The Important Role of Protein Kinases in the p53 Sestrin Signaling Pathway
Abstract
Simple Summary
Abstract
1. Introduction
- (i)
- Serine/threonine kinases are pivotal components of cellular signaling networks. These kinases catalyze the phosphorylation of hydroxyl groups (OH groups) attached to the amino acid residues serine and threonine. Their activity is typically modulated by second messengers (e.g., cyclic adenosine monophosphate (cAMP) or cyclic guanosine monophosphate (cGMP), as well as 1,2-diacylglycerol (DAG), calcium ions (Ca2+), calmodulin, phosphatidylinositol-3,4,5-trisphosphate (PIP3), and various derivatives of phospholipids). Serine/threonine kinases are critical components in cellular signaling networks, functioning as molecular switches that transmit extracellular signals and govern intracellular responses. Consequently, they are central to comprehending cell physiology and represent potential therapeutic targets for diseases associated with disrupted signaling pathways [1,5].
- (ii)
- Tyrosine kinases can be classified into two groups: membrane-bound tyrosine kinases (e.g., KIT (CD117), epidermal growth factor (EGF) receptor, insulin receptor, human epidermal growth factor receptor 2 (HER2)) and non-membrane-bound tyrosine kinases (e.g., Abelson murine leukemia viral oncogene homolog 1 (ABL1), SRC, Janus kinases). Membrane-bound tyrosine kinases include receptors with intrinsic kinase activity (where the kinase is an integral part of the receptor) and receptors with associated kinase activity (where the kinase binds to the receptor). Upon ligand binding to a membrane-bound or non-membrane-bound tyrosine kinase, conformational changes occur, promoting the formation of homodimers or heterodimers. This process triggers the autophosphorylation of specific tyrosine residues on the kinase, subsequently enabling the recruitment of additional proteins/kinases and initiating intricate signaling cascades [1,2].
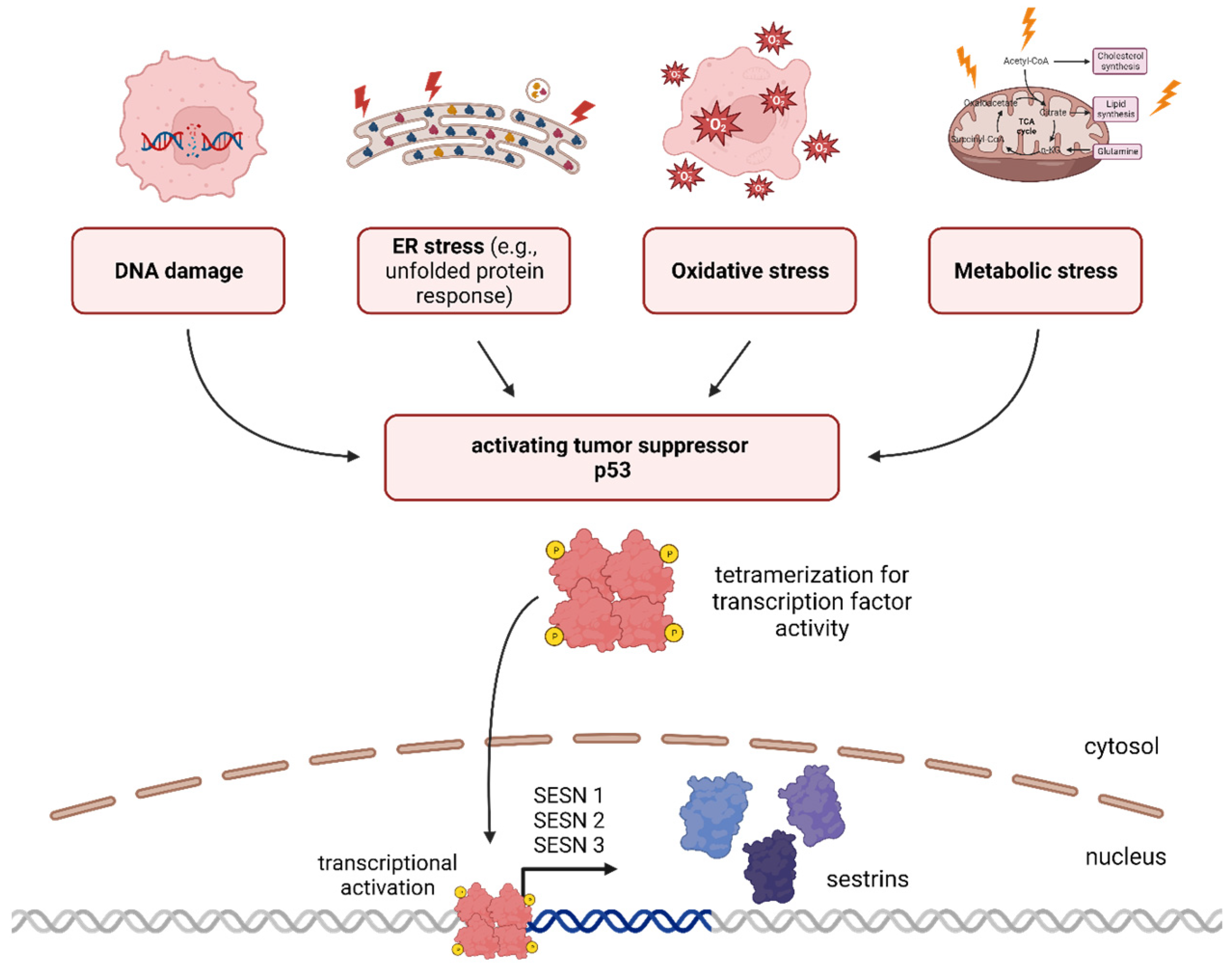
1.1. p53—Tumor Suppression and Beyond
1.2. Sestrins—The Universal Shield against Cellular Stress
2. p53/Sestrin Regulated Signaling Pathways
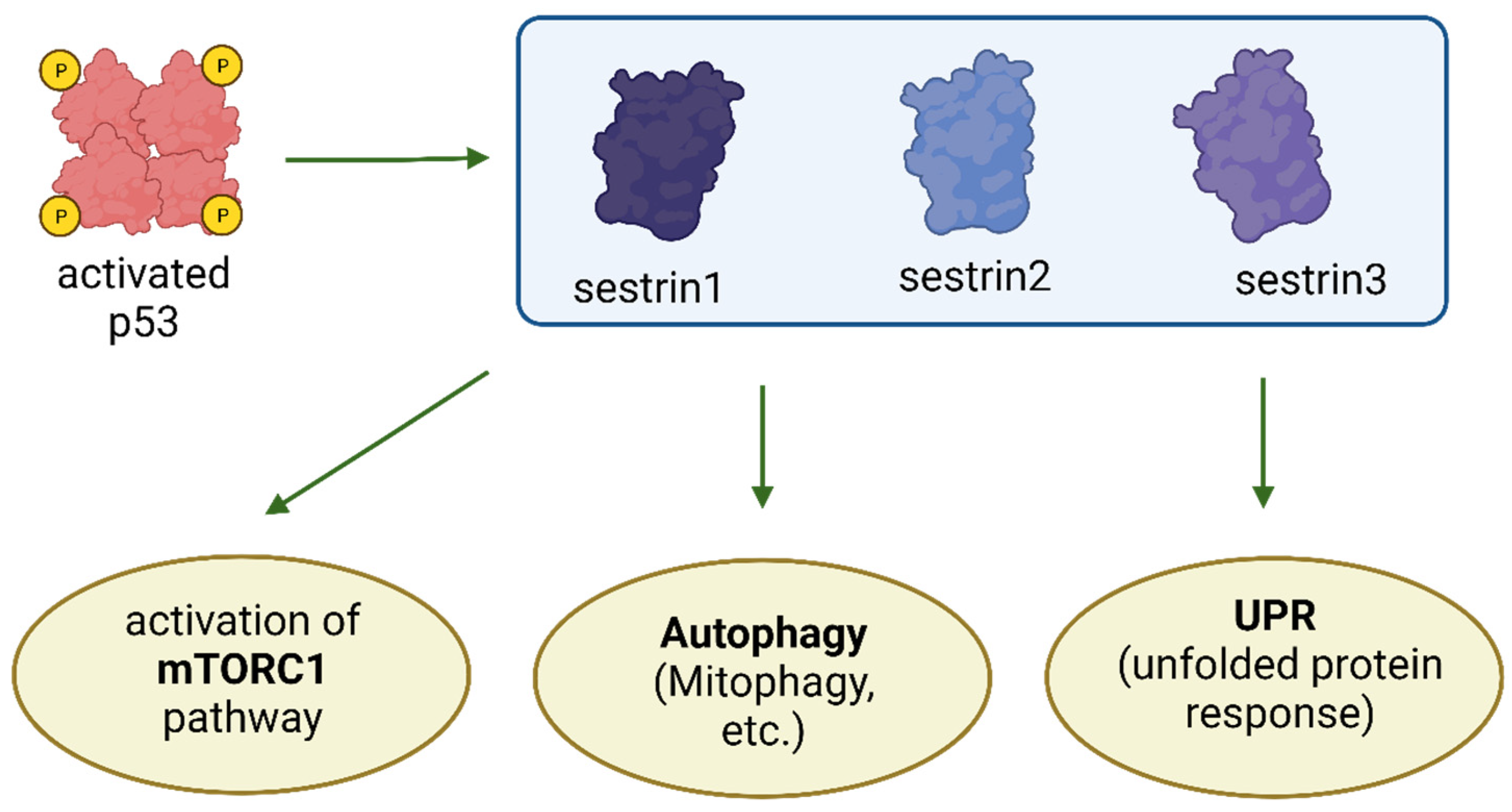
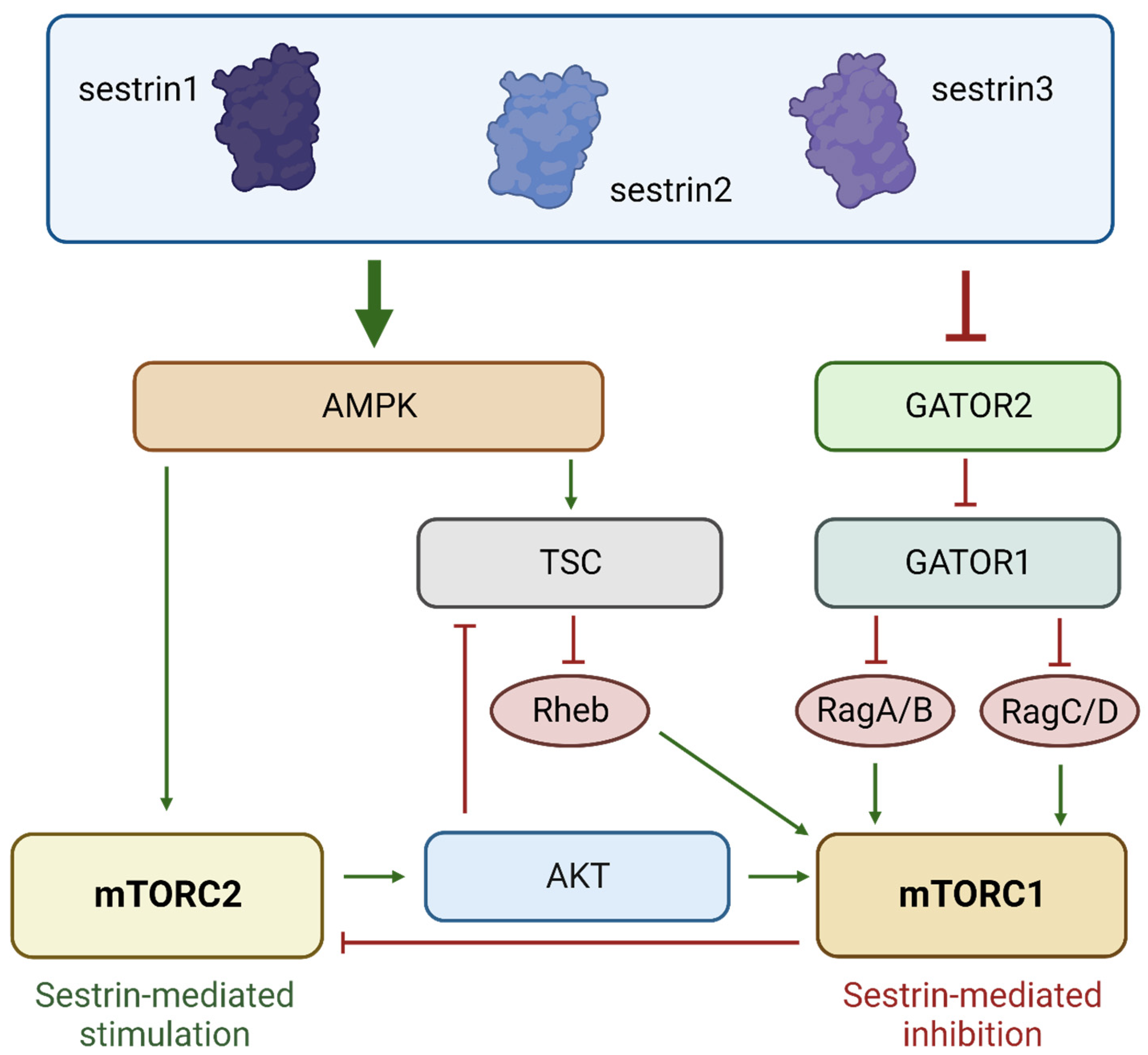
2.1. p53 Target Genes Sestrin 1, Sestrin 2, and Sestrin 3 Link Cellular Stress to mTOR Signaling
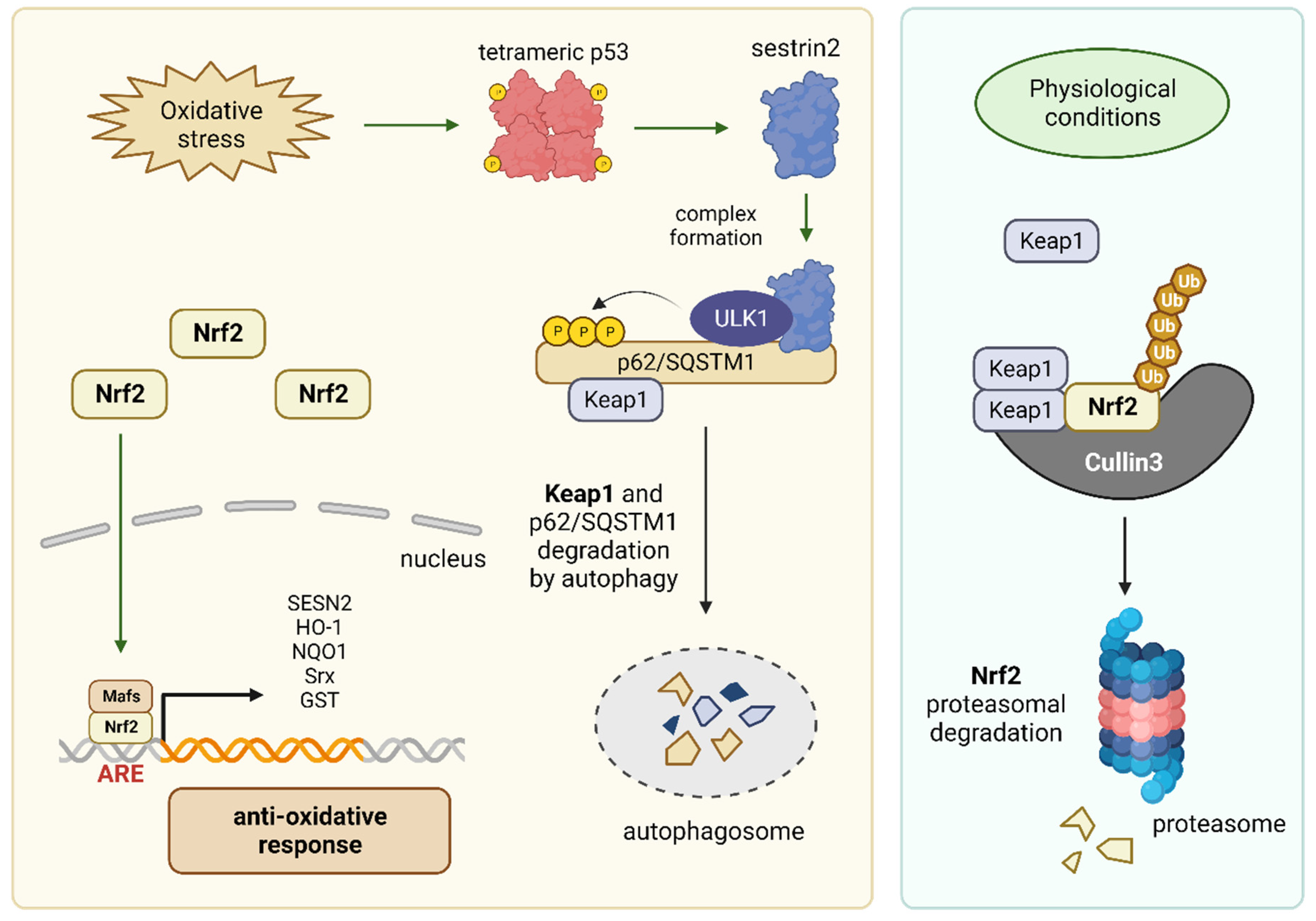
2.2. The p53 Target Gene Sestrin 2 Controls Antioxidant Defense Mechanisms
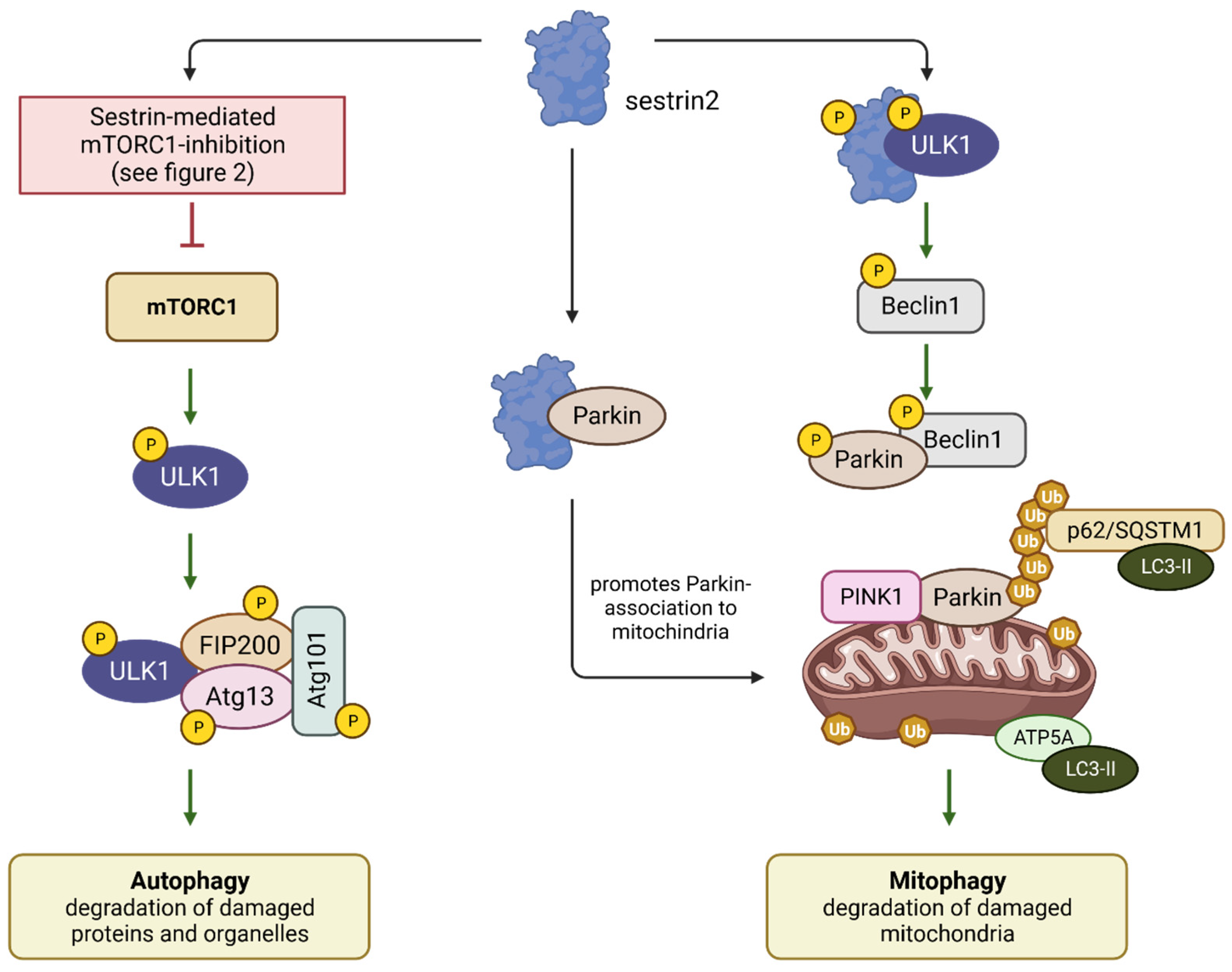
2.3. Sestrins: Initiators of Autophagy and Mitophagy
2.4. Sestrins and the UPR
3. Sestrins and Cancer
3.1. mTORC1 Triggers the Proliferation of Malignant Cells and Facilitates the Formation of Metastases
3.2. Antioxidative Defense and Cancer Prevention
3.3. Sestrin-Induced Autophagy/Mitophagy
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cohen, P. The origins of protein phosphorylation. Nat. Cell Biol. 2002, 4, E127–E130. [Google Scholar] [CrossRef]
- Hunter, T.; Cooper, J.A. Protein-tyrosine kinases. Annu. Rev. Biochem. 1985, 54, 897–930. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, L.J.; Sapkota, G.P. Functions and regulation of the serine/threonine protein kinase CK1 family: Moving beyond promiscuity. Biochem. J. 2020, 477, 4603–4621. [Google Scholar] [CrossRef]
- Cicenas, J.; Zalyte, E.; Bairoch, A.; Gaudet, P. Kinases and Cancer. Cancers 2018, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-C.; Qi, R.Z.; Paudel, H.; Zhu, H.-J. Regulation and function of protein kinases and phosphatases. Enzym. Res. 2011, 2011, 794089. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Wilder, S.; Bannasch, D.; Israeli, D.; Lehlbach, K.; Li-Weber, M.; Friedman, S.L.; Galle, P.R.; Stremmel, W.; Oren, M.; et al. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J. Exp. Med. 1998, 188, 2033–2045. [Google Scholar] [CrossRef]
- Kruiswijk, F.; Labuschagne, C.F.; Vousden, K.H. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015, 16, 393–405. [Google Scholar] [CrossRef]
- Ferraiuolo, M.; Di Agostino, S.; Blandino, G.; Strano, S. Oncogenic Intra-p53 Family Member Interactions in Human Cancers. Front. Oncol. 2016, 6, 77. [Google Scholar] [CrossRef]
- Dötsch, V.; Bernassola, F.; Coutandin, D.; Candi, E.; Melino, G. p63 and p73, the ancestors of p53. Cold Spring Harb. Perspect. Biol. 2010, 2, a004887. [Google Scholar] [CrossRef]
- Woodstock, D.L.; Sammons, M.A.; Fischer, M. p63 and p53: Collaborative Partners or Dueling Rivals? Front. Cell Dev. Biol. 2021, 9, 701986. [Google Scholar] [CrossRef]
- Surget, S.; Khoury, M.P.; Bourdon, J.-C. Uncovering the role of p53 splice variants in human malignancy: A clinical perspective. OncoTargets Ther. 2013, 7, 57–68. [Google Scholar] [CrossRef]
- Müller, M.; Schleithoff, E.S.; Stremmel, W.; Melino, G.; Krammer, P.H.; Schilling, T. One, two, three—p53, p63, p73 and chemosensitivity. Drug Resist. Updat. 2006, 9, 288–306. [Google Scholar] [CrossRef]
- Pflaum, J.; Schlosser, S.; Müller, M. p53 Family and Cellular Stress Responses in Cancer. Front. Oncol. 2014, 4, 285. [Google Scholar] [CrossRef]
- Levine, A.J. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 2020, 20, 471–480. [Google Scholar] [CrossRef]
- Bourdon, J.-C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef]
- Steffens Reinhardt, L.; Groen, K.; Newton, C.; Avery-Kiejda, K.A. The role of truncated p53 isoforms in the DNA damage response. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188882. [Google Scholar] [CrossRef]
- Senturk, S.; Yao, Z.; Camiolo, M.; Stiles, B.; Rathod, T.; Walsh, A.M.; Nemajerova, A.; Lazzara, M.J.; Altorki, N.K.; Krainer, A.; et al. p53Ψ is a transcriptionally inactive p53 isoform able to reprogram cells toward a metastatic-like state. Proc. Natl. Acad. Sci. USA 2014, 111, E3287–E3296. [Google Scholar] [CrossRef]
- Aoubala, M.; Murray-Zmijewski, F.; Khoury, M.P.; Fernandes, K.; Perrier, S.; Bernard, H.; Prats, A.-C.; Lane, D.P.; Bourdon, J.-C. p53 directly transactivates Δ133p53α, regulating cell fate outcome in response to DNA damage. Cell Death Differ. 2011, 18, 248–258. [Google Scholar] [CrossRef]
- Marcel, V.; Perrier, S.; Aoubala, M.; Ageorges, S.; Groves, M.J.; Diot, A.; Fernandes, K.; Tauro, S.; Bourdon, J.-C. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett. 2010, 584, 4463–4468. [Google Scholar] [CrossRef]
- Murray-Zmijewski, F.; Lane, D.P.; Bourdon, J.-C. p53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006, 13, 962–972. [Google Scholar] [CrossRef]
- Anbarasan, T.; Bourdon, J.-C. The Emerging Landscape of p53 Isoforms in Physiology, Cancer and Degenerative Diseases. Int. J. Mol. Sci. 2019, 20, 6257. [Google Scholar] [CrossRef]
- Zhao, L.; Sanyal, S. p53 Isoforms as Cancer Biomarkers and Therapeutic Targets. Cancers 2022, 14, 3145. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Mondal, A.M.; Horikawa, I.; Nguyen, G.H.; Kumamoto, K.; Sohn, J.J.; Bowman, E.D.; Mathe, E.A.; Schetter, A.J.; Pine, S.R.; et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat. Cell Biol. 2009, 11, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.; Garmy-Susini, B.; Ainaoui, N.; van den Berghe, L.; Peurichard, A.; Javerzat, S.; Bikfalvi, A.; Lane, D.P.; Bourdon, J.C.; Prats, A.-C. The p53 isoform, Δ133p53α, stimulates angiogenesis and tumour progression. Oncogene 2013, 32, 2150–2160. [Google Scholar] [CrossRef]
- Jänicke, R.U.; Sohn, D.; Schulze-Osthoff, K. The dark side of a tumor suppressor: Anti-apoptotic p53. Cell Death Differ. 2008, 15, 959–976. [Google Scholar] [CrossRef]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef]
- Holmberg Olausson, K.; Nistér, M.; Lindström, M.S. p53 -Dependent and -Independent Nucleolar Stress Responses. Cells 2012, 1, 774–798. [Google Scholar] [CrossRef]
- Qu, L.; Koromilas, A.E. Control of tumor suppressor p53 function by endoplasmic reticulum stress. Cell Cycle 2004, 3, 567–570. [Google Scholar] [CrossRef][Green Version]
- Hasty, P.; Campisi, J.; Sharp, Z.D. Do p53 stress responses impact organismal aging? Transl. Cancer Res. 2016, 5, 685–691. [Google Scholar] [CrossRef]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Boutelle, A.M.; Attardi, L.D. p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol. 2021, 31, 298–310. [Google Scholar] [CrossRef]
- Friedel, L.; Loewer, A. The guardian’s choice: How p53 enables context-specific decision-making in individual cells. FEBS J. 2022, 289, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Scaffidi, C.A.; Galle, P.R.; Stremmel, W.; Krammer, P.H. The role of p53 and the CD95 (APO-1/Fas) death system in chemotherapy-induced apoptosis. Eur. Cytokine Netw. 1998, 9, 685–686. [Google Scholar] [PubMed]
- Peeters, H.; Debeer, P.; Bairoch, A.; Wilquet, V.; Huysmans, C.; Parthoens, E.; Fryns, J.P.; Gewillig, M.; Nakamura, Y.; Niikawa, N.; et al. PA26 is a candidate gene for heterotaxia in humans: Identification of a novel PA26-related gene family in human and mouse. Hum. Genet. 2003, 112, 573–580. [Google Scholar] [CrossRef]
- Budanov, A.V.; Shoshani, T.; Faerman, A.; Zelin, E.; Kamer, I.; Kalinski, H.; Gorodin, S.; Fishman, A.; Chajut, A.; Einat, P.; et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene 2002, 21, 6017–6031. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Miguel, S.; Buckbinder, L.; Jean, P.; Gelbert, L.; Talbott, R.; Laidlaw, J.; Seizinger, B.; Kley, N. PA26, a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene 1999, 18, 127–137. [Google Scholar] [CrossRef]
- Budanov, A.V.; Sablina, A.A.; Feinstein, E.; Koonin, E.V.; Chumakov, P.M. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 2004, 304, 596–600. [Google Scholar] [CrossRef]
- Haidurov, A.; Budanov, A.V. Sestrin family—The stem controlling healthy ageing. Mech. Ageing Dev. 2020, 192, 111379. [Google Scholar] [CrossRef]
- Budanov, A.V.; Karin, M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef]
- Ro, S.-H.; Fay, J.; Cyuzuzo, C.I.; Jang, Y.; Lee, N.; Song, H.-S.; Harris, E.N. SESTRINs: Emerging Dynamic Stress-Sensors in Metabolic and Environmental Health. Front. Cell Dev. Biol. 2020, 8, 603421. [Google Scholar] [CrossRef]
- Lu, C.; Jiang, Y.; Xu, W.; Bao, X. Sestrin2: Multifaceted functions, molecular basis, and its implications in liver diseases. Cell Death Dis. 2023, 14, 160. [Google Scholar] [CrossRef] [PubMed]
- Hagenbuchner, J.; Kuznetsov, A.; Hermann, M.; Hausott, B.; Obexer, P.; Ausserlechner, M.J. FOXO3-induced reactive oxygen species are regulated by BCL2L11 (Bim) and SESN3. J. Cell Sci. 2012, 125, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhang, H.; Wu, Q.; Li, F.; Wang, Y.; Liu, S.; Wang, J. Sestrin 2 protects against LPS-induced acute lung injury by inducing mitophagy in alveolar macrophages. Life Sci. 2021, 267, 118941. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-J.; Bae, S.H.; Ryu, J.-C.; Kwon, Y.; Oh, J.-H.; Kwon, J.; Moon, J.-S.; Kim, K.; Miyawaki, A.; Lee, M.G.; et al. SESN2/sestrin2 suppresses sepsis by inducing mitophagy and inhibiting NLRP3 activation in macrophages. Autophagy 2016, 12, 1272–1291. [Google Scholar] [CrossRef]
- Wang, P.; Wang, L.; Lu, J.; Hu, Y.; Wang, Q.; Li, Z.; Cai, S.; Liang, L.; Guo, K.; Xie, J.; et al. SESN2 protects against doxorubicin-induced cardiomyopathy via rescuing mitophagy and improving mitochondrial function. J. Mol. Cell. Cardiol. 2019, 133, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Pedrazzini, E.; Vitale, A. Protein Biosynthesis and Maturation in the ER. Methods Mol. Biol. 2018, 1691, 179–189. [Google Scholar] [CrossRef]
- Gülow, K.; Bienert, D.; Haas, I.G. BiP is feed-back regulated by control of protein translation efficiency. J. Cell Sci. 2002, 115, 2443–2452. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, J.; Yang, N.; Huang, Y.; Hu, T.; Rao, C. Endoplasmic reticulum stress-mediated cell death in liver injury. Cell Death Dis. 2022, 13, 1051. [Google Scholar] [CrossRef]
- Bourougaa, K.; Naski, N.; Boularan, C.; Mlynarczyk, C.; Candeias, M.M.; Marullo, S.; Fåhraeus, R. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Mol. Cell 2010, 38, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Corradetti, M.N.; Guan, K.-L. Upstream of the mammalian target of rapamycin: Do all roads pass through mTOR? Oncogene 2006, 25, 6347–6360. [Google Scholar] [CrossRef]
- Kim, J.S.; Ro, S.-H.; Kim, M.; Park, H.-W.; Semple, I.A.; Park, H.; Cho, U.-S.; Wang, W.; Guan, K.-L.; Karin, M.; et al. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci. Rep. 2015, 5, 9502. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; An, S.; Ro, S.-H.; Teixeira, F.; Park, G.J.; Kim, C.; Cho, C.-S.; Kim, J.-S.; Jakob, U.; Lee, J.H.; et al. Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains. Nat. Commun. 2015, 6, 10025. [Google Scholar] [CrossRef]
- Ro, S.-H.; Semple, I.A.; Park, H.; Park, H.; Park, H.-W.; Kim, M.; Kim, J.S.; Lee, J.H. Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1. FEBS J. 2014, 281, 3816–3827. [Google Scholar] [CrossRef]
- Bae, S.H.; Sung, S.H.; Oh, S.Y.; Lim, J.M.; Lee, S.K.; Park, Y.N.; Lee, H.E.; Kang, D.; Rhee, S.G. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013, 17, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Szinyákovics, J.; Keresztes, F.; Kiss, E.A.; Falcsik, G.; Vellai, T.; Kovács, T. Potent New Targets for Autophagy Enhancement to Delay Neuronal Ageing. Cells 2023, 12, 1753. [Google Scholar] [CrossRef]
- Klionsky, D.J. Autophagy revisited: A conversation with Christian de Duve. Autophagy 2008, 4, 740–743. [Google Scholar] [CrossRef]
- Djajadikerta, A.; Keshri, S.; Pavel, M.; Prestil, R.; Ryan, L.; Rubinsztein, D.C. Autophagy Induction as a Therapeutic Strategy for Neurodegenerative Diseases. J. Mol. Biol. 2020, 432, 2799–2821. [Google Scholar] [CrossRef]
- Chan, E.Y. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci. Signal. 2009, 2, pe51. [Google Scholar] [CrossRef]
- Doblado, L.; Lueck, C.; Rey, C.; Samhan-Arias, A.K.; Prieto, I.; Stacchiotti, A.; Monsalve, M. Mitophagy in Human Diseases. Int. J. Mol. Sci. 2021, 22, 3903. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Shaha, C. SESN2 facilitates mitophagy by helping Parkin translocation through ULK1 mediated Beclin1 phosphorylation. Sci. Rep. 2018, 8, 615. [Google Scholar] [CrossRef]
- Karagöz, G.E.; Acosta-Alvear, D.; Walter, P. The Unfolded Protein Response: Detecting and Responding to Fluctuations in the Protein-Folding Capacity of the Endoplasmic Reticulum. Cold Spring Harb. Perspect. Biol. 2019, 11, a033886. [Google Scholar] [CrossRef] [PubMed]
- Read, A.; Schröder, M. The Unfolded Protein Response: An Overview. Biology 2021, 10, 384. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Parmigiani, A.; Divakaruni, A.S.; Archer, K.; Murphy, A.N.; Budanov, A.V. Sestrin2 is induced by glucose starvation via the unfolded protein response and protects cells from non-canonical necroptotic cell death. Sci. Rep. 2016, 6, 22538. [Google Scholar] [CrossRef]
- Park, H.-J.; Yang, S.-G.; Koo, D.-B. SESN2/NRF2 signaling activates as a direct downstream regulator of the PERK pathway against endoplasmic reticulum stress to improve the in vitro maturation of porcine oocytes. Free Radic. Biol. Med. 2022, 178, 413–427. [Google Scholar] [CrossRef]
- Hsieh, A.C.; Liu, Y.; Edlind, M.P.; Ingolia, N.T.; Janes, M.R.; Sher, A.; Shi, E.Y.; Stumpf, C.R.; Christensen, C.; Bonham, M.J.; et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012, 485, 55–61. [Google Scholar] [CrossRef]
- Zou, Z.; Tao, T.; Li, H.; Zhu, X. mTOR signaling pathway and mTOR inhibitors in cancer: Progress and challenges. Cell Biosci. 2020, 10, 31. [Google Scholar] [CrossRef]
- Nathan, N.; Keppler-Noreuil, K.M.; Biesecker, L.G.; Moss, J.; Darling, T.N. Mosaic Disorders of the PI3K/PTEN/AKT/TSC/mTORC1 Signaling Pathway. Dermatol. Clin. 2017, 35, 51–60. [Google Scholar] [CrossRef]
- Chen, J.-S.; Wang, Q.; Fu, X.-H.; Huang, X.-H.; Chen, X.-L.; Cao, L.-Q.; Chen, L.-Z.; Tan, H.-X.; Li, W.; Bi, J.; et al. Involvement of PI3K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: Association with MMP-9. Hepatol. Res. 2009, 39, 177–186. [Google Scholar] [CrossRef]
- Chen, S.; Fisher, R.C.; Signs, S.; Molina, L.A.; Shenoy, A.K.; Lopez, M.-C.; Baker, H.V.; Koomen, J.M.; Chen, Y.; Gittleman, H.; et al. Inhibition of PI3K/Akt/mTOR signaling in PI3KR2-overexpressing colon cancer stem cells reduces tumor growth due to apoptosis. Oncotarget 2017, 8, 50476–50488. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Chen, L.; Zhao, L.; Xu, Y.; Peng, X.; Wang, X.; Ding, L.; Jin, J.; Teng, H.; Wang, Y.; et al. Ubiquitination of Rheb governs growth factor-induced mTORC1 activation. Cell Res. 2019, 29, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lu, H.; Bai, Y. Nrf2 in cancers: A double-edged sword. Cancer Med. 2019, 8, 2252–2267. [Google Scholar] [CrossRef] [PubMed]
- Mitsuishi, Y.; Taguchi, K.; Kawatani, Y.; Shibata, T.; Nukiwa, T.; Aburatani, H.; Yamamoto, M.; Motohashi, H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 2012, 22, 66–79. [Google Scholar] [CrossRef]
- Jain, A.; Lamark, T.; Sjøttem, E.; Larsen, K.B.; Awuh, J.A.; Øvervatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef] [PubMed]
- Wild, A.C.; Moinova, H.R.; Mulcahy, R.T. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 1999, 274, 33627–33636. [Google Scholar] [CrossRef] [PubMed]
- Patergnani, S.; Missiroli, S.; Morciano, G.; Perrone, M.; Mantovani, C.M.; Anania, G.; Fiorica, F.; Pinton, P.; Giorgi, C. Understanding the Role of Autophagy in Cancer Formation and Progression Is a Real Opportunity to Treat and Cure Human Cancers. Cancers 2021, 13, 5622. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; White, E. Role of autophagy in cancer prevention. Cancer Prev. Res. 2011, 4, 973–983. [Google Scholar] [CrossRef]
- Mathew, R.; Kongara, S.; Beaudoin, B.; Karp, C.M.; Bray, K.; Degenhardt, K.; Chen, G.; Jin, S.; White, E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007, 21, 1367–1381. [Google Scholar] [CrossRef]
- Mathew, R.; Karp, C.M.; Beaudoin, B.; Vuong, N.; Chen, G.; Chen, H.-Y.; Bray, K.; Reddy, A.; Bhanot, G.; Gelinas, C.; et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009, 137, 1062–1075. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gülow, K.; Tümen, D.; Kunst, C. The Important Role of Protein Kinases in the p53 Sestrin Signaling Pathway. Cancers 2023, 15, 5390. https://doi.org/10.3390/cancers15225390
Gülow K, Tümen D, Kunst C. The Important Role of Protein Kinases in the p53 Sestrin Signaling Pathway. Cancers. 2023; 15(22):5390. https://doi.org/10.3390/cancers15225390
Chicago/Turabian StyleGülow, Karsten, Deniz Tümen, and Claudia Kunst. 2023. "The Important Role of Protein Kinases in the p53 Sestrin Signaling Pathway" Cancers 15, no. 22: 5390. https://doi.org/10.3390/cancers15225390
APA StyleGülow, K., Tümen, D., & Kunst, C. (2023). The Important Role of Protein Kinases in the p53 Sestrin Signaling Pathway. Cancers, 15(22), 5390. https://doi.org/10.3390/cancers15225390



