Revealing Pan-Histology Immunomodulatory Targets in Pediatric Central Nervous System Tumors
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of Data Source
2.2. Data Availability
2.3. Code Availability
2.4. Immune Landscape Assessment of the pCNS Transcriptome
2.5. Cancer Cell Culture and T-Cell Expansion and Activation
2.6. EZH2 Knockdown
2.7. CD8+ T-Cell Cytotoxic Assay
2.8. Western Blot Analysis
2.9. Ethics Statement
3. Results
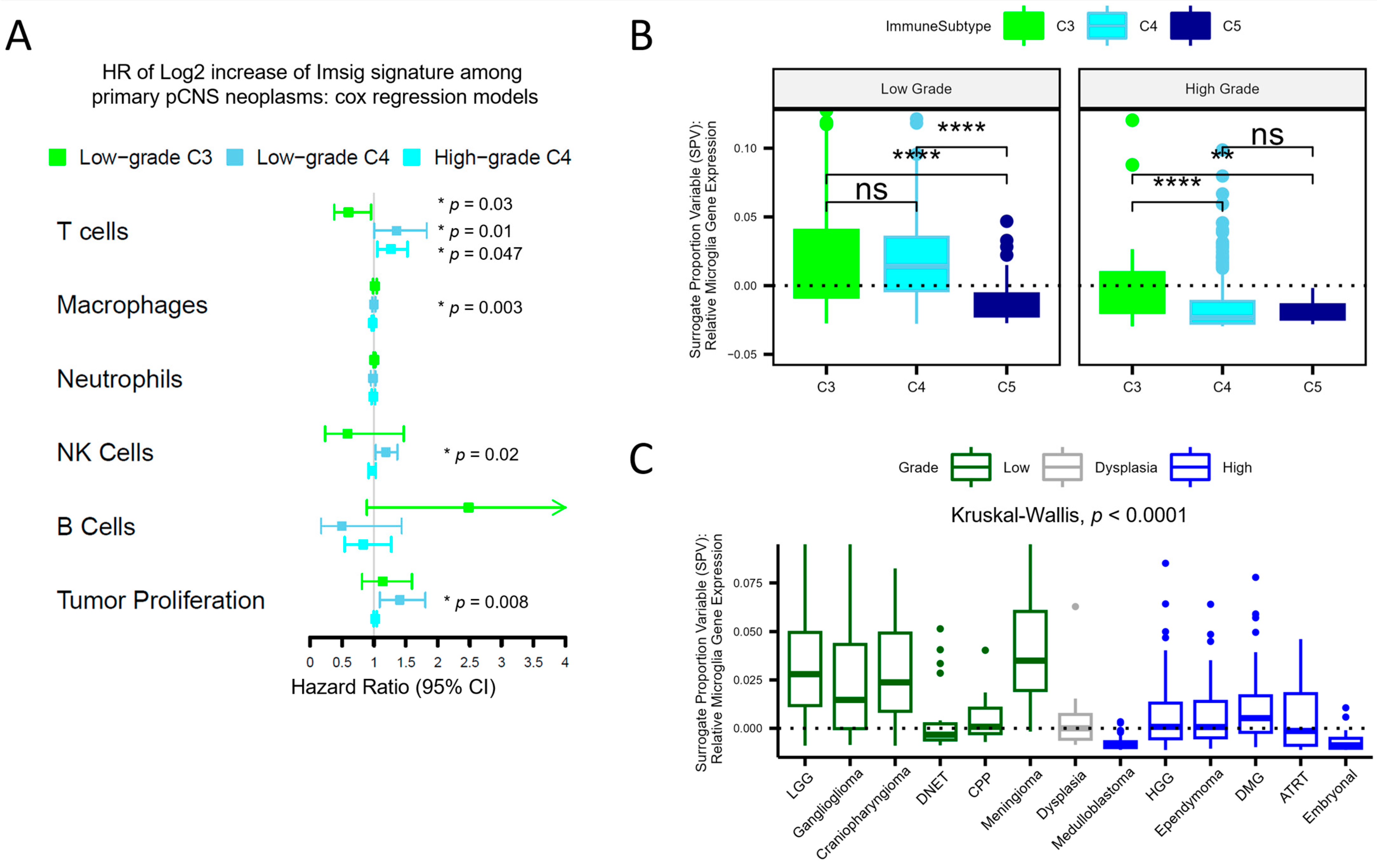
3.1. An Immunosuppressive Immune Subtype Is Common across High-Grade pCNS Malignancies and Correlates with Poor Survival
3.2. Deconvolution Provides Insight into the Cellular Composition of pCNS Tumors, Stratified by Immune Subtypes
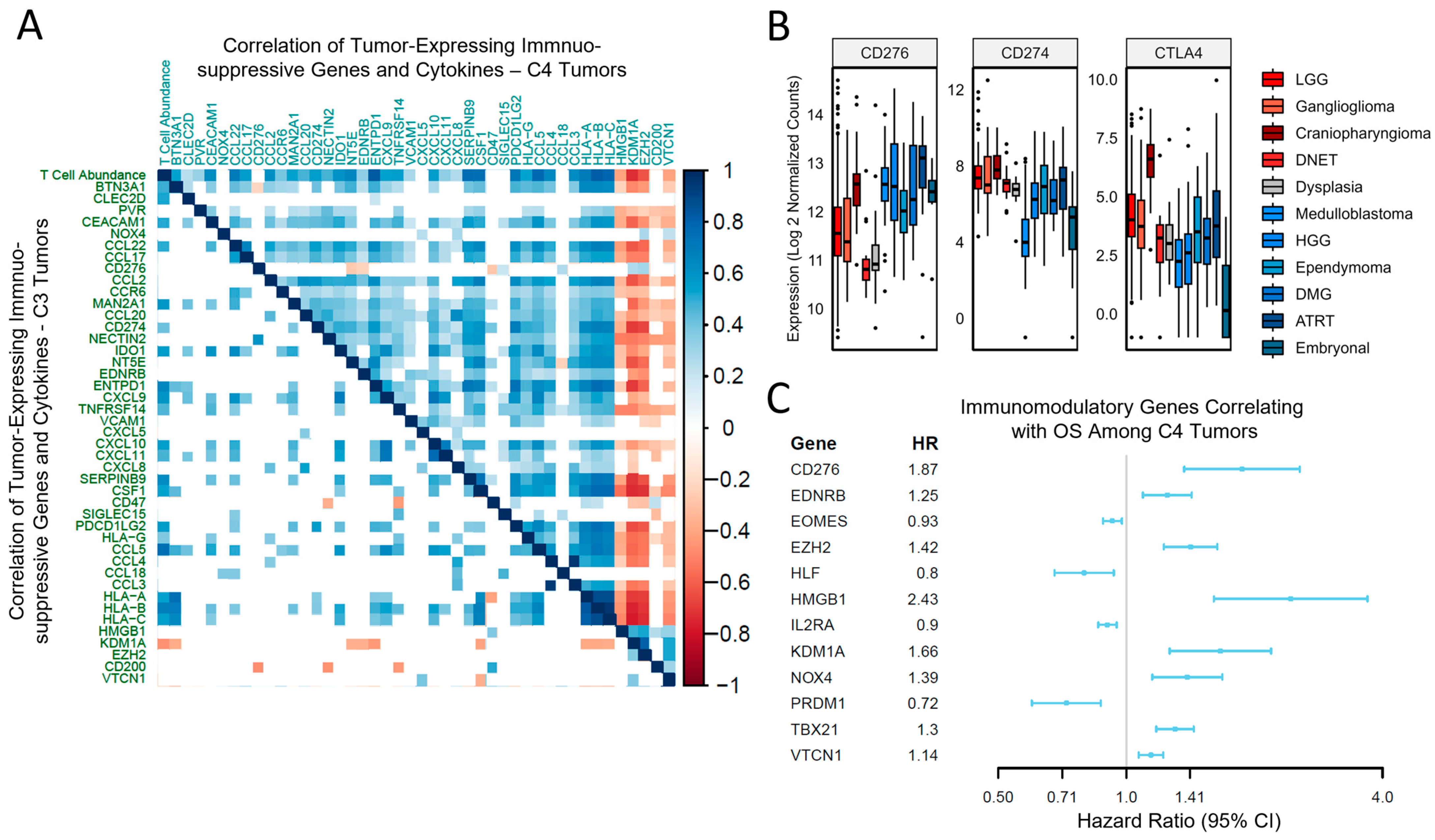
3.3. Tumor-Driven Immunomodulation Occurs via Different Mechanisms Depending on the Immune Context
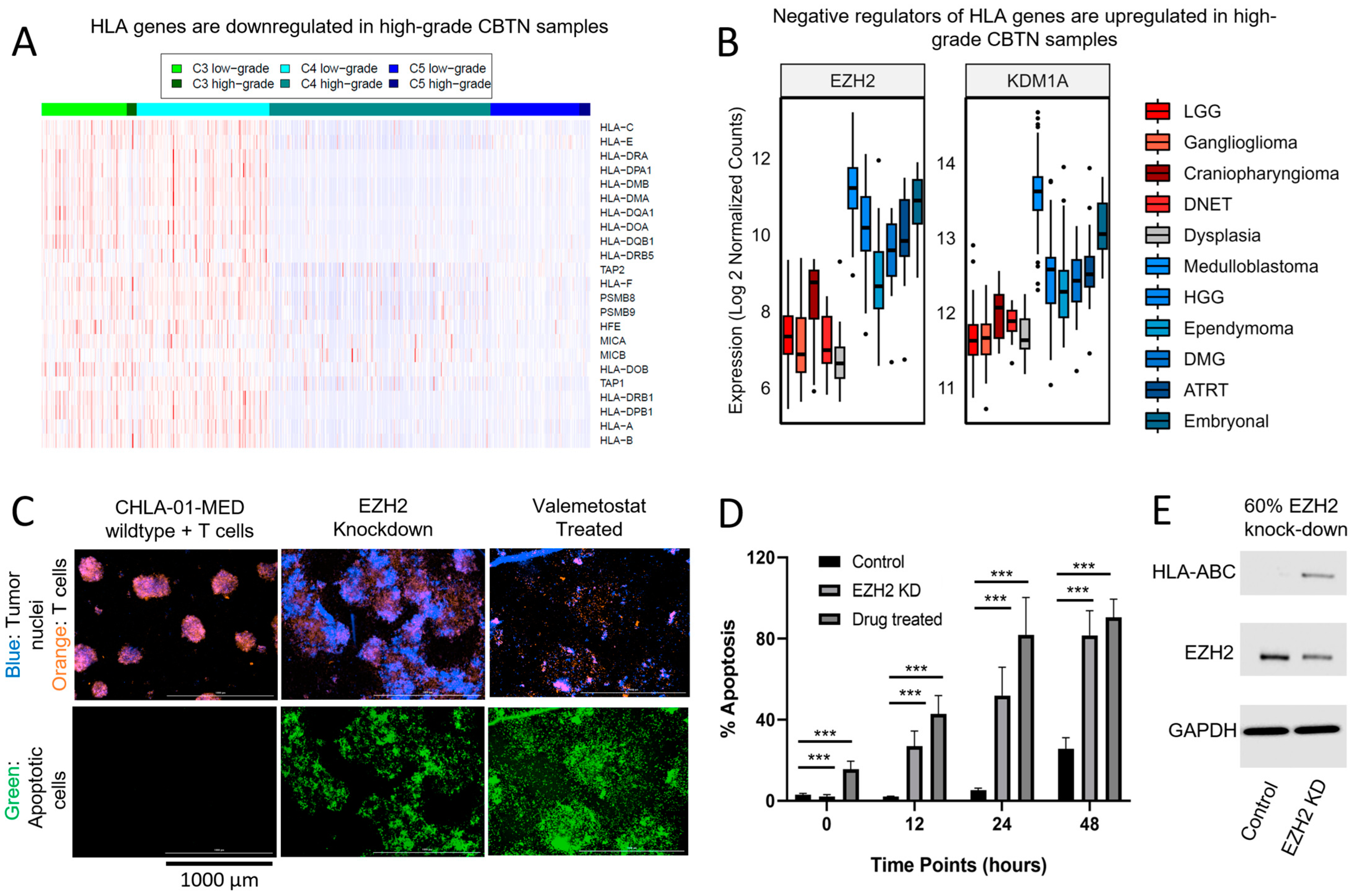
3.4. Decreased Expression of Antigen-Presenting Machinery Is Another Mechanism for Immunosuppression Observed in pCNS Malignancies and Is Modifiable Using Epigenetic Therapy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Brain Tumor Society. Quick Brain Tumor Facts. Available online: braintumor.org/brain-tumor-information/brain-tumor-facts/ (accessed on 22 March 2021).
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Xu, H.; Xu, X.; Guo, T.; Ge, W. High tumor mutation burden predicts better efficacy of immunotherapy: A pooled analysis of 103078 cancer patients. OncoImmunology 2019, 8, e1629258. [Google Scholar] [CrossRef]
- Kim, H.; Lim, K.Y.; Park, J.W.; Kang, J.; Won, J.K.; Lee, K.; Shim, Y.; Park, C.-K.; Kim, S.-K.; Park, S.H.; et al. Sporadic and Lynch syndrome-associated mismatch repair-deficient brain tumors. Lab. Investig. 2022, 102, 160–171. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Yang, T.H.O.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Ellison, D.W.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Gibbs, D.L. Robust Classification of Immune Subtype in Cancer. bioRxiv 2020. [Google Scholar] [CrossRef]
- Nirmal, A.J.; Regan, T.; Shih, B.B.; Hume, D.A.; Sims, A.H.; Freeman, T.C. Immune Cell Gene Signatures for Profiling the Microenvironment of Solid Tumors. Cancer Immunol. Res. 2018, 6, 1388–1400. [Google Scholar] [CrossRef]
- McKenzie, A.T.; Wang, M.; Hauberg, M.E.; Fullard, J.F.; Kozlenkov, A.; Keenan, A.; Hurd, Y.L.; Dracheva, S.; Casaccia, P.; Roussos, P. Brain Cell Type Specific Gene Expression and Co-expression Network Architectures. Sci. Rep. 2018, 8, 8868. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Grabovska, Y.; Mackay, A.; O’Hare, P.; Crosier, S.; Finetti, M.; Schwalbe, E.C.; Pickles, J.C.; Fairchild, A.R.; Avery, A.; Cockle, J.; et al. Pediatric pan-central nervous system tumor analysis of immune-cell infiltration identifies correlates of antitumor immunity. Nat. Commun. 2020, 11, 4324. [Google Scholar] [CrossRef] [PubMed]
- Sevenich, L. Brain-Resident Microglia and Blood-Borne Macrophages Orchestrate Central Nervous System Inflammation in Neurodegenerative Disorders and Brain Cancer. Front. Immunol. 2018, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Chikina, M.; Zaslavsky, E.; Sealfon, S.C. CellCODE: A robust latent variable approach to differential expression analysis for heterogeneous cell populations. Bioinformatics 2015, 31, 1584–1591. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Hinojosa, S.; Grant, M.; Panigrahi, A.; Zhang, H.; Caisova, V.; Bollard, C.M.; Rood, B.R. Proteogenomic discovery of neoantigens facilitates personalized multi-antigen targeted T cell immunotherapy for brain tumors. Nat. Commun. 2021, 12, 6689. [Google Scholar] [CrossRef] [PubMed]
- Burr, M.L.; Sparbier, C.E.; Chan, K.L. EZH1/2: A Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation and Enables Immune Evasion in Cancer. Cancer Cell 2019, 36, 385–401.e8. [Google Scholar] [CrossRef]
- Ren, J.; Li, N.; Pei, S.; Lian, Y.; Li, L.; Peng, Y.; Liu, Q.; Guo, J.; Wang, X.; Han, Y.; et al. Histone methyltransferase WHSC1 loss dampens MHC-I antigen presentation pathway to impair IFN-γ–stimulated antitumor immunity. J. Clin. Investig. 2022, 132, e153167. [Google Scholar] [CrossRef]
- Foster, J.B.; Madsen, P.J.; Hegde, M.; Ahmed, N.; Cole, K.A.; Maris, J.M.; Resnick, A.C.; Storm, P.B.; Waanders, A.J. Immunotherapy for pediatric brain tumors: Past and present. Neuro-Oncology 2019, 21, 1226–1238. [Google Scholar] [CrossRef]
- Hwang, E.I.; Sayour, E.J.; Flores, C.T.; Grant, G.; Wechsler-Reya, R.; Hoang-Minh, L.B.; Kieran, M.W.; Salcido, J.; Prins, R.M.; Figg, J.W.; et al. The current landscape of immunotherapy for pediatric brain tumors. Nat. Cancer 2022, 3, 11–24. [Google Scholar] [CrossRef]
- Neglia, J.P.; Robison, L.L.; Stovall, M.; Liu, Y.; Packer, R.J.; Hammond, S.; Yasui, Y.; Kasper, C.E.; Mertens, A.C.; Donaldson, S.S.; et al. New Primary Neoplasms of the Central Nervous System in Survivors of Childhood Cancer: A Report from the Childhood Cancer Survivor Study. J. Natl. Cancer Inst. 2006, 98, 1528–1537. [Google Scholar] [CrossRef]
- Bonaventura, P.; Shekarian, T.; Alcazer, V.; Valladeau-Guilemond, J.; Valsesia-Wittmann, S.; Amigorena, S.; Caux, C.; Depil, S. Cold Tumors: A Therapeutic Challenge for Immunotherapy. Front. Immunol. 2019, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell 2017, 31, 326–341. [Google Scholar] [CrossRef]
- Wang, S.S.; Bandopadhayay, P.; Jenkins, M.R. Towards Immunotherapy for Pediatric Brain Tumors. Trends Immunol. 2019, 40, 748–761. [Google Scholar] [CrossRef]
- Mohme, M.; Neidert, M.C. Tumor-Specific T Cell Activation in Malignant Brain Tumors. Front. Immunol. 2020, 11, 205. [Google Scholar] [CrossRef] [PubMed]
- Haydar, D.; Houke, H.; Chiang, J.; Yi, Z.; Odé, Z.; Caldwell, K.; Zhu, X.; Mercer, K.S.; Stripay, J.L.; Krenciute, G.; et al. Cell-surface antigen profiling of pediatric brain tumors: B7-H3 is consistently expressed and can be targeted via local or systemic CAR T-cell delivery. Neuro-Oncology 2021, 23, 999–1011. [Google Scholar] [CrossRef]
- Li, J.; Stanger, B.Z. How Tumor Cell Dedifferentiation Drives Immune Evasion and Resistance to Immunotherapy. Cancer Res. 2020, 80, 4037–4041. [Google Scholar] [CrossRef]
- Karasarides, M.; Cogdill, A.P.; Robbins, P.B.; Bowden, M.; Burton, E.M.; Butterfield, L.H.; Cesano, A.; Hammer, C.; Haymaker, C.L.; Horak, C.E.; et al. Hallmarks of Resistance to Immune-Checkpoint Inhibitors. Cancer Immunol. Res. 2022, 10, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Castel, D.; Kergrohen, T.; Tauziède-Espariat, A.; Mackay, A.; Ghermaoui, S.; Lechapt, E.; Pfister, S.M.; Kramm, C.M.; Boddaert, N.; Blauwblomme, T.; et al. Histone H3 wild-type DIPG/DMG overexpressing EZHIP extend the spectrum diffuse midline gliomas with PRC2 inhibition beyond H3-K27M mutation. Acta Neuropathol. 2020, 139, 1109–1113. [Google Scholar] [CrossRef]
- Jain, S.U.; Khazaei, S.; Marchione, D.M.; Lundgren, S.M.; Wang, X.; Weinberg, D.N.; Deshmukh, S.; Juretic, N.; Lu, C.; Allis, C.D.; et al. Histone H3.3 G34 mutations promote aberrant PRC2 activity and drive tumor progression. Proc. Natl. Acad. Sci. USA 2020, 117, 27354–27364. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, F.; Weissmann, S.; Leblanc, B.; Pandey, D.P.; Højfeldt, J.W.; Comet, I.; Zheng, C.; Johansen, J.V.; Rapin, N.; Porse, B.T.; et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 2017, 23, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Ohtani, H.; Chakravarthy, A.; De Carvalho, D.D. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer 2019, 19, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Petralia, F.; Tignor, N.; Reva, B.; Koptyra, M.; Chowdhury, S.; Rykunov, D.; Krek, A.; Ma, W.; Zhu, Y.; Ji, J.; et al. Integrated Proteogenomic Characterization across Major Histological Types of Pediatric Brain Cancer. Cell 2020, 183, 1962–1985.e31. [Google Scholar] [CrossRef]
- de Billy, E.; Pellegrino, M.; Orlando, D.; Pericoli, G.; Ferretti, R.; Businaro, P.; Ajmone-Cat, M.A.; Rossi, S.; Petrilli, L.L.; Maestro, N.; et al. Dual IGF1R/IR inhibitors in combination with GD2-CAR T-cells display a potent anti-tumor activity in diffuse midline glioma H3K27M-mutant. Neuro-Oncology 2022, 24, 1150–1163. [Google Scholar] [CrossRef]
- Kang, N.; Eccleston, M.; Clermont, P.L.; Latarani, M.; Male, D.K.; Wang, Y.; Crea, F. EZH2 inhibition: A promising strategy to prevent cancer immune editing. Epigenomics 2020, 12, 1457–1476. [Google Scholar] [CrossRef] [PubMed]
- Whelan, R.; Prince, E.; Gilani, A.; Hankinson, T. The Inflammatory Milieu of Adamantinomatous Craniopharyngioma and Its Implications for Treatment. J. Clin. Med. 2020, 9, 519. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Castro, L.N.; Liu, I.; Filbin, M. Characterizing the biology of primary brain tumors and their microenvironment via single-cell profiling methods. Neuro-Oncology 2023, 25, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, A.L.; Shah, P.P.; Lim, M. The Tumor Immune Microenvironment in Primary CNS Neoplasms: A Review of Current Knowledge and Therapeutic Approaches. Int. J. Mol. Sci. 2023, 24, 2020. [Google Scholar] [CrossRef]

| High Grade/Low Grade | C4/C3 | |||||
|---|---|---|---|---|---|---|
| Characteristic | OR 1 | 95% CI 1 | p-Value | OR1 | 95% CI 1 | p-Value |
| T-Cells | 0.69 | 0.43, 1.09 | 0.11 | 0.36 | 0.20, 0.62 | <0.001 |
| Neutrophils | 1.01 | 0.81, 1.25 | >0.9 | 0.96 | 0.77, 1.20 | 0.7 |
| NK Cells | 0.84 | 0.79, 0.90 | <0.001 | 1.22 | 1.13, 1.32 | <0.001 |
| Macrophages | 0.90 | 0.60, 1.34 | 0.6 | 2.05 | 1.28, 3.31 | 0.003 |
| Proliferation | 7.12 | 5.41, 9.62 | <0.001 | 2.85 | 2.13, 3.91 | <0.001 |
| Interferon | 1.44 | 1.08, 1.94 | 0.013 | 1.19 | 0.86, 1.68 | 0.3 |
| Grade | ||||||
| Low | — | — | ||||
| High | 3.99 | 2.19, 7.48 | <0.001 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galvin, R.T.; Jena, S.; Maeser, D.; Gruener, R.; Huang, R.S. Revealing Pan-Histology Immunomodulatory Targets in Pediatric Central Nervous System Tumors. Cancers 2023, 15, 5455. https://doi.org/10.3390/cancers15225455
Galvin RT, Jena S, Maeser D, Gruener R, Huang RS. Revealing Pan-Histology Immunomodulatory Targets in Pediatric Central Nervous System Tumors. Cancers. 2023; 15(22):5455. https://doi.org/10.3390/cancers15225455
Chicago/Turabian StyleGalvin, Robert T., Sampreeti Jena, Danielle Maeser, Robert Gruener, and R. Stephanie Huang. 2023. "Revealing Pan-Histology Immunomodulatory Targets in Pediatric Central Nervous System Tumors" Cancers 15, no. 22: 5455. https://doi.org/10.3390/cancers15225455
APA StyleGalvin, R. T., Jena, S., Maeser, D., Gruener, R., & Huang, R. S. (2023). Revealing Pan-Histology Immunomodulatory Targets in Pediatric Central Nervous System Tumors. Cancers, 15(22), 5455. https://doi.org/10.3390/cancers15225455






