Comprehensive Transcriptome Analysis Reveals the Distinct Gene Expression Patterns of Tumor Microenvironment in HPV-Associated and HPV-Non Associated Tonsillar Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Participant Characteristics
2.2. Ethical Considerations
2.3. Simultaneous Isolation of Viral DNA and Total RNA
2.4. HPV Detection, Genotyping, and HPV E Oncogenes Integration Detection
2.5. Cellular RNA Processing and Cancer Inflammation, Immunity Crosstalk, Oncogenes, and Tumor Suppressor Genes Transcriptomes
2.6. Data Analysis
2.7. Protein–Protein Interaction Network and Hub Protein Analysis
3. Results
3.1. Basic TSCC Patient’s Characteristics
3.2. Prevalence of HPV in Tonsil Squamous Cell Carcinoma (TSCC)
3.3. Cancer Inflammation and Immunity Crosstalk Gene Expression Analysis of TME in HPV-Associated and Non-Associated TSCC
3.4. Gene Expression Analysis of Oncogenes and Tumor Suppressor Genes of TME in HPV-Associated and Non-Associated TSCC
3.5. No Association between the Most Up-Regulated Gene Expression and Age
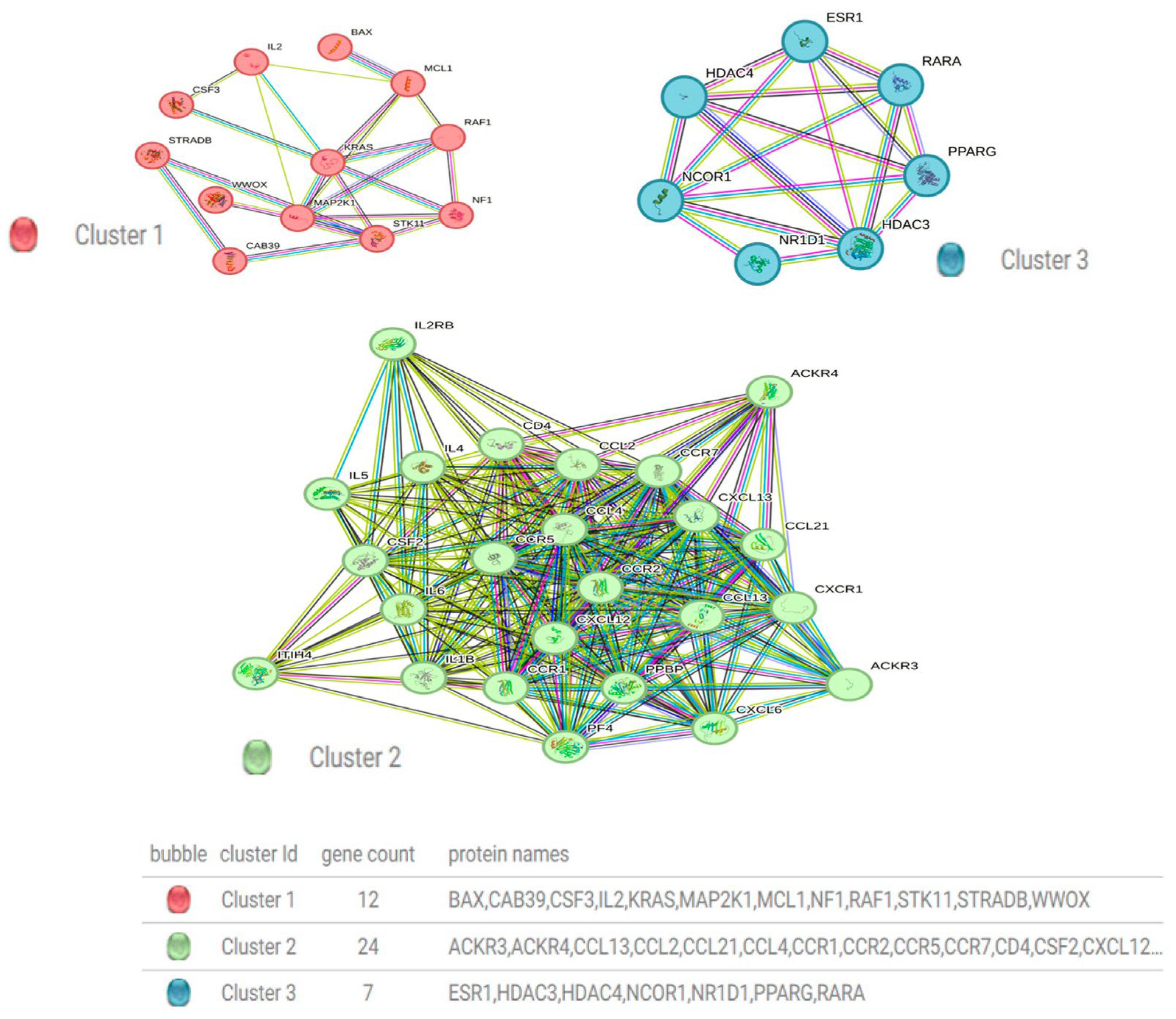
3.6. Construction of a Protein–Protein Interaction Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef]
- Global Cancer Observatory (GLOBOCAN). Available online: https://gco.iarc.fr/today/home (accessed on 11 September 2023).
- Alqahtani, W.S.; Almufareh, N.A.; Domiaty, D.M.; Albasher, G.; Alduwish, M.A.; Alkhalaf, H.; Almuzzaini, B.; Al-Marshidy, S.S.; Alfraihi, R.; Elasbali, A.M.; et al. Epidemiology of cancer in Saudi Arabia thru 2010–2019: A systematic review with constrained meta-analysis. AIMS Public Health 2020, 7, 679–696. (In English) [Google Scholar] [CrossRef] [PubMed]
- Available online: https://gco.iarc.fr/today/data/factsheets/populations/682-saudi-arabia-fact-sheets.pdf (accessed on 14 September 2023).
- Elkashty, O.A.; Elghanam, G.A.; Su, X.; Liu, Y.; Chauvin, P.J.; Tran, S.D. Cancer stem cells enrichment with surface markers CD271 and CD44 in human head and neck squamous cell carcinomas. Carcinogenesis 2020, 41, 458–466. (In English) [Google Scholar] [CrossRef] [PubMed]
- Huo, M.; Zhang, Y.; Chen, Z.; Zhang, S.; Bao, Y.; Li, T. Tumor microenvironment characterization in head and neck cancer identifies prognostic and immunotherapeutically relevant gene signatures. Sci. Rep. 2020, 10, 11163. [Google Scholar] [CrossRef] [PubMed]
- De Martin, A.; Lütge, M.; Stanossek, Y.; Engetschwiler, C.; Cupovic, J.; Brown, K.; Demmer, I.; Broglie, M.A.; Geuking, M.B.; Jochum, W.; et al. Distinct microbial communities colonize tonsillar squamous cell carcinoma. OncoImmunology 2021, 10, 1945202. [Google Scholar] [CrossRef] [PubMed]
- Jiarpinitnun, C.; Larbcharoensub, N.; Pattaranutaporn, P.; Chureemas, T.; Juengsamarn, J.; Trachu, N.; Lukerak, S.; Chansriwong, P.; Ngamphaiboon, N. Characteristics and Impact of HPV-Associated p16 Expression on Head and Neck Squamous Cell Carcinoma in Thai Patients. Asian Pac. J. Cancer Prev. 2020, 21, 1679–1687. (In English) [Google Scholar] [CrossRef]
- Xia, C.; Li, S.; Long, T.; Chen, Z.; Chan, P.K.S.; Boon, S.S. Current Updates on Cancer-Causing Types of Human Papillomaviruses (HPVs) in East, Southeast, and South Asia. Cancers 2021, 13, 2691. (In English) [Google Scholar] [CrossRef]
- Gormley, M.; Creaney, G.; Schache, A.; Ingarfield, K.; Conway, D.I. Reviewing the epidemiology of head and neck cancer: Definitions, trends and risk factors. Br. Dent. J. 2022, 233, 780–786. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. (In English) [Google Scholar] [CrossRef]
- Basukala, O.; Banks, L. The Not-So-Good, the Bad and the Ugly: HPV E5, E6 and E7 Oncoproteins in the Orchestration of Carcinogenesis. Viruses 2021, 13, 1892. (In English) [Google Scholar] [CrossRef]
- Pal, A.; Kundu, R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front. Microbiol. 2020, 10, 3116. [Google Scholar] [CrossRef] [PubMed]
- Skelin, J.; Sabol, I.; Tomaic, V. Do or Die: HPV E5, E6 and E7 in Cell Death Evasion. Pathogens 2022, 11, 1027. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Qiu, Q.; Zhou, Q.; Li, J.; Yu, M.; Li, K.; Xu, L.; Ke, X.; Xu, H.; Lu, B.; et al. Long-read sequencing unveils high-resolution HPV integration and its oncogenic progression in cervical cancer. Nat. Commun. 2022, 13, 2563. [Google Scholar] [CrossRef]
- Lee, D.J.; Kwon, M.J.; Nam, E.S.; Kwon, J.H.; Kim, J.H.; Rho, Y.-S.; Shin, H.S.; Cho, S.J. Histopathologic predictors of lymph node metastasis and prognosis in tonsillar squamous cell carcinoma. Korean J. Pathol. 2013, 47, 203–210. (In English) [Google Scholar] [CrossRef]
- Park, H.Y.; Lee, J.S.; Wee, J.H.; Kang, J.W.; Kim, E.S.; Koo, T.; Hwang, H.S.; Kim, H.J.; Kang, H.S.; Lim, H.; et al. Assessment of the Mutation Profile of Tonsillar Squamous Cell Carcinomas Using Targeted Next-Generation Sequencing. Biomedicines 2023, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Elmusrati, A.; Wang, J.; Wang, C.-Y. Tumor microenvironment and immune evasion in head and neck squamous cell carcinoma. Int. J. Oral Sci. 2021, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Höchst, B.; Knolle, P.A. Checkpoint Inhibition in Head and Neck Cancer: Immune Therapeutic Options, Limitations, and beyond. ORL 2017, 79, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Du, Y.; Fu, D. Characterization of tumor immune microenvironment and cancer therapy for head and neck squamous cell carcinoma through identification of a genomic instability-related lncRNA prognostic signature. Front. Genet. 2022, 13, 979575. (In English) [Google Scholar] [CrossRef] [PubMed]
- Peltanova, B.; Raudenska, M.; Masarik, M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: A systematic review. Mol. Cancer 2019, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Curry, J.M.; Sprandio, J.; Cognetti, D.; Luginbuhl, A.; Bar-Ad, V.; Pribitkin, E.; Tuluc, M. Tumor Microenvironment in Head and Neck Squamous Cell Carcinoma. Semin. Oncol. 2014, 41, 217–234. [Google Scholar] [CrossRef]
- Mito, I.; Takahashi, H.; Kawabata-Iwakawa, R.; Ida, S.; Tada, H.; Chikamatsu, K. Comprehensive analysis of immune cell enrichment in the tumor microenvironment of head and neck squamous cell carcinoma. Sci. Rep. 2021, 11, 16134. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, R.; Chao, Y. Deciphering Innate Immune Cell-Tumor Microenvironment Crosstalk at a Single-Cell Level. Front. Cell Dev. Biol. 2022, 10, 803947. [Google Scholar] [CrossRef] [PubMed]
- Galli, F.; Aguilera, J.V.; Palermo, B.; Markovic, S.N.; Nisticò, P.; Signore, A. Relevance of immune cell and tumor microenvironment imaging in the new era of immunotherapy. J. Exp. Clin. Cancer Res. 2020, 39, 89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fei, Y.; Wang, H.; Hu, S.; Liu, C.; Hu, R.; Du, Q. CAFs orchestrates tumor immune microenvironment—A new target in cancer therapy? Front. Pharmacol. 2023, 14, 1113378. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Huang, Q.; Deng, Y.; Guo, F.; Wang, G.; Liu, M. Crosstalk Between the Tumor Microenvironment and Cancer Cells: A Promising Predictive Biomarker for Immune Checkpoint Inhibitors. Front. Cell Dev. Biol. 2021, 9, 738373. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. (In English) [Google Scholar] [CrossRef]
- Elinav, E.; Nowarski, R.; Thaiss, C.A.; Hu, B.; Jin, C.; Flavell, R.A. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 2013, 13, 759–771. (In English) [Google Scholar] [CrossRef]
- Lee, E.Y.; Muller, W.J. Oncogenes and tumor suppressor genes. Cold Spring Harb. Perspect. Biol. 2010, 2, a003236. (In English) [Google Scholar] [CrossRef]
- Barros-Filho, M.C.; Guisier, F.; Rock, L.D.; Becker-Santos, D.D.; Sage, A.P.; Marshall, E.A.; Lam, W.L. Tumour Suppressor Genes with Oncogenic Roles in Lung Cancer. In Genes and Cancer; Guy-Joseph, L., Ed.; IntechOpen: Rijeka, Croatia, 2019; p. Ch. 3. [Google Scholar]
- Alosaimi, B.; Hamed, M.E.; Naeem, A.; Alsharef, A.A.; AlQahtani, S.Y.; AlDosari, K.M.; Alamri, A.A.; Al-Eisa, K.; Khojah, T.; Assiri, A.M.; et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine 2020, 126, 154895. (In English) [Google Scholar] [CrossRef]
- Yu, X.; Wang, Z.; Zeng, T. Essential gene expression pattern of head and neck squamous cell carcinoma revealed by tumor-specific expression rule based on single-cell RNA sequencing. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165791. [Google Scholar] [CrossRef] [PubMed]
- Knebel, M.; Körner, S.; Kühn, J.P.; Wemmert, S.; Brust, L.; Smola, S.; Wagner, M.; Bohle, R.M.; Morris, L.G.T.; Pandey, A.; et al. Prognostic impact of intra- and peritumoral immune cell subpopulations in head and neck squamous cell carcinomas—Comprehensive analysis of the TCGA-HNSC cohort and immunohistochemical validation on 101 patients. Front. Immunol. 2023, 14, 1172768. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yuan, C.; Ma, J.; Pan, Y.; Xu, H. Influence of Immune Microenvironment on Diagnosis and Prognosis of Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 604784. (In English) [Google Scholar] [CrossRef] [PubMed]
- Qiang, W.; Dai, Y.; Xing, X.; Sun, X. Identification and validation of a prognostic signature and combination drug therapy for immunotherapy of head and neck squamous cell carcinoma. Comput. Struct. Biotechnol. J. 2021, 19, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; Ye, Z.; Sun, B.; Xiao, Z. Molecular Evolutionary Landscape of the Immune Microenvironment of Head and Neck Cancer. Biomolecules 2023, 13, 1120. [Google Scholar] [CrossRef]
- Hanna, G.J.; Lizotte, P.; Cavanaugh, M.; Kuo, F.C.; Shivdasani, P.; Frieden, A.; Chau, N.G.; Schoenfeld, J.D.; Lorch, J.H.; Uppaluri, R.; et al. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. J. Clin. Investig. 2018, 3, e98811. (In English) [Google Scholar] [CrossRef]
- Saleh, K.; Eid, R.; Haddad, F.G.H.; Khalife-Saleh, N.; Kourie, H.R. New developments in the management of head and neck cancer—Impact of pembrolizumab. Ther. Clin. Risk Manag. 2018, 14, 295–303. [Google Scholar] [CrossRef]
- Erreni, M.; Bianchi, P.; Laghi, L.; Mirolo, M.; Fabbri, M.; Locati, M.; Mantovani, A.; Allavena, P. Expression of chemokines and chemokine receptors in human colon cancer. Methods Enzymol. 2009, 460, 105–121. (In English) [Google Scholar] [CrossRef]
- Sadeghi, M.; Lahdou, I.; Oweira, H.; Daniel, V.; Terness, P.; Schmidt, J.; Weiss, K.-H.; Longerich, T.; Schemmer, P.; Opelz, G.; et al. Serum levels of chemokines CCL4 and CCL5 in cirrhotic patients indicate the presence of hepatocellular carcinoma. Br. J. Cancer 2015, 113, 756–762. (In English) [Google Scholar] [CrossRef]
- Saito, S.; Kitayama, J.; Jin, Z.X.; Tsuno, N.; Kaisaki, S.; Seto, Y.; Nagawa, H. Beta-chemokine, macrophage inflammatory protein-1beta (MIP-1beta), is highly expressed in diffuse type human gastric cancers. J. Exp. Clin. Cancer Res. 2003, 22, 453–459. (In English) [Google Scholar]
- Mukaida, N.; Sasaki, S.I.; Baba, T. CCL4 Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1231, 23–32. (In English) [Google Scholar] [CrossRef] [PubMed]
- Lien, M.Y.; Tsai, H.-C.; Chang, A.-C.; Tsai, M.-H.; Hua, C.-H.; Wang, S.-W.; Tang, C.-H. Chemokine CCL4 Induces Vascular Endothelial Growth Factor C Expression and Lymphangiogenesis by miR-195-3p in Oral Squamous Cell Carcinoma. Front. Immunol. 2018, 9, 412. (In English) [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Ma, L.; Jiang, C.; Zhang, S. Expression and potential role of CCL4 in CD8+T cells in NSCLC. Clin. Transl. Oncol. 2022, 24, 2420–2431. (In English) [Google Scholar] [CrossRef] [PubMed]
- Carla, C.; Daris, F.; Cecilia, B.; Francesca, B.; Francesca, C.; Paolo, F. Angiogenesis in Head and Neck Cancer: A Review of the Literature. J. Oncol. 2012, 2012, 358472. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vassilakopoulou, M.; Psyrri, A.; Argiris, A. Targeting angiogenesis in head and neck cancer. Oral Oncol. 2015, 51, 409–415. [Google Scholar] [CrossRef]
- Micaily, I.; Johnson, J.; Argiris, A. An update on angiogenesis targeting in head and neck squamous cell carcinoma. Cancers Head Neck 2020, 5, 5. [Google Scholar] [CrossRef]
- Wilde, L.; Johnson, J.; Argiris, A. Angiogenesis and Anti-angiogenic Therapy in Head and Neck Cancer. In Molecular Determinants of Head and Neck Cancer; Burtness, B., Golemis, E.A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 439–467. [Google Scholar]
- Hwang, K.T.; Woo, J.W.; Shin, H.C.; Kim, H.S.; Ahn, S.K.; Moon, H.-G.; Han, W.; Park, I.A.; Noh, D.-Y. Prognostic influence of BCL2 expression in breast cancer. Int. J. Cancer 2012, 131, E1109–E1119. [Google Scholar] [CrossRef]
- Pereira, H.; Silva, S.; Julião, R.; Garcia, P.; Perpétua, F. Prognostic markers for colorectal cancer: Expression of P53 and BCL2. World J. Surg. 1997, 21, 210–213. [Google Scholar] [CrossRef]
- Callagy, G.M.; Webber, M.J.; Pharoah, P.D.; Caldas, C. Meta-analysis confirms BCL2 is an independent prognostic marker in breast cancer. BMC Cancer 2008, 8, 153. (In English) [Google Scholar] [CrossRef]
- Eom, Y.H.; Kim, H.S.; Lee, A.; Song, B.J.; Chae, B.J. BCL2 as a Subtype-Specific Prognostic Marker for Breast Cancer. J. Breast Cancer 2016, 19, 252–260. (In English) [Google Scholar] [CrossRef]
- Liang, Y.K.; Deng, Z.-K.; Chen, M.-T.; Qiu, S.-Q.; Xiao, Y.-S.; Qi, Y.-Z.; Xie, Q.; Wang, Z.-H.; Jia, S.-C.; Zeng, D.; et al. CXCL9 Is a Potential Biomarker of Immune Infiltration Associated with Favorable Prognosis in ER-Negative Breast Cancer. Front. Oncol. 2021, 11, 710286. (In English) [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Su, X.M.; Ke, L.N.; Huang, Y.G. CXCL9 correlates with antitumor immunity and is predictive of a favorable prognosis in uterine corpus endometrial carcinoma. Front. Oncol. 2023, 13, 1077780. (In English) [Google Scholar] [CrossRef] [PubMed]
- Kowalczuk, O.; Burzykowski, T.; Niklinska, W.E.; Kozlowski, M.; Chyczewski, L.; Niklinski, J. CXCL5 as a potential novel prognostic factor in early stage non-small cell lung cancer: Results of a study of expression levels of 23 genes. Tumor Biol. 2014, 35, 4619–4628. (In English) [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Ma, Y.B.; Lv, J.; Maimaiti, N.B.; Zhang, J.B.; Aibibula, M.B.; Gong, Z.; Ling, B. Expression of chemokine CXCL8/9/10/11/13 and its prognostic significance in head and neck cancer. Medicine 2022, 101, e29378. (In English) [Google Scholar] [CrossRef] [PubMed]
- Saintigny, P.; Massarelli, E.; Lin, S.; Ahn, Y.-H.; Chen, Y.; Goswami, S.; Erez, B.; O’Reilly, M.S.; Liu, D.; Lee, J.J.; et al. CXCR2 Expression in Tumor Cells Is a Poor Prognostic Factor and Promotes Invasion and Metastasis in Lung Adenocarcinoma. Cancer Res. 2013, 73, 571–582. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, C.; Cao, Y.; Li, Q.; Deng, H.; Gu, S.; Wu, Y.; Shen, Z. Expression profile and prognostic value of CXCR family members in head and neck squamous cell carcinoma. World J. Surg. Oncol. 2022, 20, 259. (In English) [Google Scholar] [CrossRef]
- Terabe, M.; Park, J.M.; Berzofsky, J.A. Role of IL-13 in regulation of anti-tumor immunity and tumor growth. Cancer Immunol. Immunother. 2004, 53, 79–85. [Google Scholar] [CrossRef]
- Bernstein, Z.J.; Shenoy, A.; Chen, A.; Heller, N.M.; Spangler, J.B. Engineering the IL-4/IL-13 axis for targeted immune modulation. Immunol. Rev. 2023. [Google Scholar] [CrossRef]
- Zhou, R.; Qian, S.; Gu, X.; Chen, Z.; Xiang, J. Interleukin-13 and its receptors in colorectal cancer (Review). Biomed. Rep. 2013, 1, 687–690. (In English) [Google Scholar] [CrossRef][Green Version]
- Saunders, A.S.; Bender, D.E.; Ray, A.L.; Wu, X.; Morris, K.T. Colony-stimulating factor 3 signaling in colon and rectal cancers: Immune response and CMS classification in TCGA data. PLoS ONE 2021, 16, e0247233. (In English) [Google Scholar] [CrossRef]
- Liu, X.; Liu, P.; Chernock, R.D.; Kuhs, K.A.L.; Lewis, J.S.; Li, H.; Gay, H.A.; Thorstad, W.L.; Wang, X. Impact of human papillomavirus on the tumor microenvironment in oropharyngeal squamous cell carcinoma. Int. J. Cancer 2022, 150, 521–531. (In English) [Google Scholar] [CrossRef] [PubMed]
- Tosi, A.; Parisatto, B.; Menegaldo, A.; Spinato, G.; Guido, M.; Del Mistro, A.; Bussani, R.; Zanconati, F.; Tofanelli, M.; Tirelli, G.; et al. The immune microenvironment of HPV-positive and HPV-negative oropharyngeal squamous cell carcinoma: A multiparametric quantitative and spatial analysis unveils a rationale to target treatment-naïve tumors with immune checkpoint inhibitors. J. Exp. Clin. Cancer Res. 2022, 41, 279. (In English) [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Birkeland, A.C.; Ludwig, M.L.; Spector, M.E.; Brenner, J.C. The potential for tumor suppressor gene therapy in head and neck cancer. Discov. Med. 2016, 21, 41–47. (In English) [Google Scholar] [PubMed]
- Sanchez-Martin, A.; Sanchon-Sanchez, P.; Romero, M.R.; Marin, J.J.G.; Briz, O. Impact of tumor suppressor genes inactivation on the multidrug resistance phenotype of hepatocellular carcinoma cells. Biomed. Pharmacother. 2023, 165, 115209. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Ramasubbu, K.; Banerjee, M.; Balaji, M.P.; Vinayagam, Y. Devi Rajeswari VA systematic review on the molecular and clinical association between Human Papillomavirus and Human Immunodeficiency Virus co-infection in Head, Neck and Oral squamous cell carcinoma. Rev. Med. Virol. 2023, 33, e2462. [Google Scholar] [CrossRef]
- Pazhani, J.; Veeraraghavan, V.P.; Jayaraman, S. Transcription factors: A potential therapeutic target in head and neck squamous cell carcinoma. Epigenomics 2023, 15, 57–60. [Google Scholar] [CrossRef]
- Qin, C.; Liu, S.; Zhou, S.; Wang, Q.; Xia, X.; Hu, J.; Yuan, X.; Wang, Z.; Yu, Y.; Ma, D. PIK3C2A is a prognostic biomarker that is linked to immune infiltrates in kidney renal clear cell carcinoma. Front. Immunol. 2023, 14, 1114572. (In English) [Google Scholar] [CrossRef]
- Chu, C.-A.; Wang, Y.-W.; Chen, Y.-L.; Chen, H.-W.; Chuang, J.-J.; Chang, H.-Y.; Ho, C.-L.; Chang, C.; Chow, N.-H.; Lee, C.-T. The Role of Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 in the Pathogenesis of Human Cancer. Int. J. Mol. Sci. 2021, 22, 10964. [Google Scholar] [CrossRef]
- Huang, J.; Manning, B.D. The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem. J. 2008, 412, 179–190. (In English) [Google Scholar] [CrossRef]
- Kwiatkowski, D.J.; Choueiri, T.K.; Fay, A.P.; Rini, B.I.; Thorner, A.R.; de Velasco, G.; Tyburczy, M.E.; Hamieh, L.; Albiges, L.; Agarwal, N.; et al. Mutations in TSC1, TSC2, and MTOR Are Associated with Response to Rapalogs in Patients with Metastatic Renal Cell Carcinoma. Clin. Cancer Res. 2016, 22, 2445–2452. (In English) [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, R.; Zhao, J.; Zhu, L.; Xu, J.; Ma, F.; Guo, R.; Xu, C.; Wang, W.-X.; Yi, X.; et al. Mutation profiling of TSC1 and TSC2 genes in solid tumors using comprehensive NGS panel. J. Clin. Oncol. 2018, 36 (Suppl. 15), e24244. [Google Scholar] [CrossRef]
- Bruno, M.T.; Scalia, G.; Cassaro, N.; Boemi, S. Multiple HPV 16 infection with two strains: A possible marker of neoplastic progression. BMC Cancer 2020, 20, 444. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Katki, H.A.; Hildesheim, A.; Rodríguez, A.C.; Quint, W.; Schiffman, M.; Van Doorn, L.-J.; Porras, C.; Wacholder, S.; Gonzalez, P.; et al. Human Papillomavirus Infection with Multiple Types: Pattern of Coinfection and Risk of Cervical Disease. J. Infect. Dis. 2011, 203, 910–920. [Google Scholar] [CrossRef]
- Salazar, K.L.; Zhou, H.S.; Xu, J.; Peterson, L.E.; Schwartz, M.R.; Mody, D.R.; Ge, Y. Multiple Human Papilloma Virus Infections and Their Impact on the Development of High-Risk Cervical Lesions. Acta Cytol. 2015, 59, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Depuydt, C.; Benoy, I.; Bogers, J.; Antoine, J.; Arbyn, M.; Pawlita, M. Multiple human papillomavirus infections with high viral loads are associated with cervical lesions but do not differentiate grades of cervical abnormalities. J. Clin. Microbiol. 2013, 51, 1458–1464. (In English) [Google Scholar] [CrossRef]
- Alosaimi, B.; Hampson, L.; He, X.; Maranga, I.O.; Oliver, A.W.; Hampson, I.N. Increased prevalence of JC polyomavirus in cervical carcinomas from women infected with HIV. J. Med. Virol. 2014, 86, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, K.; Carey, R.M.; Lin, X.; Seckar, T.D.; Wei, Z.; Chorath, K.; Newman, J.G.; O’Malley, B.W.; Weinstein, G.S.; Feldman, M.D.; et al. The microbiome of HPV-positive tonsil squamous cell carcinoma and neck metastasis. Oral Oncol. 2021, 117, 105305. [Google Scholar] [CrossRef]
- Qin, T.; Li, S.; Henry, L.E.; Liu, S.; Sartor, M.A. Molecular Tumor Subtypes of HPV-Positive Head and Neck Cancers: Biological Characteristics and Implications for Clinical Outcomes. Cancers 2021, 13, 2721. (In English) [Google Scholar] [CrossRef]
- Sun, Q.; Wang, L.; Zhang, C.; Hong, Z.; Han, Z. Cervical cancer heterogeneity: A constant battle against viruses and drugs. Biomark. Res. 2022, 10, 85. (In English) [Google Scholar] [CrossRef]
- Aggarwal, C.; Cohen, R.B.; Morrow, M.P.; Kraynyak, K.A.; Sylvester, A.J.; Cheung, J.; Dickerson, K.; Schulten, V.; Knoblock, D.; Gillespie, E.; et al. Immune Therapy Targeting E6/E7 Oncogenes of Human Paillomavirus Type 6 (HPV-6) Reduces or Eliminates the Need for Surgical Intervention in the Treatment of HPV-6 Associated Recurrent Respiratory Papillomatosis. Vaccines 2020, 8, 56. (In English) [Google Scholar] [CrossRef] [PubMed]
- Hoppe-Seyler, K.; Bossler, F.; Braun, J.A.; Herrmann, A.L.; Hoppe-Seyler, F. The HPV E6/E7 Oncogenes: Key Factors for Viral Carcinogenesis and Therapeutic Targets. Trends Microbiol. 2018, 26, 158–168. [Google Scholar] [CrossRef] [PubMed]

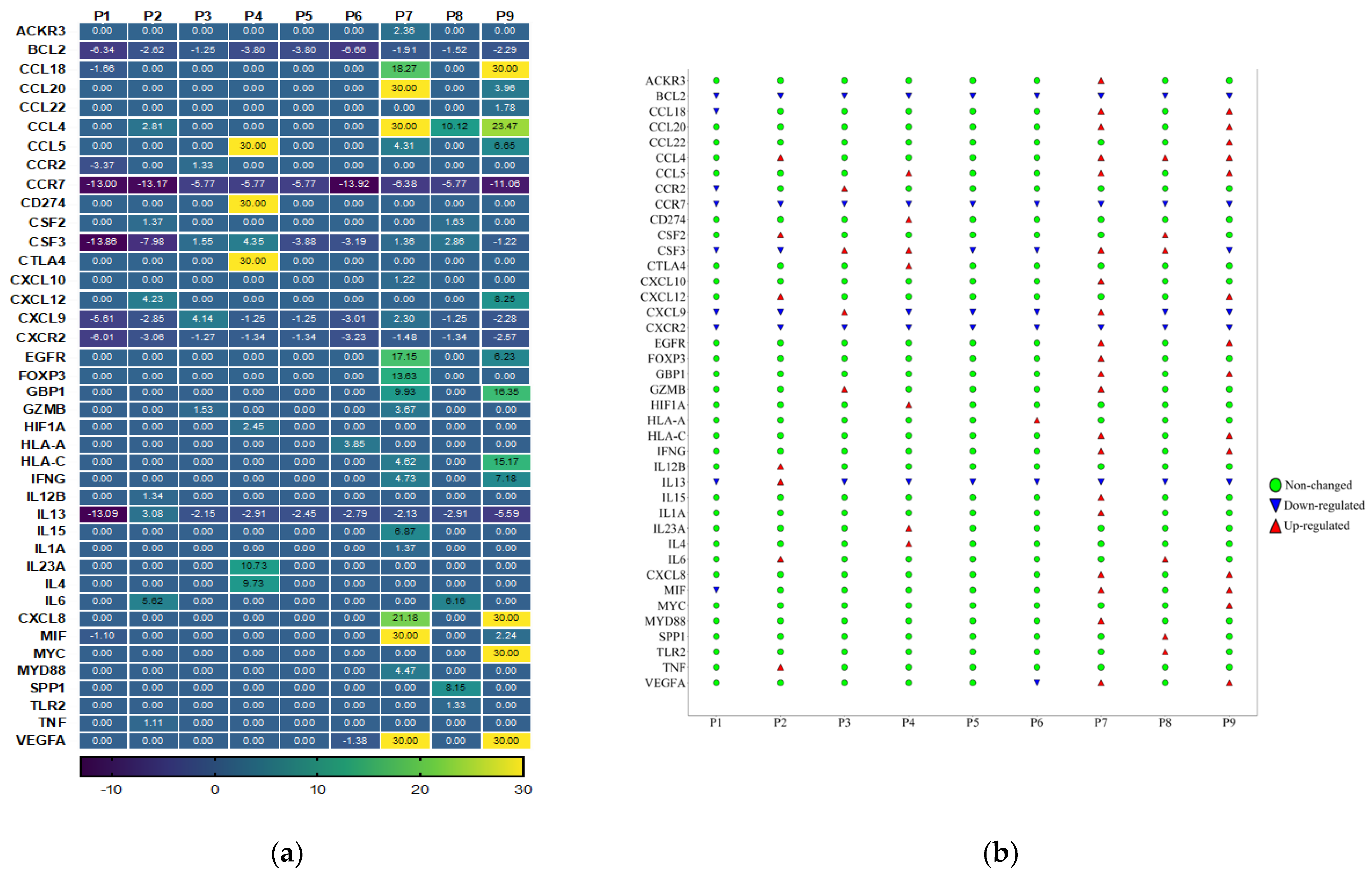
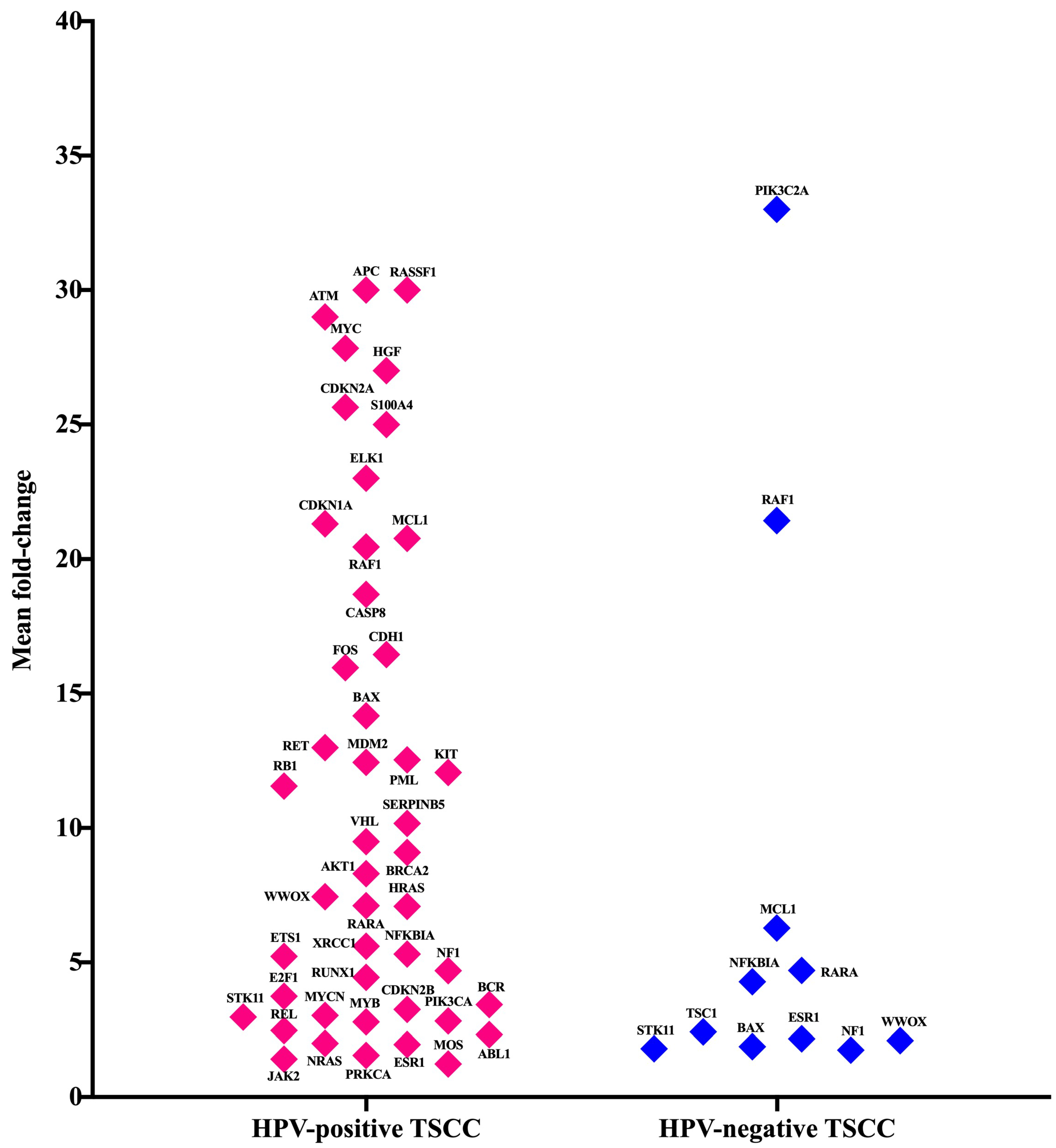
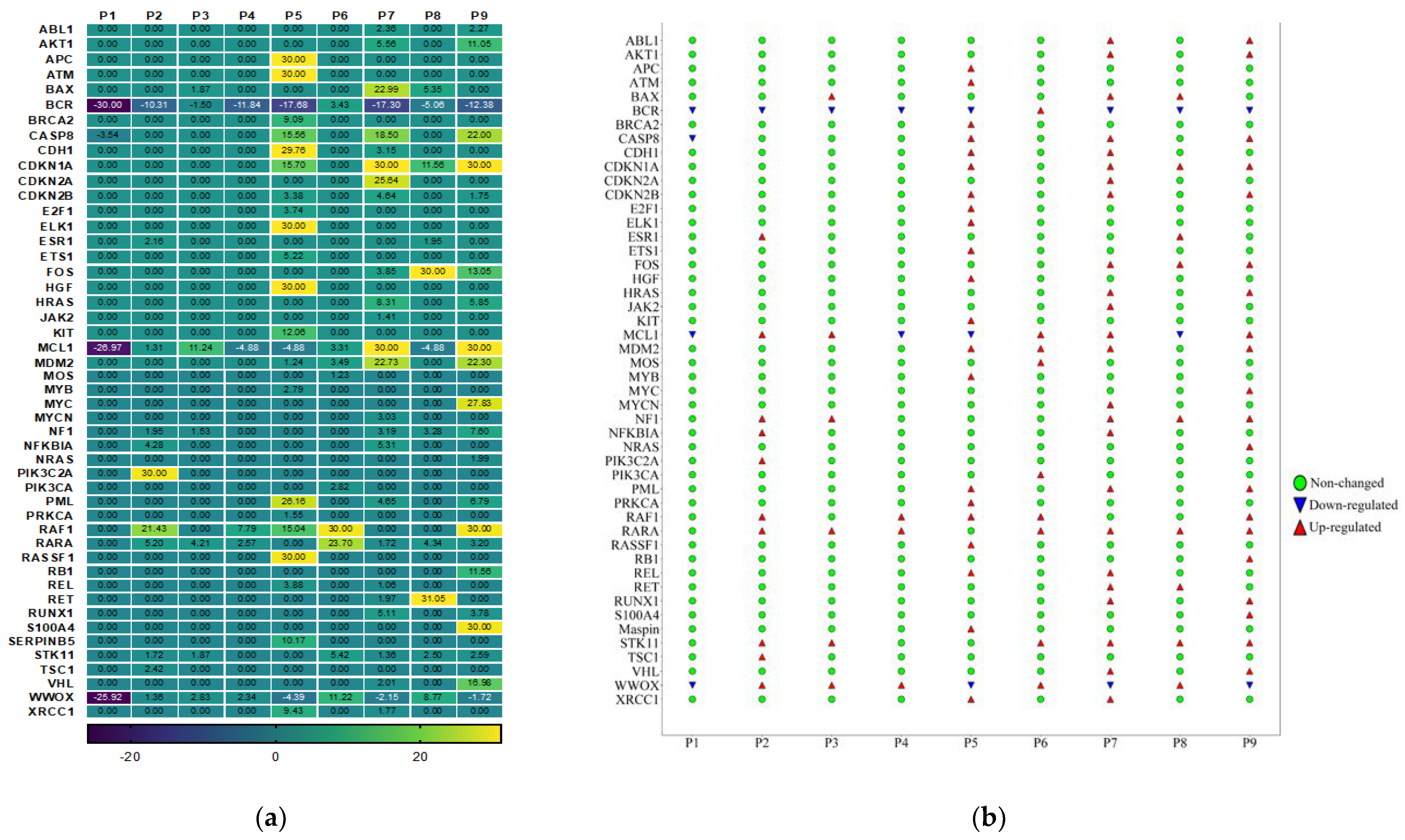
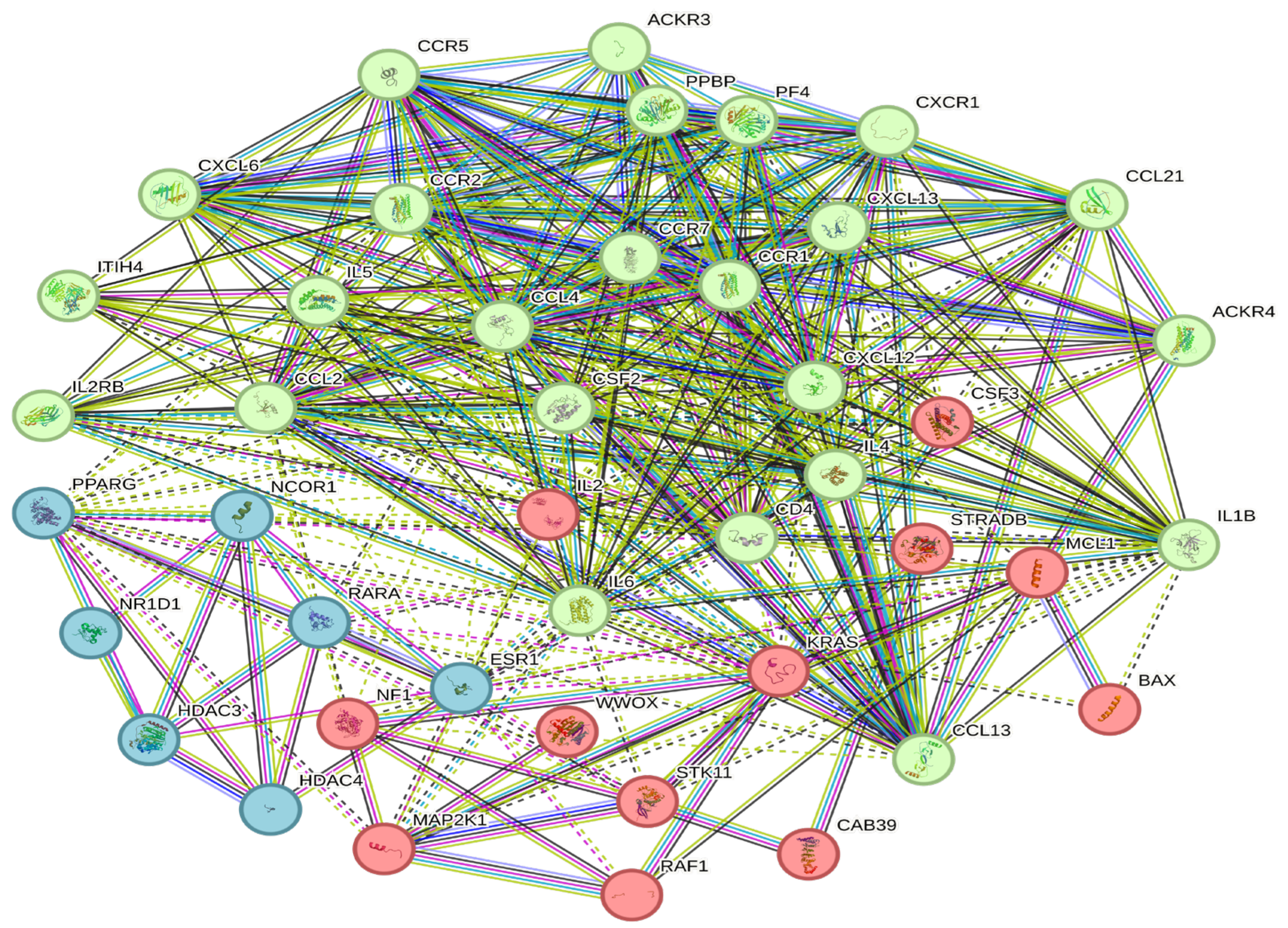
| Baseline Variables | All Patients | HPV-Positive | HPV-Negative |
|---|---|---|---|
| Demographics | n = 16 (%) | n = 13 (%) | n = 3 (%) |
| Age (years) | |||
| Median | 57 ± 12 | 52 ± 13 | 59 ± 4 |
| Range | 24–70 | 24–70 | 58–67 |
| Gender | |||
| Male | 7 (43.7%) | 5 (38.5%) | 2 (66.7%) |
| Female | 9 (56.3%) | 8 (61.5%) | 1 (33.3%) |
| Ethnicity | |||
| Saudi | 13 (81.2%) | 10 (53.8%) | 3 (100%) |
| Non-Saudi | 3 (18.8%) | 3 (23%) | - |
| Smoking | 4 (25%) | 2 (15.3%) | 2 (66.7%) |
| Comorbidities | |||
| Diabetes mellitus | 3 (18.8%) | 2 (15.3%) | 1 (33.3%) |
| Hypertension | 4 (25%) | 2 (15.3%) | 2 (66.7%) |
| Variables | n (%) |
|---|---|
| Type of HPV infection (n = 13) | |
| Single | 5 (38%) |
| Multiple | 8 (61%) |
| HPV genotypes | |
| Single infection (n = 5) | |
| HPV16 | 3 (60%) |
| HPV35 | 2 (40%) |
| Multiple infection (n = 8) | |
| Double infections | 2 (25%) |
| Triple infections | 5 (62%) |
| Quadruple infection | 1 (12%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alahmadi, R.M.; Marraiki, N.; Alswayyed, M.; Khoja, H.A.; Al-Anazi, A.E.; Alahmadi, R.M.; Alkusayer, M.M.; Alosaimi, B.; Awadalla, M. Comprehensive Transcriptome Analysis Reveals the Distinct Gene Expression Patterns of Tumor Microenvironment in HPV-Associated and HPV-Non Associated Tonsillar Squamous Cell Carcinoma. Cancers 2023, 15, 5548. https://doi.org/10.3390/cancers15235548
Alahmadi RM, Marraiki N, Alswayyed M, Khoja HA, Al-Anazi AE, Alahmadi RM, Alkusayer MM, Alosaimi B, Awadalla M. Comprehensive Transcriptome Analysis Reveals the Distinct Gene Expression Patterns of Tumor Microenvironment in HPV-Associated and HPV-Non Associated Tonsillar Squamous Cell Carcinoma. Cancers. 2023; 15(23):5548. https://doi.org/10.3390/cancers15235548
Chicago/Turabian StyleAlahmadi, Reham M., Najat Marraiki, Mohammed Alswayyed, Hatim A. Khoja, Abdullah E. Al-Anazi, Rawan M. Alahmadi, Meshael M. Alkusayer, Bandar Alosaimi, and Maaweya Awadalla. 2023. "Comprehensive Transcriptome Analysis Reveals the Distinct Gene Expression Patterns of Tumor Microenvironment in HPV-Associated and HPV-Non Associated Tonsillar Squamous Cell Carcinoma" Cancers 15, no. 23: 5548. https://doi.org/10.3390/cancers15235548
APA StyleAlahmadi, R. M., Marraiki, N., Alswayyed, M., Khoja, H. A., Al-Anazi, A. E., Alahmadi, R. M., Alkusayer, M. M., Alosaimi, B., & Awadalla, M. (2023). Comprehensive Transcriptome Analysis Reveals the Distinct Gene Expression Patterns of Tumor Microenvironment in HPV-Associated and HPV-Non Associated Tonsillar Squamous Cell Carcinoma. Cancers, 15(23), 5548. https://doi.org/10.3390/cancers15235548







