C.; Verhoeyen, E.; Martin, L.; Chassagne-Clement, C.; Salmi, L.; Mhaidly, R.; Pangault, C.; Fest, T.; Ramla, S.; Jardin, F.; et al. HSP110 sustains chronic NF-kappaB signaling in activated B-cell diffuse large B-cell lymphoma through MyD88 stabilization. Blood 2018, 132, 510–520. [Google Scholar] [CrossRef]
- Jego, G.; Hermetet, F.; Girodon, F.; Garrido, C. Chaperoning STAT3/5 by Heat Shock Proteins: Interest of Their Targeting in Cancer Therapy. Cancers 2019, 12, 21. [Google Scholar] [CrossRef]
- Sottile, M.L.; Nadin, S.B. Heat shock proteins and DNA repair mechanisms: An updated overview. Cell Stress Chaperones 2018, 23, 303–315. [Google Scholar] [CrossRef]
- Gonnella, R.; Arena, A.; Zarrella, R.; Gilardini Montani, M.S.; Santarelli, R.; Cirone, M. HSPs/STAT3 Interplay Sustains DDR and Promotes Cytokine Release by Primary Effusion Lymphoma Cells. Int. J. Mol. Sci. 2023, 24, 3933. [Google Scholar] [CrossRef]
- Aoki, Y.; Feldman, G.M.; Tosato, G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood 2003, 101, 1535–1542. [Google Scholar] [CrossRef]
- Tolani, B.; Gopalakrishnan, R.; Punj, V.; Matta, H.; Chaudhary, P.M. Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene 2014, 33, 2928–2937. [Google Scholar] [CrossRef] [PubMed]
- Arena, A.; Romeo, M.A.; Benedetti, R.; Gilardini Montani, M.S.; Cirone, M. JQ-1/bortezomib combination strongly impairs MM and PEL survival by inhibiting c-Myc and mTOR despite the activation of prosurvival mechanisms. Exp. Hematol. 2023, 119–120, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Barre, B.; Vigneron, A.; Coqueret, O. The STAT3 transcription factor is a target for the Myc and riboblastoma proteins on the Cdc25A promoter. J. Biol. Chem. 2005, 280, 15673–15681. [Google Scholar] [CrossRef] [PubMed]
- Bowman, T.; Broome, M.A.; Sinibaldi, D.; Wharton, W.; Pledger, W.J.; Sedivy, J.M.; Irby, R.; Yeatman, T.; Courtneidge, S.A.; Jove, R. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. USA 2001, 98, 7319–7324. [Google Scholar] [CrossRef] [PubMed]
- Jurkovicova, D.; Neophytou, C.M.; Gasparovic, A.C.; Goncalves, A.C. DNA Damage Response in Cancer Therapy and Resistance: Challenges and Opportunities. Int. J. Mol. Sci. 2022, 23, 14672. [Google Scholar] [CrossRef] [PubMed]
- McCarthy-Leo, C.; Darwiche, F.; Tainsky, M.A. DNA Repair Mechanisms, Protein Interactions and Therapeutic Targeting of the MRN Complex. Cancers 2022, 14, 5278. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Pan, W.; Xing, Y.; Xiao, Y.; Chen, J.; Xu, Z. Recent advances in DDR (DNA damage response) inhibitors for cancer therapy. Eur. J. Med. Chem. 2022, 230, 114109. [Google Scholar] [CrossRef] [PubMed]
- Chakafana, G.; Shonhai, A. The Role of Non-Canonical Hsp70s (Hsp110/Grp170) in Cancer. Cells 2021, 10, 254. [Google Scholar] [CrossRef]
- Zappasodi, R.; Ruggiero, G.; Guarnotta, C.; Tortoreto, M.; Tringali, C.; Cavane, A.; Cabras, A.D.; Castagnoli, L.; Venerando, B.; Zaffaroni, N.; et al. HSPH1 inhibition downregulates Bcl-6 and c-Myc and hampers the growth of human aggressive B-cell non-Hodgkin lymphoma. Blood 2015, 125, 1768–1771. [Google Scholar] [CrossRef]
- Causse, S.Z.; Marcion, G.; Chanteloup, G.; Uyanik, B.; Boudesco, C.; Grigorash, B.B.; Douhard, R.; Dias, A.M.M.; Dumetier, B.; Dondaine, L.; et al. HSP110 translocates to the nucleus upon genotoxic chemotherapy and promotes DNA repair in colorectal cancer cells. Oncogene 2019, 38, 2767–2777. [Google Scholar] [CrossRef]
- Arena, A.; Romeo, M.A.; Benedetti, R.; Gilardini Montani, M.S.; Cirone, M. Targeting c-Myc Unbalances UPR towards Cell Death and Impairs DDR in Lymphoma and Multiple Myeloma Cells. Biomedicines 2022, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Granato, M.; Lacconi, V.; Peddis, M.; Lotti, L.V.; Di Renzo, L.; Gonnella, R.; Santarelli, R.; Trivedi, P.; Frati, L.; D’Orazi, G.; et al. HSP70 inhibition by 2-phenylethynesulfonamide induces lysosomal cathepsin D release and immunogenic cell death in primary effusion lymphoma. Cell Death Dis. 2013, 4, e730. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Suarez, I.; Redwood, A.B.; Grotsky, D.A.; Neumann, M.A.; Cheng, E.H.; Stewart, C.L.; Dusso, A.; Gonzalo, S. A new pathway that regulates 53BP1 stability implicates cathepsin L and vitamin D in DNA repair. EMBO J. 2011, 30, 3383–3396. [Google Scholar] [CrossRef] [PubMed]
- Arena, A.; Gilardini Montani, M.S.; Romeo, M.A.; Benedetti, R.; Gaeta, A.; Cirone, M. DNA damage triggers an interplay between wtp53 and c-Myc affecting lymphoma cell proliferation and Kaposi sarcoma herpesvirus replication. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119168. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.N.; Luo, Y. HSP90 inhibitors and cancer: Prospects for use in targeted therapies (Review). Oncol. Rep. 2023, 49, 6. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Lurain, K.; Yarchoan, R.; Ramaswami, R. Clinical management of Kaposi sarcoma herpesvirus-associated diseases: An update on disease manifestations and treatment strategies. Expert Rev. Anti Infect. Ther. 2023, 21, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef]
- Eriksson, I.; Vainikka, L.; Persson, H.L.; Ollinger, K. Real-Time Monitoring of Lysosomal Membrane Permeabilization Using Acridine Orange. Methods Protoc. 2023, 6, 72. [Google Scholar] [CrossRef]
- Berthenet, K.; Bokhari, A.; Lagrange, A.; Marcion, G.; Boudesco, C.; Causse, S.; De Thonel, A.; Svrcek, M.; Goloudina, A.R.; Dumont, S.; et al. HSP110 promotes colorectal cancer growth through STAT3 activation. Oncogene 2017, 36, 2328–2336. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonnella, R.; Zarrella, R.; Di Crosta, M.; Benedetti, R.; Arena, A.; Santarelli, R.; Gilardini Montani, M.S.; D’Orazi, G.; Cirone, M. HSP110 Inhibition in Primary Effusion Lymphoma Cells: One Molecule, Many Pro-Survival Targets. Cancers 2023, 15, 5651. https://doi.org/10.3390/cancers15235651
Gonnella R, Zarrella R, Di Crosta M, Benedetti R, Arena A, Santarelli R, Gilardini Montani MS, D’Orazi G, Cirone M. HSP110 Inhibition in Primary Effusion Lymphoma Cells: One Molecule, Many Pro-Survival Targets. Cancers. 2023; 15(23):5651. https://doi.org/10.3390/cancers15235651
Chicago/Turabian StyleGonnella, Roberta, Roberta Zarrella, Michele Di Crosta, Rossella Benedetti, Andrea Arena, Roberta Santarelli, Maria Saveria Gilardini Montani, Gabriella D’Orazi, and Mara Cirone. 2023. "HSP110 Inhibition in Primary Effusion Lymphoma Cells: One Molecule, Many Pro-Survival Targets" Cancers 15, no. 23: 5651. https://doi.org/10.3390/cancers15235651
APA StyleGonnella, R., Zarrella, R., Di Crosta, M., Benedetti, R., Arena, A., Santarelli, R., Gilardini Montani, M. S., D’Orazi, G., & Cirone, M. (2023). HSP110 Inhibition in Primary Effusion Lymphoma Cells: One Molecule, Many Pro-Survival Targets. Cancers, 15(23), 5651. https://doi.org/10.3390/cancers15235651






