3DGAUnet: 3D Generative Adversarial Networks with a 3D U-Net Based Generator to Achieve the Accurate and Effective Synthesis of Clinical Tumor Image Data for Pancreatic Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
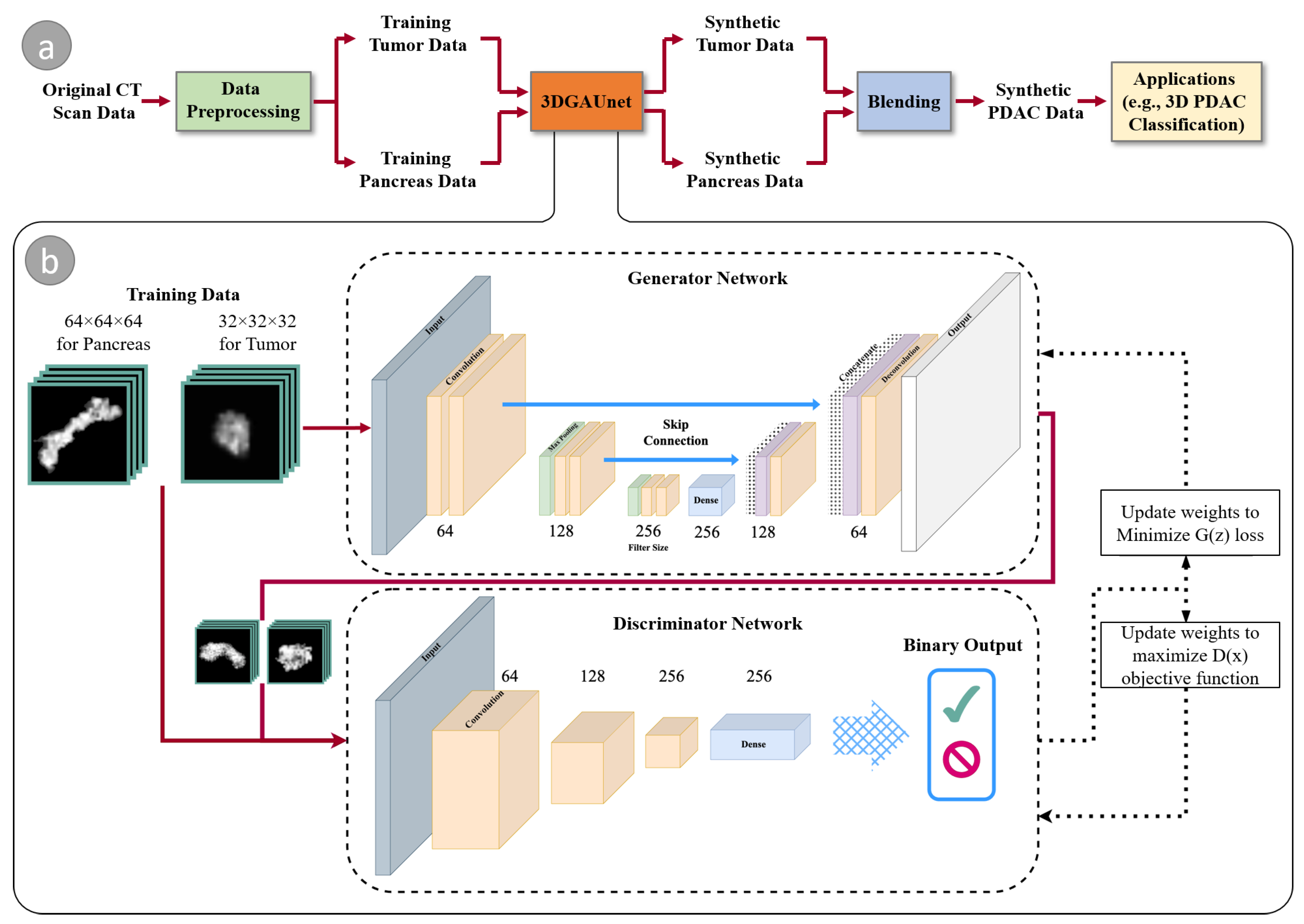
2. Materials and Methods
2.1. 3D CT Image Data Preprocessing
2.2. 3DGAUnet: 3D U-Net Based GAN Model
2.3. Blending to Create PDAC Tissues
2.4. Evaluation of Synthesized Images
2.5. 3D CNN PDAC Classifier
3. Results
3.1. 3D Volumetric Tissue Data Generation
3.2. 3D Volumetric Data Blending
3.3. Enhanced Training Dataset with Synthesized Data to Improve 3D PDAC Tumor Classification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Cancer Facts & Figures 2022; Technical Report; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Program, S.R. Surveillance, Epidemiology, and End Results (SEER) Program. SEER*Stat Database: Populations-Total U.S. (1969–2020) [Katrina/Rita Adjustment]-Linked To County Attributes-Total U.S., 1969–2020 Counties; Technical Report; National Cancer Institute, DCCPS: Bethesda, ML, USA, 2022. Available online: www.seer.cancer.gov (accessed on 1 January 2023).
- Wu, J.; Qian, T. A survey of pulmonary nodule detection, segmentation and classification in computed tomography with deep learning techniques. J. Med. Artif. Intell. 2019, 2. [Google Scholar] [CrossRef]
- Tandel, G.S.; Biswas, M.; Kakde, O.G.; Tiwari, A.; Suri, H.S.; Turk, M.; Laird, J.R.; Asare, C.K.; Ankrah, A.A.; Khanna, N.; et al. A review on a deep learning perspective in brain cancer classification. Cancers 2019, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Sharif, M.I.; Li, J.P.; Naz, J.; Rashid, I. A comprehensive review on multi-organs tumor detection based on machine learning. Pattern Recognit. Lett. 2020, 131, 30–37. [Google Scholar] [CrossRef]
- Radiya, K.; Joakimsen, H.L.; Mikalsen, K.Ø.; Aahlin, E.K.; Lindsetmo, R.O.; Mortensen, K.E. Performance and clinical applicability of machine learning in liver computed tomography imaging: A systematic review. Eur. Radiol. 2023, 33, 6689–6717. [Google Scholar] [CrossRef]
- Xue, Y.; Tong, W.; Neri, F.; Zhang, Y. PEGANs: Phased Evolutionary Generative Adversarial Networks with Self-Attention Module. Mathematics 2022, 10, 2792. [Google Scholar] [CrossRef]
- Baltruschat, I.M.; Nickisch, H.; Grass, M.; Knopp, T.; Saalbach, A. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci. Rep. 2019, 9, 6381. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Xue, X.; Jiang, Y.; Shen, Q. Deep learning for remote sensing image classification: A survey. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2018, 8, e1264. [Google Scholar] [CrossRef]
- Razzak, M.I.; Naz, S.; Zaib, A. Deep learning for medical image processing: Overview, challenges and the future. In Classification in BioApps: Automation of Decision Making; Springer: Cham, Switzerland, 2018; pp. 323–350. [Google Scholar]
- Li, S.; Song, W.; Fang, L.; Chen, Y.; Ghamisi, P.; Benediktsson, J.A. Deep learning for hyperspectral image classification: An overview. IEEE Trans. Geosci. Remote Sens. 2019, 57, 6690–6709. [Google Scholar] [CrossRef]
- Chu, L.C.; Park, S.; Kawamoto, S.; Yuille, A.L.; Hruban, R.H.; Fishman, E.K. Pancreatic cancer imaging: A new look at an old problem. Curr. Probl. Diagn. Radiol. 2021, 50, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Si, K.; Xue, Y.; Yu, X.; Zhu, X.; Li, Q.; Gong, W.; Liang, T.; Duan, S. Fully end-to-end deep-learning-based diagnosis of pancreatic tumors. Theranostics 2021, 11, 1982. [Google Scholar] [CrossRef] [PubMed]
- Foret, P.; Kleiner, A.; Mobahi, H.; Neyshabur, B. Sharpness-aware minimization for efficiently improving generalization. arXiv 2020, arXiv:2010.01412. [Google Scholar]
- Wei, Z.; Chen, Y.; Guan, Q.; Hu, H.; Zhou, Q.; Li, Z.; Xu, X.; Frangi, A.; Chen, F. Pancreatic Image Augmentation Based on Local Region Texture Synthesis for Tumor Segmentation. In Proceedings of the 31st International Conference on Artificial Neural Networks, Bristol, UK, 6–9 September 2022; Springer: Cham, Switzerland, 2022; pp. 419–431. [Google Scholar]
- Guan, Q.; Chen, Y.; Wei, Z.; Heidari, A.A.; Hu, H.; Yang, X.H.; Zheng, J.; Zhou, Q.; Chen, H.; Chen, F. Medical image augmentation for lesion detection using a texture-constrained multichannel progressive GAN. Comput. Biol. Med. 2022, 145, 105444. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, C.; Xue, T.; Freeman, B.; Tenenbaum, J. Learning a probabilistic latent space of object shapes via 3d generative-adversarial modeling. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; Volume 29. [Google Scholar]
- Antonelli, M.; Reinke, A.; Bakas, S.; Farahani, K.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; Ronneberger, O.; Summers, R.M.; et al. The medical segmentation decathlon. Nat. Commun. 2022, 13, 4128. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.; Farag, A.; Turkbey, E.B.; Lu, L.; Liu, J.; Summers, R.M. Data From Pancreas-CT (Version 2). In The Cancer Imaging Archive; NCI: Bethesda, MD, USA, 2016. [Google Scholar] [CrossRef]
- Hounsfield, G.N. Computed medical imaging. Science 1980, 210, 22–28. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Zhang, L.; Wen, T.; Shi, J. Deep Image Blending. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision, Snowmass Village, CO, USA, 1–5 March 2020; pp. 231–240. [Google Scholar]
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A platform for subject-specific image analysis, visualization, and clinical support. In Intraoperative Imaging and Image-Guided Therapy; Springer: Berlin/Heidelberg, Germany, 2013; pp. 277–289. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. Gans trained by a two time-scale update rule converge to a local nash equilibrium. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Gretton, A.; Borgwardt, K.M.; Rasch, M.J.; Schölkopf, B.; Smola, A. A kernel two-sample test. J. Mach. Learn. Res. 2012, 13, 723–773. [Google Scholar]
- Odena, A.; Olah, C.; Shlens, J. Conditional image synthesis with auxiliary classifier gans. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 2642–2651. [Google Scholar]
- Tran, D.; Bourdev, L.; Fergus, R.; Torresani, L.; Paluri, M. Learning spatiotemporal features with 3d convolutional networks. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 4489–4497. [Google Scholar]




| Tissue | Model | FID-Sag | FID-Ax | FID-Cor | PSNR-Sag | PSNR-Ax | PSNR-Cor |
|---|---|---|---|---|---|---|---|
| Tumor | 3D-GAN | 249.32 | 262.18 | 244.27 | 20.10 | 18.63 | 19.49 |
| 3DGAUNet | 198.23 | 202.44 | 188.66 | 16.52 | 17.76 | 17.16 | |
| Pancreas | 3D-GAN | 293.62 | 342.60 | 335.20 | 18.20 | 16.31 | 14.05 |
| 3DGAUNet | 287.75 | 435.72 | 327.41 | 12.73 | 7.21 | 9.42 |
| Tissue | Model | F3D | MMD2 | MS-SSIM |
|---|---|---|---|---|
| Tumor | 3DGAN | 472.64 | 5571.90 | 0.86 |
| 3DGAUNet | 271.31 | 5327.32 | 0.81 | |
| Pancreas | 3DGAN | 889.40 | 8924.39 | 0.83 |
| 3DGAUNet | 872.33 | 9122.40 | 0.77 |
| Blending Methods | FID-Sag | FID-Ax | FID-Cor |
|---|---|---|---|
| Blend I | 42.10 | 40.26 | 32.94 |
| Blend II | 21.01 | 35.82 | 12.62 |
| Blend III | 13.21 | 13.88 | 10.06 |
| Training Set | |
|---|---|
| Config I | 139 True PDAC |
| 203 True Healthy Pancreas | |
| Config II | 139 True + 114 synthesized PDAC |
| 203 True + 50 synthesized Healthy Pancreas |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Tang, H.; Baine, M.J.; Hollingsworth, M.A.; Du, H.; Zheng, D.; Zhang, C.; Yu, H. 3DGAUnet: 3D Generative Adversarial Networks with a 3D U-Net Based Generator to Achieve the Accurate and Effective Synthesis of Clinical Tumor Image Data for Pancreatic Cancer. Cancers 2023, 15, 5496. https://doi.org/10.3390/cancers15235496
Shi Y, Tang H, Baine MJ, Hollingsworth MA, Du H, Zheng D, Zhang C, Yu H. 3DGAUnet: 3D Generative Adversarial Networks with a 3D U-Net Based Generator to Achieve the Accurate and Effective Synthesis of Clinical Tumor Image Data for Pancreatic Cancer. Cancers. 2023; 15(23):5496. https://doi.org/10.3390/cancers15235496
Chicago/Turabian StyleShi, Yu, Hannah Tang, Michael J. Baine, Michael A. Hollingsworth, Huijing Du, Dandan Zheng, Chi Zhang, and Hongfeng Yu. 2023. "3DGAUnet: 3D Generative Adversarial Networks with a 3D U-Net Based Generator to Achieve the Accurate and Effective Synthesis of Clinical Tumor Image Data for Pancreatic Cancer" Cancers 15, no. 23: 5496. https://doi.org/10.3390/cancers15235496
APA StyleShi, Y., Tang, H., Baine, M. J., Hollingsworth, M. A., Du, H., Zheng, D., Zhang, C., & Yu, H. (2023). 3DGAUnet: 3D Generative Adversarial Networks with a 3D U-Net Based Generator to Achieve the Accurate and Effective Synthesis of Clinical Tumor Image Data for Pancreatic Cancer. Cancers, 15(23), 5496. https://doi.org/10.3390/cancers15235496






