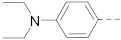
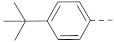
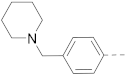
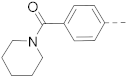
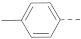
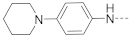
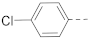
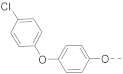
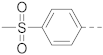
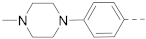
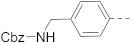
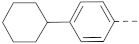
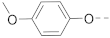
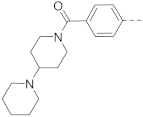
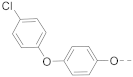
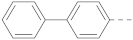
Simple Summary
Chromosomal translocations involving the mixed lineage leukemia (MLL) gene generate potent fusion oncogenes and cause acute myeloid leukemia or lymphocytic leukemia, which account for ~75% infant and 5–10% child/adult acute leukemia cases with a poor prognosis (5-year survival rates < 45%). Protein–protein interactions between the most frequent MLL fusion partner proteins AF9/ENL and AF4 or histone methyltransferase DOT1L are critical to malignant gene expression and are therefore a potential drug target for cancer. Compound screening followed by medicinal chemistry studies identified several novel small-molecule inhibitors showing strong inhibition of these protein–protein interactions, significant suppression of characteristic gene expression, and robust cellular anticancer activities with negligible cytotoxicity. These compounds are useful chemical probes for biological studies of these protein–protein interactions, as well as pharmacological leads for further drug development against MLL-rearranged and other leukemias.
Abstract
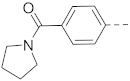
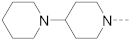
Chromosomal translocations involving the mixed lineage leukemia (MLL) gene cause 5–10% acute leukemias with poor clinical outcomes. Protein–protein interactions (PPI) between the most frequent MLL fusion partner proteins AF9/ENL and AF4 or histone methyltransferase DOT1L are drug targets for MLL-rearranged (MLL-r) leukemia. Several benzothiophene-carboxamide compounds were identified as novel inhibitors of these PPIs with IC50 values as low as 1.6 μM. Structure–activity relationship studies of 77 benzothiophene and related indole and benzofuran compounds show that a 4-piperidin-1-ylphenyl or 4-pyrrolidin-1-ylphenyl substituent is essential for the activity. The inhibitors suppressed expression of MLL target genes HoxA9, Meis1 and Myc, and selectively inhibited proliferation of MLL-r and other acute myeloid leukemia cells with EC50 values as low as 4.7 μM. These inhibitors are useful chemical probes for biological studies of AF9/ENL, as well as pharmacological leads for further drug development against MLL-r and other leukemias.
1. Introduction
Acute leukemias in ~75% infant and 5–10% child/adult patients are caused by chromosomal translocations of mixed lineage leukemia (MLL, also known as MLL1 or KMT2A) gene, which generate oncogenic fusion MLL (onco-MLL). MLL-rearranged (MLL-r) leukemias can be clinically characterized as acute lymphocytic leukemia (ALL) or acute myeloid leukemia (AML) with a poor prognosis [1,2,3,4,5]. Five-year survival rates for MLL-r ALL patients are <40%, in contrast to ~90% for other pediatric ALLs [6,7,8,9,10]. The situation is even worse for very young patients with five-year survivals of <20% [9]. MLL-r AML has 5-year survival rates of ~45%, similar to other AMLs [11,12]. More effective and/or less toxic targeted therapies are needed for patients with MLL-r leukemias [13,14,15,16].
The onco-MLL in these MLL-r leukemias consists of the N-terminal ~1400 amino acid residues of MLL, which function as a transcription factor and recognize MLL-target genes, in-frame fused with one of >70 other proteins [17,18,19]. However, only several fusion partners are predominant, including transcription cofactor AF4 (~35%), AF9 (25%) and its paralog ENL (10%) [1,2,11,17]. These proteins associate with each other and recruit other proteins (e.g., positive transcription elongation factor or P-TEFb) to form super elongation complexes (SEC) [20,21,22], which promote gene transcription elongation. Thus, certain MLL target genes, such as HoxA9 and Myc [23,24,25], are constitutively or excessively expressed, causing leukemia.
The C-terminal AHD domain (~70 residues) of AF9 and highly homologous ENL is a novel and validated drug target for MLL-r leukemia [11,26]. Although it is disordered by itself, AF9/ENL AHD forms a structured protein complex with a consensus peptide segment of LxVxIxLxxV/L in AF4 or its paralog AFF4 with a high affinity [27,28]. Such protein–protein interaction (PPI) is essential for the oncoprotein MLL-AF9/-ENL or MLL-AF4 to recruit SEC for aberrant gene expression. Moreover, similar interactions between AF9/ENL AHD and histone-H3 lysine-79 (H3K79) methyltransferase DOT1L (which also contains several LxVxIxLxxV/L sequences) can recruit DOT1L to MLL target gene loci, causing genome-wide H3K79 hypermethylation, which has been observed and characteristic to MLL-r leukemia [29]. Knockdown or pharmacological inhibition of DOT1L was found to selectively inhibit MLL-r leukemia in cells, animal models and clinical trials [30,31,32]. Therefore, disruption of the PPIs between AF9/ENL and AF4/AFF4 or DOT1L is a potentially useful therapy for MLL-r leukemia by suppressing SEC-mediated gene expression and DOT1L-caused H3K79 methylation [26].
Two series of compounds have been reported to inhibit the AF9/ENL-DOT1L interactions, including our previously disclosed compound SYC-1456 [26] and a series of 7-mer peptidomimetic compounds derived from DOT1L, such as Cpd-10 [33,34] (Chart 1). These compounds can inhibit AF9-DOT1L and other related PPIs and selectively suppress aberrant gene expression and cell proliferation of MLL-r leukemia, showing on-target activities, as well as a need for additional inhibitor discovery and development. Here, we report the discovery, synthesis, structure–activity relationships (SAR) and biological activities of a new chemo-type of inhibitors of the PPIs between AF9/ENL and DOT1L or AF4.
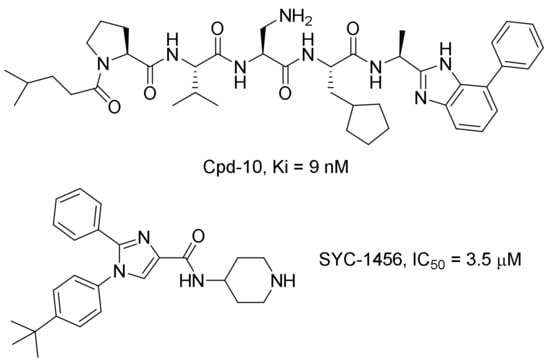
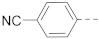
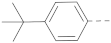
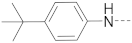
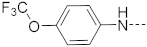
Chart 1.
Known inhibitors of the AF9/ENL-DOT1L interaction.
2. Results and Discussion
2.1. Discovery of Novel Inhibitors of the AF9-DOT1L Interaction
An AlphaLisa assay we previously developed [26] was used for compound screening for inhibitors of the AF9-DOT1L interaction. The PPI takes the DOT1L-peptide coated donor beads and AF9 AHD coated acceptor beads together. The donor beads are illuminated with a laser beam (680 nm) to generate singlet oxygen radicals, which activate the adjacent acceptor beads to produce luminescence at 615 nm. When a compound can disrupt such PPI and separate the donor and acceptor beads, the highly unstable radicals are rapidly quenched by water without generating luminescence. With this method, indole-carboxamide compounds 1 and 2 were found to be novel inhibitors of the AF9-DOT1L interaction, which can inhibit the PPI dose-dependently with an IC50 value of 3.3 and 4.5 μM, respectively (Table 1 and Supplementary Materials Figure S1), which showed comparable activities to SYC-1456 (IC50: 3.5 μM) [26]. Moreover, another reported inhibitor, Cpd-10 (Chart 1), was synthesized and found to exhibit an IC50 of 2.1 μM in our AlphaLisa assay conditions (Figure S1). These results support medicinal chemistry optimization based on the novel chemical scaffold in compounds 1 and 2.

Table 1.
Structures and activities of 6-substituted indole compounds against the AF9-DOT1L interaction.
2.2. Synthesis
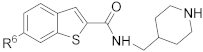
Medicinal chemistry based on the structures of these two compounds was next performed to investigate structure–activity relationships, as well as to find compounds with improved potency. Three series of indole- and closely related benzothiophene- and benzofuran-carboxamide compounds were synthesized for the study.
The general synthesis of the indole-carboxamide compounds is shown in Scheme 1. A 5- or 6-bromo-substituted indole-2-carboxylic acid ethyl ester 78 was hydrolyzed and then reacted with tert-butyloxycarbonyl (BOC)-protected 4-aminomethylpiperidine to give an indole-2-carboxamide intermediate compound 79, which was subjected to a Suzuki coupling reaction, followed by deprotection of the BOC, to give the target compounds 1, 3–11 and 15–22. To synthesize indole-3-carboxamide compounds, 5- or 6-bromo- or iodo-substituted indole 80 was reacted with trifluoroacetic anhydride to give 3-trifluoacetyl-indole 81, which was hydrolyzed to provide the corresponding indole-3-carboxylic acid 82. A similar amidation and Suzuki coupling reaction for compound 82 gave, upon BOC deprotection, the target compounds 2, 12–14 and 23.
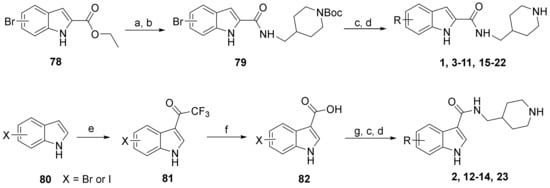
Scheme 1.
Synthesis of compounds 1–23. Reagents and conditions: (a) NaOH, THF-H2O, 50 °C, 5 h; (b) 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate, N,N-diisopropylethylamine, DMF, 4-aminomethyl-1-Boc-piperidine, 12 h; (c) aryl boronic acid or aryl 4,4,5,5-tetramethyl-1,3,2-dioxaborolane, tetrakis(triphenylphosphine)palladium, Na2CO3, 1,4-dioxane-H2O, 100 °C; (d) HCl (4 N in 1,4-dioxane), CH2Cl2, 0 °C; (e) trifluoroacetic anhydride, 0 °C to room temperature, 2 h; (f) NaOH (20% aq.), 80 °C, 2 days; (g) 4-aminomethyl-1-Boc-piperidine, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide, 1-hydroxybenzotriazole, triethylamine, CH2Cl2.
The synthesis of benzothiophene and benzofuran compounds is shown in Scheme 2. A bromo-substituted benzothiophene- or benzofuran-2-carboxylic acid 83 was coupled with BOC-protected 4-aminomethylpiperidine to provide 84, which was subjected to a Suzuki coupling reaction with an aryl boronic acid/ester, or a Buchwald-Hartwig amination reaction with an amine/phenol to give, after deprotection of the BOC, the target compounds 24–28, 31, 32, 35–50, 53–56, 58–62 and 67–77. The carboxylic acid 85, the Suzuki coupling product from compound 84, reacted with an amine to yield, upon removal of the BOC, the final compounds 29, 30, 33, 34, 51–52 and 63–66. Similar transformations from benzofuran-3-carboxylic acid 86 gave compound 57.
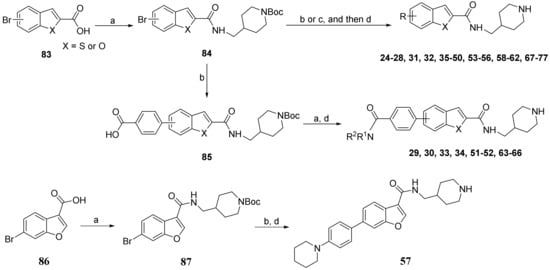
Scheme 2.
Synthesis of compounds 24–77. Reagents and conditions: (a) 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate, N,N-diisopropylethylamine, DMF, 4-aminomethyl-1-Boc-piperidine or another amine,12 h; (b) aryl boronic acid or aryl 4,4,5,5-tetramethyl-1,3,2-dioxaborolane, tetrakis(triphenylphosphine)palladium, Na2CO3, 1,4-dioxane-H2O, 100 °C; (c) a amine or phenol, tris(dibenzylideneacetone)dipalladium, 2-dicyclohexylphosphino-2′,6′-dimethoxybiphenyl, t-BuOH, toluene, 100 °C; (d) HCl (4 N in 1,4-dioxane), CH2Cl2, 0 °C.
2.3. Structure–Activity Relationships
Because of the high costs for the AlphaLisa assay, inhibitory activities of all compounds were first screened at 5 µM and the IC50 values for those exhibiting >50% inhibition were determined.
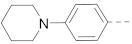
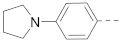
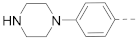
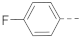
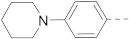
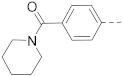
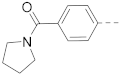
The structures and inhibitory activities of 6-substituted indole compounds, including compounds 1 and 2 (IC50 = 3.3 and 4.5 µM) with a piperidin-1-ylphenyl group, are summarized in Table 1. Ring-contracted compound 3 with a pyrrolidin-1-ylphenyl substituent was found to be equally potent, with an IC50 of 2.8 µM. Compound 4 without an N atom in the 6-substituent exhibited reduced activity (31% inhibition at 5 µM) and compound 5 having two N atoms is also weaker (40% inhibition). Replacing the piperidin-1-yl group with a thiophene in compound 6 (14% inhibition at 5 µM) significantly decreased activity. Changing to a 2-hydroxyethyl in compound 7 or to an acetyl group in 8 also resulted in a lowered potency. Compounds 9 and 10 with meta-substituted tert-butyl- and cyano-phenyl R6, respectively, are very weak (17% and 0% inhibition at 5 µM). Compound 11 with a smaller thiophene group at this position is almost inactive. In addition, three analogs of compound 2 with a 3-carboxylamide sidechain were synthesized. Compound 12 with a 4-pyrrolidin-1-ylphenyl R6 group was found to exhibit slightly improved activity with an IC50 of 2.9 µM. Compound 13 with another similar morpholin-4-yl group showed reduced activity (29% inhibition at 5 µM), while compound 14 with a polar aminomethyl group was almost inactive (11% inhibition at 5 µM).
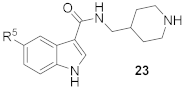
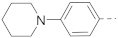
Compounds 15–23 (Table 2) with a variety of groups at the 5-position of the indole core were investigated. Compounds 15 and 16 with the 4-piperidin-1-ylphenyl and 4-pyrrolidin-1-ylphenyl R5 group, respectively, showed comparable activities (IC50 = 3.6 and 2.7 µM) to compounds 1–3 and 12. Other groups in compounds 17–22 showed less potent activities. While having a 4-piperidin-1-ylphenyl R5 group, compound 23 with a 3-carboxylamide sidechain showed reduced activity (43% inhibition at 5 µM).

Table 2.
Structures and activities of 5-substituted indole compounds against the AF9-DOT1L interaction.
Results of the above indole compounds show that in most cases, 4-piperidin-1-ylphenyl or similar 4-pyrrolidin-1-ylphenyl group contributes favorably to inhibition of the AF9-DOT1L interaction, regardless whether they are in the 6- or 5-position. However, despite their strong biochemical activities, these indole-containing inhibitors showed only modest activities to inhibit the proliferation of MLL-r leukemia cells (described below), which might be due to poor cell permeability or uptake of these compounds.
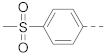
Next, a structurally similar, less polar benzothiophene core was used to replace indole. The structures and inhibitory activities of 6-substituted benzothiophene compounds are shown in Table 3. Compound 24 with a 4-piperidin-1-ylphenyl group was found to be a strong inhibitor of the AF9-DOT1L interaction with an IC50 of 1.6 µM, showing ~2× activity compared to the indole analog 1. Compound 25 with a 4-pyrrolidin-1-ylphenyl had weaker activity (IC50 = 5.3 µM). Changing to a morpholine in compound 26 (40% inhibition at 5 µM) or a diethylamino group in 27 (25% inhibition at 5 µM) considerably reduced inhibitory activity. Adding a -CH2- (in compound 28) or a carbonyl (in 29 and 30) between the piperidinyl/pyrrolidine and phenyl groups lost activity (0–8% inhibition at 5 µM). Replacing the phenyl group with a non-aromatic piperidine ring yielded very weak compounds 31 and 32 (14% and 20% inhibition at 5 µM). Inserting a carbonylphenyl into these two compounds produced inactive compounds 33 and 34. Compound 35 with an additional -NH- between the 4-piperidin-1-ylphenyl group and benzothiophene core was found to be a strong inhibitor (IC50 = 2.3 µM). Compound 36 having a 4-(4-chlorophenoxy)phenoxy R6 group is a moderate inhibitor (IC50~5 µM). In addition, a variety of substituted phenyl R6 groups in compounds 37–49 with different steric and electronic properties resulted in inactive to moderate inhibitors.

Table 3.
Structures and activities of 6-substituted benzothiophene compounds against the AF9-DOT1L interaction.
5-Substituted benzothiophene compounds 50–56 were synthesized and tested for SAR studies. As shown in Table 4, similar SAR results were observed with compound 50 having a 4-piperidin-1-ylphenyl R5 group showing the strongest activity (IC50 = 2.5 µM), while other compounds showed no to moderate activities.

Table 4.
Structures and activities of 5-substituted benzothiophene compounds against the AF9-DOT1L interaction.
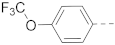
Moreover, benzofuran compounds 57–77, which possess subtle structural (e.g., angles between substituents), electronic or hydrophobic differences from indole or benzothiophene analogs, were synthesized for the SAR studies. As shown in Table 5, similar SARs were observed, but benzofuran inhibitors appeared to be less potent. The strongest benzofuran compounds 57 with a 4-piperidin-1-ylphenyl and 74 with a 4-piperidin-1-ylphenylamino R6 group had IC50 values of 7.2 and 4.6 µM, respectively. Replacing the 4-piperidin-1-ylphenyl group with a closely related morpholine (in compound 58), piperazine (in 59/60), cyclohexyl (in 61), or diethylamino group (in 62) significantly reduced the inhibitory activity. Activities of compounds 63–66 indicate that an amide-containing R6 group is disfavored. Other R6 groups in compounds 67–73 and 75–77 also showed no to modest activities against the AF9-DOT1L interaction.

Table 5.
Structures and activities of 6-substituted benzofuran compounds against the AF9-DOT1L interaction.
2.4. Activity Confirmation by a Pull-Down Assay
A pull-down assay [26] was used to confirm that the strong inhibitors could disrupt the AF9-DOT1L interaction. Streptavidin agarose beads coated with biotinylated DOT1L peptide were used to pull-down the protein AF9 AHD in the solution, followed by thorough washing, SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and Western blotting to visualize and quantitate the protein. As shown in Figure 1, inhibitors 24 and 50 (IC50 = 1.6 and 2.5 µM) can dose-dependently reduce AF9 AHD, while inactive compound 37 did not significantly affect the binding of the protein to the DOT1L-coated beads even at 50 µM. This alternative, non-optical method confirmed that compounds 24 and 50 can inhibit the PPI between AF9 and DOT1L.
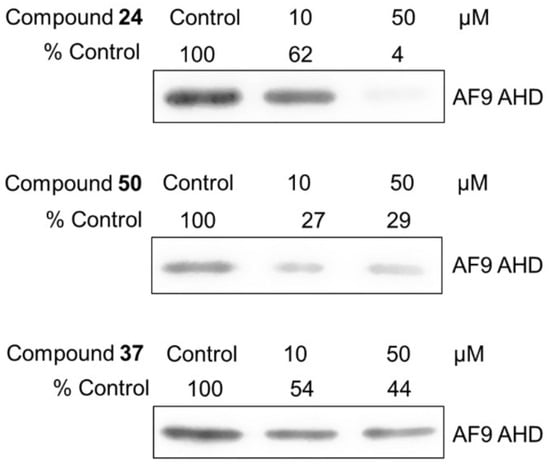
Figure 1.
Pull-down assay indicated that compounds 24 and 50 significantly reduced the amounts of AF9 AHD bound to DOT1L peptide-coated resins in a dose-dependent manner, while inactive compound 37 did not significantly affect the AF9 AHD levels. The uncropped bolts are shown in Supplementary Materials.
2.5. Activity to Block the AF9/ENL-AF4 Interactions
Transcription cofactor AF4, which is the most frequent (~35%) fusion protein in MLL1-r leukemia, is another binding partner of AF9/ENL [35,36,37]. The AF9/ENL-AF4 interactions are essential for the formation of SEC, which causes aberrant gene expression in MLL-r leukemia and other AMLs [20,28]. Using our previously developed AlphaLisa assays [26], selected compounds were tested for their ability to inhibit the PPIs between AF9 or ENL and AF4. As summarized in Table 6, inhibitors 16, 24, 25 and 50 of the AF9-DOT1L interaction (IC50 = 1.6–5.3 µM) were found to exhibit comparable activities to block the AF9-AF4 and ENL-AF4 interactions with IC50 values of 1.9–7.9 µM, while inactive compounds 22 and 37 did not inhibit these PPIs. These results are consistent with our previous finding that an inhibitor of the AF9-DOT1L interaction is broadly active against PPIs between AF9/ENL and AF4 [26].

Table 6.
Activities (IC50, μM) against the AF9/ENL AHD-AF4 interactions.
2.6. Activity to Suppress Oncogenic Gene Expression
The major function of the SEC is to promote transcription elongation for its bound genes [20,21,22]. Because of the high affinity of the PPIs between AF9/ENL and AF4, onco-MLL (e.g., MLL-AF4 or MLL-AF9/-ENL) recruits SEC to MLL target genes, facilitates their expression, and eventually causes leukemia initiation [27,28]. Next, we investigated whether selected inhibitors of these PPIs can inhibit the expression of HoxA9, Meis1 and Myc, which are MLL target genes and their high expression is characteristic of MLL-r leukemia. In addition, high expression of Myc is also observed in a broad range of AMLs and other cancers and is considered a driving factor for oncogenesis [38,39,40,41]. To this end, RNAs from the control and compound-treated Molm-13 cells (harboring MLL-AF9) were extracted and subjected to quantitative PCR. As shown in Figure 2, potent inhibitors 24 and 50 were able to reduce the expression of HoxA9, Meis1 and Myc. However, inactive compound 37 did not significantly affect the expression of these MLL target genes.
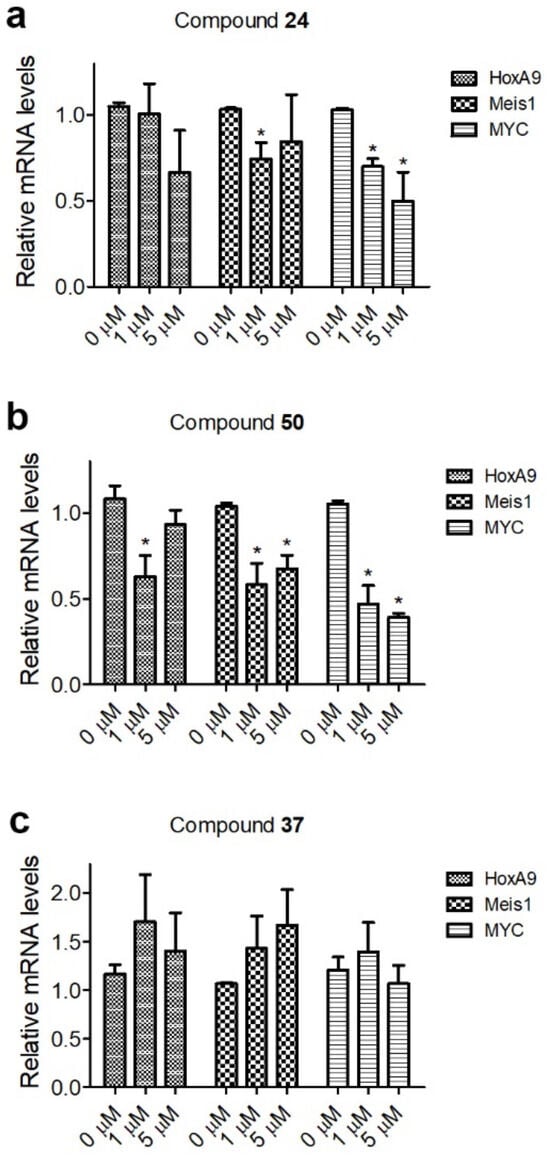
Figure 2.
RT-qPCR results of treatment of Molm-13 cells with compounds 24 (a), 50 (b) or 37 (c) at the designated concentrations for 4 days, showing 24 and 50, but not inactive compound 37, inhibited expression of representative MLL target genes HoxA9, Meis1 and Myc at 1 and 5 µM in Molm-13 cells. (* p < 0.05).
2.7. Antitumor Activity
The antitumor activity of selected compounds was evaluated against the proliferation of a panel of MLL-r leukemia and other cancer cells. The results are shown in Table 7. MV4;11 and Molm-13 leukemia cells contain MLL-AF4 and MLL-AF9 fusion oncogenes, respectively. NB4 and HL60 are AML cells without MLL translocation. Hela (cervical cancer) is a solid tumor cell line. Indole-containing compounds 2, 12 and 15 were found to exhibit modest to no activities against proliferation of MLL-r leukemia MV4;11 and Molm-13 cells, despite their strong inhibitory activities against the AF9-DOT1L interaction (IC50 = 2.9–4.5 μM) (Figure S2). They did not significantly affect the growth of other cancerous cells. Presumably due to improved cell permeability or uptake, benzothiophene-containing inhibitor 24 (IC50 = 1.6 μM) showed more potent activities against proliferation of MLL-r leukemia MV4;11 and Molm-13 and AML NB4 and HL60 cells with EC50 values of 9.6, 4.7, 5.3 and 10 μM. Benzothiophene compound 25 had generally reduced antitumor activities with EC50s of 8.7–26 μM, in line with its reduced biochemical activity (IC50 = 5.3 μM). Another benzothiophene inhibitor 50 (IC50 = 2.5 μM) exhibited comparable antitumor activities (EC50 = 5.1–9.4 μM) to compound 24. The anti-proliferation activity of these compounds against other AML cells (i.e., NB4 and HL60) might be due to their inhibition of SEC-mediated gene expression (e.g., Myc), as observed in our previous studies [26]. In addition, these three compounds were found to have weak to no activities against the proliferation of Hela cells, showing good selectivity. Benzofuran compound 57 (IC50 = 7.2 μM) moderately inhibited the proliferation of MLL-r leukemia and AML cells with EC50s of 9.7–21 μM. It did not affect the growth of Hela cells. Moreover, inactive indole, benzothiophene and benzofuran compounds 22, 37 and 72 were included in this assay as negative controls. None of these three compounds inhibited tumor cell proliferation, largely excluding possible off-target effects for these series of compounds.

Table 7.
Antitumor activity (EC50, μM) of selected compounds.
3. Materials and Methods
All chemicals for synthesis were purchased from Aldrich (Milwaukee, WI, USA) or Alfa Aesar (Ward Hill, MA, USA). Unless otherwise stated, all solvents and reagents were used as received. All reactions were performed using a Teflon-coated magnetic stir bar at the indicated temperature and were conducted under an inert atmosphere when stated. The identity of the synthesized compounds was characterized by 1H and 13C NMR on a Varian (Palo Alto, CA, USA) 400-MR spectrometer and mass spectrometer (Shimadzu LCMS-2020, Shimadzu Kyoto, Japan). Chemical shifts were reported in parts per million (ppm, δ) downfield from tetramethylsilane. Proton coupling patterns are described as singlet (s), doublet (d), triplet (t), quartet (q), multiplet (m), and broad (br). The identity of the potent inhibitors was confirmed with high resolution mass spectra (HRMS) using an Agilent 6550 iFunnel quadrupole-time-of-flight (Q-TOF) mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) with electrospray ionization (ESI). The purities of the final compounds were determined to be >95% with a Shimadzu Prominence HPLC (Shimadzu, Tokyo, Japan) using a Zorbax C18 (or C8) column (4.6 × 250 mm) (Agilent Technologies, Santa Clara, CA, USA) monitored by UV at 254 nm.
3.1. Chemical Synthesis
Synthesis of compounds 1, 5, 6, 7, 11, 14, 15, 16, 17, 18, 19, 21 and 22 was reported in our previous publications [42].
3.1.1. General Synthetic Procedure-A for Compounds 3, 4, 8–10 and 20
To a solution of 5- or 6-bromoindole-2-carboxylic ester 78 (2.0 g, 7.46 mmol) in THF/H2O (10/10 mL), sodium hydroxide (0.90 g, 22.4 mmol) was added slowly. The resulting mixture was stirred and heated at 50 °C for 5 h. After cooling down to room temperature, the solution was removed in a vacuum. The resulting residues were diluted with H2O (10 mL) and then acidified with 3 N hydrochloride acid solution to give a white precipitate, which was filtered, washed with cold H2O and dried under a vacuum overnight to give the corresponding acid as a white solid, which was used directly for the next step.
The reaction mixture containing crude indole-2-carboxylic acid (1.5 g, 6.25 mmol), 4-aminomethyl-1-Boc-piperidine (1.47 g, 6. 88 mmol), or 4-amino-1-Boc-piperidine (1.38 g, 6.89 mmol), HATU (2. 85 g, 7.5 mmol) and N, N-diisopropylethylamine (1.84 mL, 12.5 mmol) in DMF (20 mL) was stirred for 12 h at room temperature before quenched with H2O (30 mL). The crude product was extracted with ethyl acetate (3 × 30 mL) and the resulting organic phase was washed with brine (3 × 10 mL) and dried over Na2SO4. Up on removal of the organic solvents, it was purified with column chromatography (silica gel, hexanes: ethyl acetate from 1.5:1 to 1:1), to give compound 79 in a yield of 62–85%.
The reaction mixture containing indole-2-carboxamide 79 (0.2 mmol), aryl boronic acid or aryl-4,4,5,5-tetramethyl-1,3,2-dioxaborolane (0.24 mmol), tetrakis(triphenylphosphine)palladium (11.6 mg, 0.01 mmol), and sodium carbonate (42 mg, 0.4 mmol) in p-dioxane/H2O (5/1, v/v, 6 mL) was heated at 100 °C for 16 h. Upon cooling, it was diluted with brine (5 mL), and the crude product was extracted with ethyl acetate (3 × 15 mL). The organic layers were washed with brine (3 × 10 mL), dried over Na2SO4, and concentrated. The residue was purified with column chromatography (silica gel, hexanes: ethyl acetate from 8:1 to 1:1) to give the corresponding Suzuki coupling product (58–83% yield), which was deprotected in a solution of dichloromethane (3 mL) containing 4N HCl in p-dioxane (0.2 mL) at 0 °C to room temperature for 6 h. Upon filtration, the powder was washed with cold water to give the final compounds 3, 4, 8, 9, 10 and 20 as a hydrochloric salt (~100% yield).
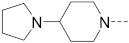
6-(4-(1-Pyrrolinylphenyl)-N-(piperidin-4-ylmethyl)-1H-indole-2-carboxamide hydrochloride (3)
1H NMR (400 MHz, DMSO-d6) δ 11.66 (s, 1H), 9.02 (s, 1H), 8.71 (s, 2H), 7.64 (d, J = 8.3 Hz, 1H), 7.61–7.54 (m, 3H), 7.31 (d, J = 8.3 Hz, 1H), 7.15 (s, 1H), 7.00 (s, 2H), 3.39 (s, 4H), 3.29–3.15 (m, 4H), 2.82 (dd, J = 21.3, 10.4 Hz, 2H), 2.03 (s, 4H), 1.83 (d, J = 12.3 Hz, 3H), 1.41 (dd, J = 24.3, 12.7 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 145.1, 137.6, 136.0, 132.5, 127.9, 126.3, 122.2, 119.4, 115.6, 109.4, 103.1, 50.6, 43.92, 43.2, 34.2, 26.7, 24.9. MS (ESI): [M + H]+ 403.2.
6-(4-Cyclohexylphenyl)-N-(piperidin-4-ylmethyl)-1H-indole-2-carboxamide hydrochloride (4)
1H NMR (400 MHz, DMSO-d6) δ 11.72 (s, 1H), 9.05 (d, J = 8.8 Hz, 1H), 8.82–8.68 (m, 2H), 7.66 (d, J = 8.4 Hz, 1H), 7.62 (s, 1H), 7.54 (d, J = 8.0 Hz, 2H), 7.32 (d, J = 8.8 Hz, 1H), 7.29 (d, J = 8.0 Hz, 2H), 7.18 (s, 1H), 3.23 (dd, J = 14.8, 9.2 Hz, 4H), 2.82 (q, J = 11.2 Hz, 2H), 1.90–1.66 (m, 9H), 1.49–1.31 (m, 7H). 13C NMR (100 MHz, DMSO-d6) δ 161.1, 146.2, 138.8, 137.1, 135.7, 132.3, 127.2, 126.6, 126.3, 121.8, 119.2, 109.8, 102.6, 43.4, 42.8, 34.0, 33.8, 26.4, 26.3, 25.6, 24.7. MS (ESI): [M + H]+ 416.3.
6-(4-Acetophenyl)-N-(piperidin-4-ylmethyl)-1H-indole-2-carboxamide hydrochloride (8)
1H NMR (400 MHz, DMSO-d6) δ 11.82 (s, 1H), 8.98 (d, J = 10.0 Hz, 1H), 8.76 (t, J = 5.6 Hz, 1H), 8.68 (d, J = 10.0 Hz, 1H), 8.04 (d, J = 8.4 Hz, 2H), 7.81 (d, J = 8.4 Hz, 2H), 7.73 (d, J = 8.8 Hz, 2H), 7.43 (dd, J = 8.4, 1.6 Hz, 1H), 7.20 (d, J = 1.6 Hz, 1H), 3.23 (dd, J = 15.6, 9.6 Hz, 4H), 2.83 (dd, J = 22.8, 11.6 Hz, 2H), 2.61 (s, 3H), 1.84 (d, J = 12.0 Hz, 3H), 1.42 (dd, J = 23.2, 11.2 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 197.5, 161.0, 145.7, 136.9, 135.1, 134.2, 133.0, 129.0, 127.2, 126.8, 122.2, 119.3, 110.6, 102.7, 43.6, 42.8, 33.8, 26.8, 26.3. MS (ESI): [M+H]+ 375.2.
6-(3-(tert-Butyl)phenyl)-N-(piperidin-4-ylmethyl)-1H-indole-2-carboxamide hydrochloride (9)
1H NMR (400 MHz, DMSO-d6) δ 11.69 (s, 1H), 9.05 (s, 1H), 8.74 (s, 2H), 7.68 (d, J = 8.4 Hz, 1H), 7.63 (d, J = 11.6 Hz, 2H), 7.46–7.31 (m, 4H), 7.19 (s, 1H), 3.30–3.18 (m, 4H), 2.83 (dd, J = 23.2, 12.0 Hz, 2H), 1.84 (d, J = 12.0 Hz, 3H), 1.42 (d, J = 12.8 Hz, 2H), 1.34 (s, 9H). 13C NMR (100 MHz, DMSO-d6) δ 161.1, 151.2, 141.1, 137.0, 136.3, 132.4, 128.6, 126.4, 124.1, 123.8, 123.6, 121.9, 119.5, 110.2, 102.6, 43.5, 42.8, 34.5, 33.8, 31.2, 26.3. MS (ESI): [M + H]+ 389.2.
6-(3-Cyanophenyl)-N-(piperidin-4-ylmethyl)-1H-indole-2-carboxamide hydrochloride (10)
1H NMR (400 MHz, DMSO-d6) δ 11.87 (s, 1H), 9.08 (d, J = 9.6 Hz, 1H), 8.80 (t, J = 5.8 Hz, 2H), 8.11 (s, 1H), 8.00 (d, J = 7.6 Hz, 1H), 7.79 (d, J = 7.6 Hz, 1H), 7.76–7.63 (m, 3H), 7.41 (d, J = 8.4 Hz, 1H), 7.22 (s, 1H), 3.23 (dd, J = 14.0, 8.8 Hz, 4H), 2.83 (q, J = 11.6 Hz, 2H), 1.84 (d, J = 12.8 Hz, 3H), 1.43 (q, J = 11.6 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.0, 142.4, 136.9, 133.4, 133.0, 131.6, 130.4, 130.2, 130.2, 127.1, 122.2, 119.2, 118.9, 112.0, 110.5, 102.7, 43.5, 42.8, 33.8, 26.3. MS (ESI): [M + H]+ 358.2.
5-(3-Fluoro-4-cyanophenyl)-N-(piperidin-4-ylmethyl)-1H-indole-2-carboxamide hydrochloride (20)
1H NMR (400 MHz, DMSO-d6) δ 11.82 (s, 1H), 8.79–8.68 (m, 2H), 8.42 (dt, J = 10.8, 8.4 Hz, 1H), 8.12 (s, 1H), 7.96 (t, J = 7.6 Hz, 1H), 7.90 (dd, J = 11.2, 1.2 Hz, 1H), 7.78 (dd, J = 8.4, 1.6 Hz, 1H), 7.64 (dd, J = 8.8, 1.6 Hz, 1H), 7.53 (d, J = 8.8 Hz, 1H), 7.25 (d, J = 1.2 Hz, 1H), 3.28–3.20 (m, 4H), 2.91–2.79 (m, 2H), 1.84 (d, J = 12.0 Hz, 3H), 1.38 (q, J = 12.0 Hz, 2H). MS (ESI): [M + H]+ 377.2.
3.1.2. Synthetic Methods for Compounds 2, 12, 13 and 23
To a solution of 5- or 6-substituted indole 80 (3 mmol) in DMF (10 mL) was slowly added to trifluoroacetyl anhydride (0.61 mL, 4.5 mmol) at 0 °C. The resulting mixture was stirred at room temperature for 2 h before quenched with H2O (10 mL) to give a white powder, which was filtered and washed with cold water to give compound 81, which was stirred in a 20% NaOH solution (20 mL) at 50 °C for 2 days. The reaction was slowly acidified with 3 N HCl solution to give a white powder, which was filtered and washed with cold water to give the corresponding indole-3-carboxylic acid, which was directly used without further purification as the starting compound for the general synthetic procedure-A (described above) to give the final compounds 2, 12, 13 and 23.
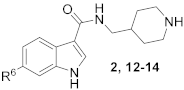
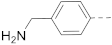
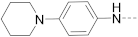
6-(4-(Piperidin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-1H-indole-3-carboxamide hydrochloride (2)
1H NMR (400 MHz, DMSO-d6) δ 11.77 (s, 1H), 8.93 (d, J = 10.6 Hz, 1H), 8.64 (d, J = 8.6 Hz, 1H), 8.18 (d, J = 8.5 Hz, 1H), 8.08 (dd, J = 25.9, 12.8 Hz, 2H), 7.92 (d, J = 7.8 Hz, 2H), 7.84 (d, J = 8.5 Hz, 2H), 7.69 (s, 1H), 7.42 (d, J = 8.4 Hz, 1H), 3.22 (d, J = 12.1 Hz, 2H), 3.18–3.07 (m, 2H), 2.79 (dd, J = 23.1, 11.7 Hz, 3H), 1.80 (d, J = 12.2 Hz, 4H), 1.48–1.24 (m, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.0, 142.7, 142.3, 137.1, 132.9, 129.4, 128.4, 126.6, 122.6, 122.0, 120.1, 111.0, 110.5, 66.8, 55.4, 43.7, 43.2, 34.6, 34.4, 26.8, 23.3, 21.3. HRMS (ESI+) calcd for C26H32N4O [M + H]+, 417.2654; found, 417.2650.
6-(4-(Pyrrolidin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-1H-indole-3-carboxamide hydrochloride (12)
1H NMR (400 MHz, DMSO-d6) δ 11.63 (s, 1H), 8.96 (d, J = 10.7 Hz, 1H), 8.66 (d, J = 10.1 Hz, 1H), 8.19–7.88 (m, 3H), 7.69–7.46 (m, 3H), 7.34 (d, J = 8.2 Hz, 1H), 7.02 (d, J = 36.4 Hz, 2H), 3.53 (s, 2H), 3.36 (s, 3H), 3.22 (d, J = 11.9 Hz, 2H), 3.14 (s, 2H), 2.79 (dd, J = 22.3, 11.9 Hz, 2H), 2.00 (s, 3H), 1.80 (d, J = 12.2 Hz, 3H), 1.37 (dd, J = 23.6, 11.7 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 165.1, 145.0, 137.3, 134.5, 128.6, 128.1, 127.9, 125.4, 121.7, 119.6, 115.6, 110.9, 109.1, 66.8, 43.6, 43.3, 34.4, 26.8, 24.9. HRMS (ESI+) calcd for C25H30N4O [M + H]+, 403.2498; found, 403.2480.
6-(4-(Morpholinophenyl)-N-(piperidin-4-ylmethyl)-1H-indole-3-carboxamide hydrochloride (13)
1H NMR (400 MHz, DMSO-d6) δ 11.62 (s, 1H), 8.86 (d, J = 9.9 Hz, 1H), 8.55 (d, J = 11.1 Hz, 1H), 8.13 (d, J = 8.4 Hz, 1H), 8.04 (d, J = 2.8 Hz, 2H), 7.68–7.52 (m, 3H), 7.36 (d, J = 8.4 Hz, 1H), 7.21 (d, J = 7.9 Hz, 2H), 3.99–3.75 (m, 5H), 3.32–3.19 (m, 5H), 3.15 (t, J = 5.1 Hz, 2H), 2.81 (dd, J = 23.0, 11.1 Hz, 2H), 1.81 (d, J = 11.2 Hz, 3H), 1.48–1.29 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 165.1, 137.3, 134.2, 128.7, 127.8, 125.6, 121.8, 119.8, 117.2, 111.0, 110.0, 109.4, 66.0, 50.0, 43.6, 43.3, 34.4, 26.8. MS (ESI): [M + H]+ 419.2.
5-(4-(Piperidin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-1H-indole-3-carboxamide hydrochloride (23)
1H NMR (400 MHz, DMSO-d6) δ 11.75 (s, 1H), 8.91 (s, 1H), 8.64 (s, 1H), 8.41 (s, 1H), 8.19–8.04 (m, 2H), 7.92 (d, J = 7.5 Hz, 2H), 7.80 (d, J = 8.6 Hz, 2H), 7.48 (dt, J = 8.6, 4.9 Hz, 2H), 3.55 (d, J = 11.6 Hz, 10H), 3.22 (d, J = 11.5 Hz, 2H), 3.16 (t, J = 5.7 Hz, 2H), 2.80 (dd, J = 22.4, 11.4 Hz, 2H), 1.80 (d, J = 11.8 Hz, 3H), 1.38 (dd, J = 23.4, 11.3 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 165.0, 136.4, 131.6, 129.2, 128.4, 127.1, 122.4, 121.7, 119.8, 112.9, 111.3, 66.8, 55.4, 43.7, 43.3, 34.4, 26.8, 23.4. MS (ESI): [M + H]+ 417.3.
3.1.3. Synthetic Methods for Compounds 24–28, 37–50, 53, 54, 58–62, and 67–73
General synthetic procedure-A was used for the synthesis of compounds 24–28, 37–50, 53, 54, 58–62 and 67–73, starting from a Br- or I-substituted benzothiophene- or benzofuran-2-carboxylic acid 83.
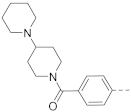
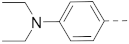
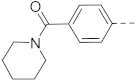
6-(4-(Piperidin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (24)
1H NMR (500 MHz, CD3OD) δ 8.23 (s, 1H), 8.05 (s, 1H), 8.02 (d, J = 8.4 Hz, 1H), 7.97–7.93 (m, 2H), 7.84–7.80 (m, 2H), 7.74 (dd, J = 8.4, 1.6 Hz, 1H), 3.72–3.66 (m, 4H), 3.46–3.41 (m, 2H), 3.39–3.36 (m, 2H), 3.05–2.97 (m, 2H), 2.17–2.08 (m, 4H), 2.05–1.98 (m, 3H), 1.88–1.80 (m, 2H), 1.60–1.50 (m, 2H). 13C NMR (126 MHz, CD3OD) δ 164.9, 143.7, 143.2, 143.1, 140.9, 140.5, 138. 7, 130.3, 126.9, 126.3, 125.5, 122.8, 122.0, 58.3, 45.5, 44.9, 35.4, 27.7, 24.8, 22.2. HRMS (ESI+) calcd for C26H31N3O3S [M + H]+, 434.2266; found, 434.2261.
6-(4-(Pyrrolindin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (25)
1H NMR (500 MHz, DMSO-d6) δ 9.06 (br, 1H), 8.98–8.90 (m, 1H), 8.76 (br, 1H), 8.23 (s, 1H), 8.13 (s, 1H), 7.93 (d, J = 8.4 Hz, 1H), 7.74–7.65 (m, 3H), 6.89–6.80 (m, 2H), 3.39–3.31 (m, 4H), 3.28–3.21 (m, 2H), 3.19 (t, J = 6.0 Hz, 2H), 2.88–2.78 (m, 2H), 2.06–1.95 (m, 4H), 1.88–1.78 (m, 3H), 1.48–1.35 (m, 2H). 13C NMR (126 MHz, DMSO-d6) δ 161.6, 141.2, 139.3, 138.2, 137.3, 127.6, 125.2, 124.5, 123.2, 118.9, 43.9, 42.6, 33.6, 30.6, 26.2, 24.6. HRMS (ESI+) calcd for C25H29N3OS [M + H]+, 420.2110; found, 420.2096.
6-(4-(Morpholino)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (26)
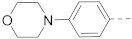
1H NMR (400 MHz, DMSO-d6) δ 8.98–8.88 (m, 2H), 8.68–8.55 (m, 1H), 8.26 (s, 1H), 8.12 (s, 1H), 7.94 (d, J = 8.4 Hz, 1H), 7.75–7.65 (m, 3H), 7.15 (d, J = 8.2 Hz, 2H), 3.83–3.75 (m, 4H), 3.28–3.16 (m, 8H), 2.89–2.75 (m, 2H), 1.88–1.75 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 141.1, 139.7, 137.9, 137.7, 127.6, 125.3, 124.5, 123.5, 119.5, 116.0, 65.7, 48.7, 44.0, 42.8, 33.6, 26.2. MS (ESI): [M + H]+ 436.2.
6-(4-(Diethylamino)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (27)
1H NMR (400 MHz, DMSO-d6) δ 10.60–10.45 (m, 1H), 8.97 (t, J = 5.6 Hz, 1H), 8.87 (br, 1H), 8.58 (br, 1H), 8.39 (s, 1H), 8.16 (s, 1H), 8.02 (d, J = 8.4 Hz, 1H), 7.87 (d, J = 8.2 Hz, 2H), 7.83–7.71 (m, 3H), 3.29–3.15 (m, 4H), 3.12–2.99 (m, 4H), 2.92–2.76 (m, 2H), 1.92–1.75 (m, 3H), 1.44–1.34 (m, 2H), 1.26 (t, J = 7.2 Hz, 6H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 141.1, 140.7, 140.4, 138.7, 137.2, 131.8, 129.7, 127.3, 125.6, 124.4, 124.0, 120.8, 45.7, 44.0, 42.8, 33.7, 26.3, 8.3. MS (ESI): [M + H]+ 422.2.
6-(4-(Piperidin-1-ylmethyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (28)
1H NMR (400 MHz, DMSO-d6) δ 10.69 (br, 1H), 9.05–8.93 (m, 2H), 8.73–8.60 (m, 1H), 8.38 (s, 1H), 8.17 (s, 1H), 8.02 (d, J = 8.4 Hz, 1H), 7.86 (d, J = 8.1 Hz, 2H), 7.78 (d, J = 8.4 Hz, 1H), 7.72 (d, J = 8.1 Hz, 2H), 4.35–4.23 (m, 2H), 3.32–3.14 (m, 6H), 2.91–2.73 (m, 4H), 1.90–1.62 (m, 8H), 1.45–1.27 (m, 3H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 141.1, 140.7, 140.5, 138.7, 137.2, 132.1, 129.2, 127.2, 125.6, 124.5, 124.1, 120.8, 58.4, 51.6, 44.0, 42.8, 33.7, 26.3, 22.1, 21.4. MS (ESI): [M + H]+ 448.2.
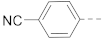
6-(4-(Carbamoyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (37)
1H NMR (400 MHz, DMSO-d6) δ 9.05–8.96 (m, 2H), 8.75–8.65 (m, 1H), 8.42 (s, 1H), 8.18 (s, 1H), 8.09–7.95 (m, 4H), 7.86 (d, J = 8.4 Hz, 2H), 7.80 (dd, J = 8.4, 1.4 Hz, 1H), 7.41 (s, 1H), 3.30–3.16 (m, 4H), 2.89–2.75 (m, 2H), 1.95–1.80 (m, 3H), 1.48–1.32 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 167.4, 161.6, 142.2, 141.1, 140.8, 138.8, 137.1, 133.2, 128.2, 126.7, 125.6, 124.5, 124.1, 121.0, 44.0, 42.8, 33.7, 26.3. MS (ESI): [M + H]+ 394.2.
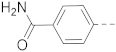
6-(4-(Acetamido)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (38)
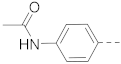
1H NMR (400 MHz, DMSO-d6) δ 10.12 (s, 1H), 8.91 (t, J = 5.3 Hz, 1H), 8.86–8.75 (m, 1H), 8.58–8.42 (m, 1H), 8.28 (s, 1H), 8.12 (s, 1H), 7.97 (d, J = 8.4 Hz, 1H), 7.70–7.50 (m, 5H), 3.30–3.14 (m, 4H), 2.90–2.74 (m, 2H), 2.07 (s, 3H), 1.89–1.75 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 168.4, 161.7, 141.1, 140.0, 139.1, 138.0, 134.0, 127.2, 126.3, 125.4, 124.5, 123.7, 119.9, 119.3, 44.0, 42.8, 33.7, 26.3, 24.1. MS (ESI): [M + H]+ 408.2.
6-(4-(Hydroxylmethyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (39)
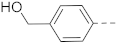
1H NMR (400 MHz, DMSO-d6) δ 8.95–8.88 (m, 1H), 8.80–8.68 (m, 1H), 8.49–8.37 (m, 1H), 8.32 (s, 1H), 8.12 (s, 1H), 8.03–7.96 (m, 1H), 7.80–7.70 (m, 3H), 7.42 (d, J = 7.6 Hz, 2H), 5.25 (br, 1H), 4.55 (s, 2H), 3.24–3.13 (m, 4H), 2.89–2.75 (m, 3H), 1.89–1.78 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.7, 142.2, 141.1, 140.2, 138.2, 138.1, 138.0, 127.1, 126.7, 125.5, 124.4, 124.0, 120.4, 62.6, 44.0, 42.9, 33.7, 26.3. MS (ESI): [M + H]+ 381.2.
6-(4-(Tert-butyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (40)
1H NMR (400 MHz, DMSO-d6) δ 8.95–8.85 (m, 1H), 8.73 (br, 1H), 8.48 (br, 1H), 8.28 (s, 1H), 8.11 (s, 1H), 7.97 (d, J = 8.4 Hz, 1H), 7.75–7.65 (m, 3H), 7.48 (d, J = 8.4 Hz, 2H), 3.27–3.13 (m, 4H), 2.82 (t, J = 12.1 Hz, 2H), 1.89–1.75 (m, 3H), 1.40–1.30 (m, 2H), 1.30 (s, 9H). 13C NMR (100 MHz, DMSO-d6) δ 161.7, 150.2, 141.1, 140.1, 138.2, 138.1, 136.8, 126.7, 125.8, 125.5, 124.5, 124.0, 120.3, 44.0, 42.8, 34.3, 33.7, 31.1, 26.3. MS (ESI): [M + H]+ 407.2.
6-(4-(Cyanomethyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (41)
1H NMR (400 MHz, DMSO-d6) δ 8.98–8.85 (m, 2H), 8.65–8.55 (m, 1H), 8.35–8.28 (m, 1H), 8.14 (d, J = 4.9 Hz, 1H), 8.03–7.94 (m, 1H), 7.80 (d, J = 8.3 Hz, 1H), 7.76–7.68 (m, 2H), 7.46 (d, J = 8.3 Hz, 1H), 7.37 (d, J = 8.3 Hz, 1H), 4.10 (s, 1H), 3.42 (s, 1H), 3.29–3.16 (m, 4H), 2.88–2.75 (m, 2H), 1.88–1.75 (m, 3H), 1.46–1.29 (m, 2H). MS (ESI): [M + H]+ 390.2.
6-(4-Cyano-3-fluorophenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (42)
1H NMR (400 MHz, DMSO-d6) δ 9.05–8.96 (m, 1H), 8.88–8.80 (m, 1H), 8.60–8.47 (m, 2H), 8.17 (s, 1H), 8.11–7.97 (m, 3H), 7.89–7.84 (m, 2H), 3.28–3.15 (m, 4H), 2.90–2.78 (m, 2H), 1.88–1.76 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 162.9 (d, J = 254.7 Hz), 161.4, 147.2 (d, J = 8.5 Hz), 141.8, 140.9, 139.7, 134.7 (d, J = 1.9 Hz), 134.3, 125.8, 124.4, 124.1, 123.8 (d, J = 2.8 Hz), 121.8, 114.6 (d, J = 20.7 Hz), 114.1, 98.7 (d, J = 15.3 Hz), 44.0, 42.8, 33.6, 26.3. MS (ESI): [M + H]+ 394.1.
6-(4-(Methylsulfonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (43)
1H NMR (400 MHz, DMSO-d6) δ 8.96 (t, J = 6.0 Hz, 1H), 8.78–8.70 (m, 1H), 8.50–8.35 (m, 2H), 8.16 (s, 1H), 8.09–7.99 (m, 5H), 7.83 (dd, J = 8.4, 1.7 Hz, 1H), 3.28–3.25 (m, 5H), 3.20 (t, J = 6.1 Hz, 2H), 2.88–2.78 (m, 2H), 1.89–1.80 (m, 3H), 1.42–1.30 (m, 2H). 13C NMR (150 MHz, DMSO-d6) δ 162.0, 145.1, 141.7, 141.5, 140.2, 139.7, 136.7, 128.4, 128.2, 126.2, 124.9, 124.8, 122.0, 44.5, 44.0, 43.4, 34.1, 26.8. MS (ESI): [M + H]+ 429.1.
6-(4-Trifluoromethoxylphenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (44)
1H NMR (400 MHz, DMSO-d6) δ 9.05–8.95 (m, 2H), 8.75–8.60 (m, 1H), 8.36 (s, 1H), 8.17 (s, 1H), 8.02 (d, J = 8.4 Hz, 1H), 7.89 (d, J = 8.7 Hz, 2H), 7.75 (d, J = 8.4 Hz, 1H), 7.48 (d, J = 8.4 Hz, 2H), 3.28–3.10 (m, 4H), 2.89–2.76 (m, 2H), 1.89–1.75 (m, 3H), 1.45–1.32 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 148.0, 141.0, 140.7, 139.0, 138.7, 136.6, 128.9, 125.6, 124.4, 124.1, 121.5, 120.9, 44.0, 42.8, 33.7, 26.3. MS (ESI): [M + H]+ 435.1.
6-(4-Methoxylphenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (45)
1H NMR (400 MHz, DMSO-d6) δ 9.18–9.06 (m, 1H), 9.00 (t, J = 5.3 Hz, 1H), 8.88–8.75 (m, 1H), 8.25 (s, 1H), 8.17 (s, 1H), 7.94 (d, J = 8.4 Hz, 1H), 7.74–7.65 (m, 3H), 7.03 (d, J = 8.7 Hz, 2H), 3.79 (s, 3H), 3.27–3.11 (m, 4H), 2.89–2.72 (m, 2H), 1.89–1.75 (m, 3H), 1.51–1.28 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.7, 159.1, 141.1, 139.9, 137.9, 137.8, 131.9, 128.1, 125.4, 124.6, 123.7, 119.8, 114.5, 55.2, 44.0, 42.7, 33.7, 26.3. MS (ESI): [M + H]+ 381.2.
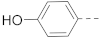
6-(4-Hydroxylphenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (46)
1H NMR (400 MHz, DMSO-d6) δ 9.61 (s, 1H), 8.92 (t, J = 5.1 Hz, 1H), 8.85–8.78 (m, 1H), 8.58–8.46 (m, 1H), 8.24 (s, 1H), 8.12 (s, 1H), 7.98 (d, J = 8.5 Hz, 1H), 7.66 (d, J = 8.4 Hz, 1H), 7.27 (t, J = 7.8 Hz, 1H), 7.16 (d, J = 7.8 Hz, 1H), 7.11 (s, 1H), 6.80 (d, J = 8.0 Hz, 1H), 3.27–3.18 (m, 4H), 2.88–2.74 (m, 2H), 1.89–1.78 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.7, 157.9, 141.1, 141.0, 140.3, 138.4, 138.4, 130.0, 125.4, 124.4, 124.1, 120.5, 117.8, 114.7, 113.8, 44.0, 42.8, 33.7, 26.3. MS (ESI): [M + H]+ 367.1.
6-(4-Methylphenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (47)
1H NMR (400 MHz, DMSO-d6) δ 8.99–8.85 (m, 2H), 8.65–8.55 (m, 1H), 8.30 (s, 1H), 8.13 (s, 1H), 7.98 (d, J = 8.4 Hz, 1H), 7.72 (dd, J = 8.4, 1.6 Hz, 1H), 7.66 (d, J = 8.1 Hz, 2H), 7.29 (d, J = 8.0 Hz, 2H), 3.29–3.15 (m, 4H), 2.89–2.75 (m, 2H), 2.35 (s, 3H), 1.89–1.75 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.7, 141.1, 140.1, 138.1, 138.1, 137.1, 136.7, 129.6, 126.8, 125.4, 124.5, 123.9, 120.2, 44.0, 42.8, 33.7, 26.3, 20.7. MS (ESI): [M + H]+ 365.2.
6-(4-Chlorophenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (48)
1H NMR (400 MHz, DMSO-d6) δ 8.96–8.90 (m, 1H), 8.82–8.72 (m, 1H), 8.50–8.40 (m, 1H), 8.35 (s, 1H), 8.13 (s, 1H), 8.01 (d, J = 8.5 Hz, 1H), 7.80 (d, J = 8.5 Hz, 2H), 7.76–7.72 (m, 1H), 7.54 (d, J = 8.5 Hz, 2H), 3.28–3.17 (m, 4H), 2.90–2.74 (m, 2H), 1.88–1.78 (m, 3H), 1.46–1.28 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 141.1, 140.6, 138.6, 138.5, 136.8, 132.6, 129.0, 128.8, 125.6, 124.4, 124.0, 120.7, 44.0, 42.9, 33.7, 26.3. MS (ESI): [M + H]+ 385.1.
6-(4-Fluorophenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (49)
1H NMR (400 MHz, DMSO-d6) δ 9.02–8.89 (m, 2H), 8.63 (br, 1H), 8.31 (s, 1H), 8.15 (s, 1H), 7.99 (d, J = 8.4 Hz, 1H), 7.85–7.76 (m, 2H), 7.72 (d, J = 8.4 Hz, 1H), 7.35–7.28 (m, 2H), 3.26–3.15 (m, 4H), 2.88–2.75 (m, 2H), 1.89–1.75 (m, 3H), 1.45–1.32 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 162.0 (d, J = 244.0 Hz), 161.6, 141.0, 140.4, 138.3, 137.1, 136.1 (d, J = 3.0 Hz), 129.0 (d, J = 8.2 Hz), 125.5, 124.4, 124.0, 120.6, 115.8 (d, J = 21.4 Hz), 44.0, 42.8, 33.7, 26.3. MS (ESI): [M + H]+ 369.1.
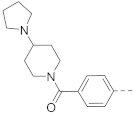
5-(4-(Piperidin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (50)
1H NMR (400 MHz, DMSO-d6) δ 9.15–9.02 (m, 2H), 8.85–8.70 (m, 1H), 8.25–8.19 (m, 2H), 8.10 (d, J = 8.5 Hz, 1H), 7.99–7.85 (m, 3H), 7.79–7.75 (m, 1H), 7.69 (d, J = 9.0 Hz, 1H), 6.89 (d, J = 9.0 Hz, 1H), 3.40–3.34 (m, 2H), 3.33–3.13 (m, 6H), 2.89–2.75 (m, 2H), 2.11–1.63 (m, 9H), 1.49–1.40 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 141.0, 139.9, 139.7, 135.9, 134.5, 128.3, 125.2, 125.0, 123.5, 123.0, 122.8, 116.1, 56.3, 44.1, 42.8, 33.7, 26.3, 23.2, 22.9. HRMS (ESI+) calcd for C26H31N3O3S [M + H]+, 434.2266; found, 434.2263.
5-(4-Cyanophenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (53)
1H NMR (400 MHz, DMSO-d6) δ 9.05 (t, J = 5.9 Hz, 1H), 8.99–8.90 (m, 1H), 8.70–8.56 (m, 1H), 8.30 (d, J = 1.5 Hz, 1H), 8.20 (s, 1H), 8.14 (d, J = 8.5 Hz, 1H), 8.01–7.92 (m, 4H), 7.83 (dd, J = 8.5, 1.8 Hz, 1H), 3.27–3.16 (m, 4H), 2.89–2.70 (m, 2H), 1.89–1.70 (m, 3H), 1.46–1.28 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.5, 144.4, 141.2, 140.5, 139.9, 135.3, 132.9, 127.9, 125.2, 125.0, 123.6, 123.6, 118.9, 110.0, 44.1, 42.8, 33.7, 26.3. MS (ESI): [M + H]+ 376.1.
5-(4-(Tert-butyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (54)
1H NMR (400 MHz, DMSO-d6) δ 8.97 (t, J = 5.9 Hz, 1H), 8.90–8.80 (m, 1H), 8.60–8.50 (m, 1H), 8.18–8.11 (m, 2H), 8.07 (d, J = 8.5 Hz, 1H), 7.73 (dd, J = 8.5, 1.6 Hz, 1H), 7.66 (d, J = 8.4 Hz, 2H), 7.50 (d, J = 8.4 Hz, 2H), 3.28–3.14 (m, 4H), 2.89–2.75 (m, 2H), 1.91–1.75 (m, 3H), 1.45–1.34 (m, 2H), 1.32 (s, 9H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 150.0, 140.7, 139.9, 139.1, 137.2, 137.1, 126.7, 125.8, 125.2, 125.0, 123.3, 122.6, 44.0, 42.8, 34.3, 33.7, 31.1, 26.3. MS (ESI): [M + H]+ 407.2.
6-(4-Morpholinophenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (58)
1H NMR (400 MHz, DMSO-d6) δ 9.05–8.95 (m, 1H), 8.87 (t, J = 5.8 Hz, 1H), 8.69–8.60 (m, 1H), 7.85–7.73 (m, 2H), 7.67 (d, J = 8.4 Hz, 2H), 7.61 (d, J = 8.2 Hz, 1H), 7.56 (s, 1H), 7.19–7.01 (m, 2H), 3.85–3.70 (m, 4H), 3.29–3.15 (m, 8H), 2.89–2.72 (m, 2H), 1.89–1.75 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.3, 155.1, 149.3, 139.1, 127.7, 125.7, 122.9, 122.4, 115.9, 109.4, 108.5, 65.8, 48.6, 43.4, 42.8, 33.6, 26.2. MS (ESI): [M + H]+ 420.2.
6-(4-(Piperazin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (59)
1H NMR (400 MHz, DMSO-d6) δ 9.35–9.25 (m, 2H), 8.99–8.90 (m, 1H), 8.88–8.80 (m, 1H), 8.70–8.58 (m, 1H), 7.85–7.74 (m, 2H), 7.70–7.54 (m, 4H), 7.10 (d, J = 8.4 Hz, 2H), 3.44 (d, J = 3.6 Hz, 4H), 3.22 (t, J = 11.0 Hz, 8H), 2.81 (dd, J = 17.8, 6.6 Hz, 2H), 1.83 (dd, J = 18.5, 8.7 Hz, 3H), 1.38 (dd, J = 20.2, 8.6 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.3, 155.1, 149.5, 149.3, 139.0, 131.0, 127.7, 125.7, 122.9, 122.4, 116.2, 109.3, 108.6, 45.1, 43.4, 42.7, 42.4, 33.6, 26.2. MS (ESI): [M + H]+ 419.2.
6-(4-(4-Methylpiperazin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (60)
1H NMR (400 MHz, DMSO-d6) δ 10.89 (br, 1H), 8.95–8.80 (m, 2H), 8.65–8.50 (m, 1H), 7.86–7.72 (m, 2H), 7.68–7.54 (m, 4H), 7.11 (d, J = 8.4 Hz, 2H), 3.95–3.85 (m, 2H), 3.52–3.45 (m, 2H), 3.27–3.09 (m, 8H), 2.86–2.75 (m, 5H), 1.90–1.75 (m, 3H), 1.42–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.3, 155.1, 149.3, 149.1, 139.0, 131.0, 127.7, 125.7, 122.9, 122.4, 116.2, 109.3, 108.6, 51.9, 45.2, 43.4, 42.8, 41.9, 33.6, 26.2. MS (ESI): [M + H]+ 433.3.
6-(4-Cyclohexylphenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (61)
1H NMR (400 MHz, DMSO-d6) δ 8.87 (t, J = 5.7 Hz, 1H), 8.78 (br, 1H), 8.47 (br, 1H), 7.90–7.76 (m, 2H), 7.68–7.60 (m, 3H), 7.56 (s, 1H), 7.33 (d, J = 8.0 Hz, 2H), 3.29–3.15 (m, 4H), 2.89–2.76 (m, 2H), 2.58–2.50 (m, 1H), 1.89–1.73 (m, 7H), 1.72–1.67 (m, 1H), 1.48–1.18 (m, 7H). 13C NMR (100 MHz, DMSO-d6) δ 158.3, 155.0, 149.5, 147.2, 139.4, 137.5, 127.4, 127.1, 126.2, 123.0, 122.9, 109.4, 109.3, 43.5, 42.9, 33.9, 33.7, 26.4, 26.3, 25.6. MS (ESI): [M + H]+ 417.2.
6-(4-(Diethylamino)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (62)
1H NMR (400 MHz, DMSO-d6) δ 10.41 (br, 1H), 8.90 (t, J = 5.9 Hz, 1H), 8.88–8.75 (m, 1H), 8.55–8.40 (m, 1H), 7.95 (s, 1H), 7.90–7.79 (m, 3H), 7.74–7.65 (m, 3H), 7.60 (s, 1H), 4.33 (d, J = 5.3 Hz, 2H), 3.27–3.16 (m, 6H), 2.85–2.75 (m, 2H), 1.89–1.75 (m, 3H), 1.45–1.35 (m, 2H), 1.29–1.20 (m, 6H). 13C NMR (100 MHz, DMSO-d6) δ 158.6, 155.4, 150.2, 141.0, 138.7, 132.2, 130.1, 127.8, 127.3, 123.6, 123.4, 110.1, 109.7, 46.1, 43.9, 43.2, 40.6, 40.4, 40.1, 34.1, 26.7, 8.7. MS (ESI): [M + H]+ 406.2.
6-((1,1′-Biphenyl)-4-yl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (67)
1H NMR (400 MHz, DMSO-d6) δ 8.90 (t, J = 5.9 Hz, 1H), 8.85–8.75 (m, 1H), 8.55–8.45 (m, 1H), 7.95 (s, 1H), 7.88–7.84 (m, 3H), 7.82–7.75 (m, 2H), 7.74–7.68 (m, 2H), 7.60 (s, 1H), 7.53–7.46 (m, 2H), 7.40–7.35 (m, 1H), 3.28–3.13 (m, 4H), 2.89–2.75 (m, 2H), 1.91–1.70 (m, 3H), 1.44–1.27 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.3, 155.0, 149.7, 139.5, 139.4, 138.8, 138.7, 129.1, 127.7, 127.3, 126.6, 126.6, 123.1, 122.9, 109.5, 109.4, 43.5, 42.8, 33.6, 26.3. MS (ESI): [M + H]+ 411.2.
6-((1,1′-Biphenyl)-3-yl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (68)
1H NMR (400 MHz, DMSO-d6) δ 8.95–8.78 (m, 2H), 8.59–8.48 (m, 1H), 8.05–7.95 (m, 2H), 7.86 (d, J = 8.2 Hz, 1H), 7.79–7.71 (m, 4H), 7.69–7.65 (m, 1H), 7.62–7.55 (m, 2H), 7.55–7.45 (m, 2H), 7.43–7.35 (m, 1H), 3.29–3.13 (m, 4H), 2.88–2.75 (m, 2H), 1.89–1.75 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.3, 155.0, 149.7, 141.1, 140.6, 140.1, 139.3, 129.7, 129.0, 127.7, 127.0, 126.6, 126.3, 126.2, 125.6, 123.3, 123.1, 110.0, 109.4, 43.5, 42.8, 33.7, 26.3. MS (ESI): [M + H]+ 411.2.
6-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (69)
1H NMR (400 MHz, DMSO-d6) δ 8.92–8.80 (m 2H), 8.60–8.50 (m, 1H), 7.82–7.76 (m, 2H), 7.60–7.55 (m, 2H), 7.35 (s, 1H), 7.25–7.16 (m, 3H), 7.09 (s, 1H), 6.95 (d, J = 8.3 Hz, 1H), 4.28 (s, 4H), 3.28–3.13 (m, 4H), 2.88–2.75 (m, 2H), 1.91–1.65 (m, 3H), 1.46–1.28 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.7, 144.1, 143.8, 139.3, 139.2, 133.5, 126.4, 123.3, 123.1, 120.4, 118.0, 117.5, 115.9, 109.8, 109.4, 64.6, 64.5, 43.9, 43.2, 34.1, 26.7. MS (ESI): [M + H]+ 393.2.
6-(4-(Methylsulfonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (70)
1H NMR (400 MHz, DMSO-d6) δ 8.92 (t, J = 6.1 Hz, 1H), 8.81–8.73 (m, 1H), 8.50–8.39 (m, 1H), 8.06–7.98 (m, 5H), 7.90 (d, J = 8.2 Hz, 1H), 7.73 (d, J = 8.3 Hz, 1H), 7.62 (s, 1H), 3.29–3.19 (m, 7H), 2.89–2.75 (m, 2H), 1.93–1.74 (m, 3H), 1.45–1.27 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.2, 154.8, 150.2, 144.8, 139.8, 137.3, 128.0, 127.7, 127.5, 123.4, 123.3, 110.4, 109.3, 43.6, 43.5, 42.8, 33.6, 26.3. MS (ESI): [M + H]+ 413.1.
6-(4-(Benzyloxycarbonylaminomethyl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (71)
1H NMR (400 MHz, DMSO-d6) δ 9.06–8.95 (m, 1H), 8.94–8.90 (m, 1H), 8.78–8.62 (m, 1H), 7.95–7.79 (m, 3H), 7.69 (d, J = 8.0 Hz, 2H), 7.65–7.57 (m, 2H), 7.43–7.25 (m, 7H), 5.05 (s, 2H), 4.25 (d, J = 6.0 Hz, 2H), 3.72–3.62 (m, 1H), 3.50–3.40 (m, 1H), 3.28–3.18 (m, 4H), 2.88–2.72 (m, 2H), 1.88–1.72 (m, 3H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.2, 156.4, 155.0, 149.6, 139.3, 139.1, 138.4, 137.1, 128.4, 127.8, 127.7, 127.7, 127.0, 126.4, 123.0, 122.9, 109.4, 109.3, 65.4, 43.5, 43.5, 42.7, 33.6, 26.2. MS (ESI): [M + H]+ 498.2.
6-(4-(Aminomethyl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (72)
1H NMR (400 MHz, DMSO-d6) δ 9.05–8.95 (m, 1H), 8.92 (t, J = 5.9 Hz, 1H), 8.75–8.65 (m, 1H), 8.50 (br, 3H), 7.91 (s, 1H), 7.88–7.75 (m, 3H), 7.69–7.65 (m, 1H), 7.64–7.58 (m, 3H), 4.11–4.02 (m, 2H), 3.29–3.15 (m, 4H), 2.89–2.73 (m, 2H), 1.89–1.70 (m, 3H), 1.43–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.3, 155.0, 149.8, 139.9, 138.6, 133.6, 129.7, 127.2, 126.7, 123.2, 123.0, 109.7, 109.3, 43.5, 42.8, 41.9, 33.7, 26.6. MS (ESI): [M + H]+ 364.2.
6-(4-Cyanophenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (73)
1H NMR (400 MHz, DMSO-d6) δ 8.92 (t, J = 5.7 Hz, 1H), 8.81–8.70 (m, 1H), 8.52–8.37 (m, 1H), 8.01 (s, 1H), 7.99–7.92 (m, 3H), 7.89 (d, J = 8.2 Hz, 1H), 7.73 (d, J = 8.3 Hz, 1H), 7.61 (s, 1H), 3.28–3.10 (m, 4H), 2.88–2.75 (m, 2H), 1.89–1.75 (m, 3H), 1.42–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.2, 154.9, 150.2, 144.3, 137.2, 133.0, 128.0, 127.6, 123.4, 123.2, 118.9, 110.3, 110.2, 109.3, 43.5, 42.9, 33.6, 26.3. MS (ESI): [M + H]+ 360.2.
3.1.4. Synthetic Methods for Compounds 29, 30, 33, 34, 51–52, and 63–66
An amidation and a Suzuki coupling reaction, as described in the General synthetic procedure-A, were used for synthesis of benzothiophene- or benzofuran-2-carboxamide 85. Compounds 29, 30, 33, 34, 51–52, and 63–66 were prepared using an additional amidation reaction followed by BOC deprotection, following the General synthetic procedure-A, as a hydrochloric salt.
6-(4-(Piperidine-1-carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (29)
1H NMR (400 MHz, DMSO-d6) δ 9.05–8.90 (m, 2H), 8.71–8.59 (m, 1H), 8.38 (s, 1H), 8.16 (s, 1H), 8.02 (d, J = 8.3 Hz, 1H), 7.83 (d, J = 6.7 Hz, 2H), 7.78 (d, J = 8.4 Hz, 1H), 7.47 (d, J = 6.7 Hz, 2H), 3.65–3.58 (m, 2H), 3.36–3.18 (m, 6H), 2.85–2.75 (m, 2H), 1.89–1.75 (m, 3H), 1.63–1.36 (m, 8H). 13C NMR (100 MHz, DMSO-d6) δ 168.6, 161.6, 141.1, 140.6, 140.4, 138.7, 137.3, 135.6, 127.4, 127.0, 125.6, 124.5, 124.1, 120.8, 48.8, 44.0, 42.8, 33.7, 26.3, 24.7, 24.1. MS (ESI): [M + H]+ 462.2.
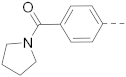
6-(4-(Pyrroline-1-carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (30)
1H NMR (400 MHz, DMSO-d6) δ 8.97 (t, J = 5.8 Hz, 1H), 8.94–8.84 (m, 1H), 8.65–8.55 (m, 1H), 8.39 (s, 1H), 8.16 (s, 1H), 8.02 (d, J = 8.4 Hz, 1H), 7.88–7.75 (m, 3H), 7.63 (d, J = 8.1 Hz, 2H), 3.50–3.41 (m, 4H), 3.28–3.18 (m, 4H), 2.85–2.78 (m, 2H), 1.93–1.77 (m, 7H), 1.45–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 168.3, 162.0, 141.5, 141.2, 141.1, 139.1, 137.7, 136.7, 128.3, 127.2, 126.0, 124.9, 124.5, 121.3, 49.4, 46.4, 44.5, 43.3, 34.1, 26.7, 25.0, 24.4. MS (ESI): [M + H]+ 448.2.
6-(4-(4-(Piperidin-1-yl)piperidin-1-carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (33)
1H NMR (400 MHz, DMSO-d6) δ 10.68–10.55 (m, 1H), 9.05–8.95 (m, 2H), 8.78–8.65 (m, 1H), 8.39 (s, 1H), 8.18 (s, 1H), 8.02 (d, J = 8.3 Hz, 1H), 7.84 (d, J = 7.8 Hz, 2H), 7.78 (d, J = 8.3 Hz, 1H), 7.52 (d, J = 7.8 Hz, 2H), 3.69–3.33 (m, 6H), 3.30–3.16 (m, 4H), 2.96–2.75 (m, 5H), 2.24–2.03 (m, 2H), 1.93–1.66 (m, 10H), 1.45–1.32 (m, 3H). 13C NMR (100 MHz, DMSO-d6) δ 168.6, 161.6, 141.1, 140.8, 140.7, 138.7, 137.2, 134.9, 127.6, 127.0, 125.6, 124.5, 124.1, 120.9, 70.5, 62.3, 60.2, 48.9, 44.0, 42.8, 33.7, 26.3, 22.5, 21.6. MS (ESI): [M + H]+ 545.3.
6-(4-(4-(Pyrrolidin-1-yl)piperidin-1-carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (34)
1H NMR (400 MHz, DMSO-d6) δ 11.21 (br, 1H), 9.05–8.95 (m, 2H), 8.76–8.56 (m, 1H), 8.39 (s, 1H), 8.17 (s, 1H), 8.02 (d, J = 8.3 Hz, 1H), 7.85 (d, J = 7.9 Hz, 2H), 7.78 (d, J = 8.3 Hz, 1H), 7.50 (d, J = 7.8 Hz, 2H), 3.75–3.65 (m, 2H), 3.52–3.32 (m, 4H), 3.28–3.16 (m, 4H), 3.09–2.98 (m, 2H), 2.88–2.75 (m, 3H), 2.15–1.66 (m, 11H), 1.48–1.34 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 168.7, 161.6, 141.1, 140.8, 140.7, 138.7, 137.2, 134.9, 127.6, 127.1, 125.6, 124.5, 124.1, 120.9, 60.5, 50.3, 44.0, 42.8, 33.7, 26.3, 22.7. MS (ESI): [M + H]+ 531.3.
5-(4-(Piperidine-1-carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (51)
1H NMR (400 MHz, DMSO-d6) δ 8.99 (t, J = 5.8 Hz, 1H), 8.85–8.75 (m, 1H), 8.52–8.40 (m, 1H), 8.24 (s, 1H), 8.16 (s, 1H), 8.11 (d, J = 8.4 Hz, 1H), 7.85–7.75 (m, 3H), 7.48 (d, J = 8.2 Hz, 2H), 3.66–3.47 (m, 2H), 3.31–3.13 (m, 6H), 2.88–2.75 (m, 2H), 1.90–1.76 (m, 3H), 1.68–1.34 (m, 8H). 13C NMR (100 MHz, DMSO-d6) δ 168.6, 161.6, 140.9, 140.7, 139.9, 139.6, 136.5, 135.5, 127.4, 127.0, 125.2, 124.9, 123.4, 123.0, 44.0, 42.9, 33.7, 26.3, 24.1. MS (ESI): [M + H]+ 462.2.
5-(4-(Pyrroline-1-carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (52)
1H NMR (400 MHz, DMSO-d6) δ 9.05–8.95 (m, 1H), 8.87–8.75 (m, 1H), 8.56–8.45 (m, 1H), 8.24 (s, 1H), 8.17 (s, 1H), 8.11 (d, J = 8.5 Hz, 1H), 7.85–7.75 (m, 3H), 7.63 (d, J = 8.1 Hz, 2H), 3.54–3.42 (m, 4H), 3.28–3.16 (m, 4H), 2.90–2.78 (m, 2H), 1.91–1.77 (m, 7H), 1.49–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 167.9, 161.6, 141.0, 140.9, 139.9, 139.7, 136.4, 136.1, 127.8, 126.7, 125.2, 124.9, 123.4, 123.0, 48.9, 46.0, 44.0, 42.8, 33.7, 26.3, 26.0, 23.9. MS (ESI): [M + H]+ 448.2.
6-(4-((Pyrrolidine-1-yl)carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (63)
1H NMR (400 MHz, DMSO-d6) δ 9.18–9.10 (m, 1H), 8.95 (t, J = 5.6 Hz, 1H), 8.89–8.80 (m, 1H), 7.94 (s, 1H), 7.85 (d, J = 8.2 Hz, 1H), 7.80 (d, J = 7.6 Hz, 2H), 7.68 (d, J = 8.3 Hz, 1H), 7.66–7.55 (m, 3H), 3.51–3.34 (m, 4H), 3.28–3.15 (m, 4H), 2.84–2.74 (m, 2H), 1.94–1.70 (m, 7H), 1.46–1.32 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 167.9, 158.2, 154.9, 149.8, 141.0, 138.4, 136.3, 127.9, 126.9, 123.2, 123.0, 109.8, 109.3, 46.0, 43.5, 42.7, 33.7, 26.2, 26.0, 23.9. MS (ESI): [M + H]+ 432.2.
6-(4-((Piperidine-1-yl)carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (64)
1H NMR (400 MHz, DMSO-d6) δ 8.98–8.88 (m, 2H), 8.68–8.55 (m, 1H), 7.93 (s, 1H), 7.86–7.78 (m, 3H), 7.74–7.57 (m, 2H), 7.47 (d, J = 7.9 Hz, 2H), 3.35–3.14 (m, 8H), 2.84–2.74 (m, 2H), 1.89–1.75 (m, 3H), 1.65–1.53 (m, 6H), 1.45–1.32 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 168.6, 158.2, 154.9, 149.8, 140.6, 138.4, 135.7, 127.5, 127.1, 126.8, 123.2, 123.0, 109.7, 109.3, 43.5, 42.8, 33.6, 26.2, 24.7, 24.1, 23.2. MS (ESI): [M + H]+ 446.2.
6-(4-((Azepane-1-yl)carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (65)
1H NMR (400 MHz, DMSO-d6) δ 8.98–8.90 (m, 2H), 8.68–8.55 (m, 1H), 7.93 (s, 1H), 7.85 (d, J = 8.2 Hz, 1H), 7.80 (d, J = 8.0 Hz, 2H), 7.71–7.53 (m, 2H), 7.46 (d, J = 8.0 Hz, 2H), 3.36–3.17 (m, 8H), 2.85–2.75 (m, 2H), 1.89–1.70 (m, 5H), 1.66–1.50 (m, 6H), 1.42–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 169.9, 158.2, 154.9, 149.8, 140.2, 138.4, 136.6, 127.1, 127.0, 126.8, 123.1, 123.0, 109.7, 109.3, 51.0, 45.4, 43.5, 42.8, 33.6, 27.2, 26.8, 26.2, 26.0, 25.8. MS (ESI): [M + H]+ 460.3.
6-(4-((1,4′-Bipiperidine-1-yl)carbonyl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (66)
1H NMR (400 MHz, DMSO-d6) δ 10.49–10.40 (m, 1H), 8.98–8.85 (m, 2H), 8.64–8.50 (m, 1H), 7.95 (s, 1H), 7.88–7.82 (m, 3H), 7.69 (d, J = 8.3 Hz, 1H), 7.61 (s, 1H), 7.53 (d, J = 7.9 Hz, 2H), 3.40–3.19 (m, 9H), 2.99–2.71 (m, 6H), 2.20–2.20 (m, 2H), 1.87–1.65 (m, 10H), 1.45–1.30 (m, 3H). 13C NMR (100 MHz, DMSO-d6) δ 168.6, 158.2, 154.9, 149.8, 141.0, 138.3, 134.9, 127.6, 127.1, 126.9, 123.2, 123.0, 109.8, 109.3, 70.5, 62.3, 60.2, 49.0, 43.5, 42.8, 33.6, 26.2, 22.5, 21.6. MS (ESI): [M + H]+ 529.3.
Compounds 31, 32, 35, 36, 55, 56 and 74–77 were synthesized using the general synthetic procedure-A, starting from benzothiophene- or benzofuran-2-carboxamide 84, as a hydrochloric salt.
6-((4-(Piperidin-1-yl)piperidin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (31)
1H NMR (400 MHz, DMSO-d6) δ 10.37 (br, 1H), 8.96–8.85 (m, 1H), 8.76 (t, J = 6.0 Hz, 1H), 8.66–8.49 (m, 1H), 7.97 (s, 1H), 7.75 (d, J = 8.9 Hz, 1H), 7.53 (s, 1H), 7.23 (d, J = 8.7 Hz, 1H), 3.99–3.92 (m, 2H), 3.45–3.36 (m, 2H), 3.28–3.20 (m, 2H), 3.15 (t, J = 5.6 Hz, 2H), 2.95–2.80 (m, 6H), 2.24–2.15 (m, 2H), 1.89–1.75 (m, 10H), 1.73–1.65 (m, 1H), 1.45–1.32 (m, 3H). 13C NMR (100 MHz, DMSO-d6) δ 161.9, 148.3, 142.3, 139.0, 135.2, 125.5, 124.5, 119.4, 116.2, 62.2, 50.6, 48.9, 43.9, 42.7, 33.7, 26.3, 25.2, 22.5, 21.6. MS (ESI): [M + H]+ 441.3.
6-((4-(Pyrrolidin-1-yl)piperidin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (32)
1H NMR (400 MHz, DMSO-d6) δ 10.87 (br, 1H), 8.88–8.80 (m, 1H), 8.75 (t, J = 5.8 Hz, 1H), 8.60–8.50 (m, 1H), 7.96 (s, 1H), 7.74 (d, J = 9.0 Hz, 1H), 7.53 (s, 1H), 7.23 (d, J = 8.6 Hz, 1H), 3.99–3.90 (m, 2H), 3.54–3.44 (m, 2H), 3.33–3.20 (m, 3H), 3.15 (t, J = 5.8 Hz, 2H), 3.10–3.00 (m, 2H), 2.89–2.75 (m, 4H), 2.18–2.10 (m, 2H), 1.99–1.76 (m, 9H), 1.50–1.25 (m, 2H). MS (ESI): [M + H]+ 427.2.
6-((4-(Piperidin-1-yl)phenyl)amino)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (35)
1H NMR (400 MHz, DMSO-d6) δ 12.58 (br, 1H), 9.13–8.87 (m, 2H), 8.84–8.57 (m, 2H), 8.02 (s, 1H), 7.85–7.72 (m, 3H), 7.67 (s, 1H), 7.25 (d, J = 8.9 Hz, 2H), 7.18 (dd, J = 8.7, 1.9 Hz, 1H), 3.52–3.32 (m, 4H), 3.26–3.20 (m, 4H), 2.88–2.74 (m, 2H), 2.23–2.05 (m, 2H), 1.92–1.53 (m, 7H), 1.45–1.28 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.9, 143.9, 142.1, 141.3, 136.7, 135.0, 133.1, 125.8, 124.6, 122.6, 117.3, 117.0, 109.6, 108.4, 56.1, 43.9, 42.7, 33.7, 26.3, 22.9, 20.8. MS (ESI): [M + H]+ 449.2.
6-(4-(4-Chlorophenoxyl)phenoxyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (36)
1H NMR (400 MHz, DMSO-d6) δ 8.92–8.80 (m, 2H), 8.61–8.45 (m, 1H), 8.08 (s, 1H), 7.93 (d, J = 8.7 Hz, 1H), 7.61 (s, 1H), 7.43 (d, J = 8.8 Hz, 2H), 7.17–7.01 (m, 7H), 3.27–3.14 (m, 4H), 2.88–2.75 (m, 2H), 1.89–1.75 (m, 3H), 1.39–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 161.6, 156.2, 156.0, 152.3, 152.0, 141.8, 139.1, 135.1, 129.9, 127.0, 126.5, 124.3, 120.8, 119.7, 117.2, 111.0, 44.0, 42.8, 33.7, 26.3. MS (ESI): [M + H]+ 493.1.
5-(4-(Tert-butyl)phenyl)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (55)
1H NMR (400 MHz, DMSO-d6) δ 9.03–8.92 (m, 1H), 8.83 (t, J = 5.8 Hz, 1H), 8.69–8.60 (m, 1H), 7.96 (s, 1H), 7.79 (d, J = 8.8 Hz, 1H), 7.49 (d, J = 2.0 Hz, 1H), 7.26 (d, J = 8.6 Hz, 2H), 7.17 (dd, J = 8.8, 2.1 Hz, 1H), 7.05 (d, J = 8.6 Hz, 2H), 3.31–3.10 (m, 4H), 2.88–2.75 (m, 2H), 1.89–1.75 (m, 3H), 1.45–1.35 (m, 2H), 1.25 (s, 9H). 13C NMR (100 MHz, DMSO-d6) δ 161.8, 142.3, 141.7, 140.8, 140.3, 140.3, 131.4, 125.8, 124.5, 123.3, 118.4, 116.9, 109.6, 44.0, 42.8, 33.8, 33.7, 31.3, 26.3. MS (ESI): [M + H]+ 422.2.
5-((4-(Trifluoromethoxyl)phenyl)amino)-N-(piperidin-4-ylmethyl)-benzo[b]thiophene-2-carboxamide hydrochloride (56)
1H NMR (400 MHz, DMSO-d6) δ 8.86–8.75 (m, 2H), 8.58–8.43 (m, 2H), 7.98 (s, 1H), 7.86 (d, J = 8.7 Hz, 1H), 7.59 (s, 1H), 7.25–7.10 (m, 5H), 3.27–3.13 (m, 4H), 2.88–2.78 (m, 2H), 1.88–1.76 (m, 3H), 1.45–1.30 (m, 2H). MS (ESI): [M + H]+ 450.1.
6-((4-(Piperidin-1-yl)phenyl)amino)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (74)
1H NMR (400 MHz, DMSO-d6) δ 12.39 (br, 1H), 9.01–8.83 (m, 2H), 8.72–8.48 (m, 2H), 7.79–7.68 (m, 2H), 7.59 (d, J = 8.5 Hz, 1H), 7.43 (s, 1H), 7.33–7.16 (m, 3H), 7.08 (d, J = 8.5 Hz, 1H), 3.40–3.06 (m, 8H), 2.86–2.72 (m, 2H), 2.19–2.05 (m, 2H), 1.87–1.56 (m, 7H), 1.39–1.25 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.5, 156.1, 147.3, 146.9, 145.7, 133.9, 122.9, 120.9, 118.1, 117.3, 113.5, 109.9, 94.7, 50.5, 45.4, 42.8, 33.7, 26.2, 25.5, 23.9. MS (ESI): [M + H]+ 433.3.
6-((4-Trifuluoromethoxyl)phenyl)amino)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (75)
1H NMR (400 MHz, DMSO-d6) δ 8.86–8.78 (m, 1H), 8.76 (s, 1H), 8.67 (t, J = 5.8 Hz, 1H), 8.57–8.43 (m, 1H), 7.59 (d, J = 8.6 Hz, 1H), 7.42 (s, 1H), 7.28–7.15 (m, 4H), 7.06 (d, J = 8.5 Hz, 1H), 3.27–3.10 (m, 4H), 2.86–2.75 (m, 2H), 1.88–1.73 (m, 3H), 1.41–1.26 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.4, 155.6, 147.8, 142.8, 142.3, 123.2, 122.3, 120.1, 117.9, 115.3, 109.7, 98.0, 43.3, 42.8, 33.7, 26.2. MS (ESI): [M + H]+ 434.2.
6-(4-Methoxylphenoxyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (76)
1H NMR (400 MHz, DMSO-d6) δ 9.08–8.95 (m, 1H), 8.81–8.63 (m, 2H), 7.70 (d, J = 8.5 Hz, 1H), 7.52 (s, 1H), 7.10 (s, 1H), 7.08–6.92 (m, 5H), 3.75 (s, 3H), 3.27–3.07 (m, 4H), 2.88–2.72 (m, 2H), 1.88–1.72 (m, 3H), 1.46–1.28 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.1, 157.7, 155.8, 154.9, 149.3, 149.2, 123.5, 122.2, 120.8, 115.2, 114.7, 109.4, 100.4, 55.5, 43.3, 42.7, 33.6, 26.2. MS (ESI): [M + H]+ 381.2.
6-(4-(4-Cholorophenoxyl)phenoxyl)-N-(piperidin-4-ylmethyl)-benzofuran-2-carboxamide hydrochloride (77)
1H NMR (400 MHz, DMSO-d6) δ 8.94–8.77 (m, 2H), 8.62–8.42 (m, 1H), 7.75 (d, J = 8.5 Hz, 1H), 7.53 (s, 1H), 7.42 (d, J = 8.5 Hz, 2H), 7.25 (s, 1H), 7.15–6.98 (m, 7H), 3.28–3.15 (m, 4H), 2.88–2.75 (m, 2H), 1.89–1.72 (m, 3H), 1.40–1.30 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 158.1, 156.6, 156.2, 154.8, 152.5, 151.9, 149.4, 129.8, 126.9, 123.7, 122.8, 120.8, 120.6, 119.7, 115.5, 109.4, 101.5, 43.3, 42.8, 33.6, 26.2. MS (ESI): [M + H]+ 477.2.
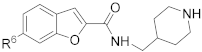
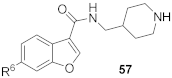
6-(4-(Piperidin-1-yl)phenyl)-N-(piperidin-4-ylmethyl)-benzofuran-3-carboxamide hydrochloride (57)
It was synthesized using the general synthetic procedure-A, starting from 6-bromo-benzofuran-3-carboxylic acid 86, as a hydrochloric salt. 1H NMR (400 MHz, DMSO-d6) δ 9.01–8.89 (m, 1H), 8.75–8.60 (m, 4H), 8.13 (d, J = 8.2 Hz, 1H), 8.03–7.89 (m, 4H), 7.71 (d, J = 8.3 Hz, 1H), 3.55–3.50 (m, 2H), 3.34–3.07 (m, 6H), 2.85–2.75 (m, 2H), 2.10–1.58 (m, 8H), 1.47–1.28 (m, 3H). 13C NMR (100 MHz, DMSO-d6) δ 162.0, 155.4, 148.3, 148.1, 135.9, 128.3, 127.0, 123.4, 122.9, 122.3, 116.9, 114.9, 109.8, 66.4, 43.4, 42.8, 33.8, 26.3, 25.0, 23.1. MS (ESI): [M + H]+ 418.2.
3.2. Plasmids and Peptides
The cDNA for the human AF9 AHD domain (475–568) and ENL AHD domain (489–559) was synthesized (by Genscript, Piscataway, NJ, USA) and inserted into the pMAL c5X expression plasmid. The DOT1L peptide (Biotin-AHX-NKLPVSIPLASVVLPSRAERARST) and AF4 peptide (Biotin-AHX-QSLMVKITLDLLSRIPQPPGK) for the Alpha assay were purchased from Genscript.
3.3. Protein Expression and Purification
The AF9 or ENL expression plasmid was used to transform E. coli BL21(DE3) strain (Novagen, Madison, WI, USA), and protein expression was induced by adding 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16 °C overnight. The cells were collected and lysed using a French press (GlenMills, Clifton, NJ, USA) in a lysis buffer (50 mM HEPES, 200 mM NaCl, 1 mM DTT, pH 7.4). Upon centrifugation, the supernatant was applied to an amylose resin column (GE Healthcare) and the recombinant protein with a N-terminal maltose-binding protein (MBP) tag was eluted with 20 mM HEPES, 10 mM Maltose, pH 7.4, which was further purified to be > 95% (SDS-PAGE) using a size exclusion column (HiLoad 16/60 Superdex 200, GE Healthcare, Chicago, IL, USA).
3.4. AlphaLisa Assays
Alpha assays were developed in our previous publication [26], using a Perkin-Elmer AlphaLisa anti-MBP kit, which contains streptavidin donor beads and anti-MBP acceptor beads. Briefly, the assay was performed using a MBP-protein (5 nM), a biotinylated peptide (40 nM), and increasing concentrations of a compound in 25 µL buffer (PBS with 0.5% BSA, pH 7.5) in 384-well plates according to the manufacturer’s protocol and measured with a Tecan SPARK microplate reader. Data were imported into Prism (version 5.0), and IC50 values from 3 independent experiments with standard deviation were obtained by using a standard dose-response curve fitting.
3.5. Pull-Down Assay
The pull-down assays were conducted following our previous method [26]. Briefly, the biotinylated DOT1L peptide (0.25 mg/mL) was incubated with streptavidin agarose beads (20 μL) in PBS containing 0.1% Triton X-100 (400 µL) for 3h to yield, after washing, DOT1L-coated beads. MBP-tagged AF9 AHD (0.2 µM, 400 µL) was pre-incubated with a compound for 1 h before adding to the beads. After 6 h incubation at 4 °C, the beads were collected, thoroughly washed, and subjected to SDS-PAGE and analyzed by Western blot using an MBP antibody (#2396, Cell signaling, Danvers, MA, USA).
3.6. RNA Extraction and Quantitative Real-Time PCR
RNA extraction and RT-qPCR were performed following our previous method [26]. 105 cells/mL were incubated with a compound for 4 days and the RNA was extracted using an RNeasy mini kit (#74104, Qiagen, Germantown, MD, USA). 100–1000 ng of total RNA was reverse transcribed using iScript™ Reverse Transcription Supermix (Bio-Rad, Hercules, CA, USA), using the manufacturer’s protocol. Quantitative real-time PCR was carried out using Fast SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s instructions. Measurements were performed in triplicate using GAPDH as the reference gene. Real-time PCR was performed using the Biosystems Step One Plus detection system. The following sequences of primers were used:
MYC (forward: 5′-CACCGAGTCGTAGTCGAGGT-3′; reverse: 5′-TTTCGGGTAGTGGAAAACCA-3′);
HoxA9 (forward: 5′-TACGTGGACTCGTTCCTGCT-3′; reverse: 5′-CGTCGCCTTGGACTGGAAG-3′);
Meis1 (forward: 5′-CCAGCATCTAACACACCCTTAC-3′; reverse: 5′-TATGTTGCTGACCGTCCATTAC -3′);
GAPDH (forward: 5′-GCGAGATCCCTCCAAAATCAA-3′; reverse: 5′-GTTCACACCCATGACGAACAT-3′)
3.7. Antiproliferation Assay
Proliferation inhibition assays were performed using an XTT assay kit (Biotium, Fremont, CA, USA) following our previous methods [26]. The antiproliferation EC50 values were determined using Prism 5 and the reported results were the mean values of at least three independent experiments. The incubation time for all compounds was 7 days.
3.8. Statistical Analysis
At least three independent experiments were carried out to generate each dataset. The significance of experimental differences was evaluated using the Student’s t test (Prism 5.0, GraphPad Software, Boston, MA, USA). The results are expressed as the mean ± SEM.
4. Conclusions
PPIs between AF9/ENL and AF4 or DOT1L are a potential drug target for MLL-r leukemia, as well as other cancers (e.g., AML) driven by SEC-mediated aberrant gene expression. Several indole-carboxamide compounds were identified as novel inhibitors of the AF9-DOT1L interaction. Synthesis and structure–activity relationship studies of 77 compounds show that a 4-piperidin-1-ylphenyl or 4-pyrrolidin-1-ylphenyl R5 or R6 substituent is essential for these indole- and closely related benzothiophene- and benzofuran-carboxamide compounds to have strong inhibitory activity. The activities of these inhibitors with IC50 values as low as 1.6 μM were further confirmed using a pull-down assay. Inhibitors of the AF9-DOT1L interaction also blocked the PPIs between AF9/ENL and AF4 with comparable IC50s. Selected inhibitors were found to suppress expression of MLL target genes HoxA9, Meis1 and Myc and inhibit proliferation of MLL-r leukemia and other AML cells with EC50s as low as 4.7 μM, while they were inactive against solid tumor Hela cells. This antitumor activity profile is consistent with the roles of AF9/ENL in these tumors, suggesting that their activities are on-target. In conclusion, the identified benzothiophene compounds 24 and 50 are not only useful chemical probes for biological studies of AF9/ENL or SEC, but they also represent novel lead compounds for further drug development against MLL-r leukemia and related cancers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15215283/s1, Figure S1. Dose-dependent inhibition of AF9 AHD-DOT1L interaction. Figure S2. Dose-dependent inhibition of different cancer cell growth.
Author Contributions
Methodology, X.L., S.N. and X.W.; Validation, X.L.; Investigation, X.L., S.N., X.W., J.Z., Y.Y., F.W., C.B.M., M.A.-U.-Z. and B.K.M.; Writing—original draft, Y.S.; Writing—review & editing, X.L., S.N. and Y.S.; Visualization, Y.S.; Supervision, Y.S.; Project administration, Y.S.; Funding acquisition, Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant (R01CA266057) from the United States National Institute of Health/National Cancer Institute and a grant (RP220232) from the Cancer Prevention and Research Institute of Texas to Y.S.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
MLL, mixed lineage leukemia; PPI, protein–protein interaction; MLL-r, mixed lineage leukemia-rearranged; onco-MLL, oncogenic fusion MLL; ALL, acute lymphocytic leukemia; AML, acute myeloid leukemia; SEC, super elongation complexes; H3K79, histone-H3 lysine-79; SAR, structure–activity relationship; BOC, tert-butyloxycarbonyl; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
References
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef]
- Daser, A.; Rabbitts, T. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2005; pp. 175–188. [Google Scholar]
- Winters, A.C.; Bernt, K.M. MLL-rearranged leukemias—An update on science and clinical approaches. Front. Pediatr. 2017, 5, 4. [Google Scholar] [CrossRef]
- Shih, L.; Liang, D.; Fu, J.; Wu, J.; Wang, P.; Lin, T.; Dunn, P.; Kuo, M.; Tang, T.; Lin, T. Characterization of fusion partner genes in 114 patients with de novo acute myeloid leukemia and MLL rearrangement. Leukemia 2006, 20, 218–223. [Google Scholar] [CrossRef]
- Tamai, H.; Inokuchi, K. 11q23/MLL acute leukemia: Update of clinical aspects. J. Clin. Exp. Hematop. 2010, 50, 91–98. [Google Scholar] [CrossRef]
- Chen, C.-S.; Sorensen, P.; Domer, P.H.; Reaman, G.H.; Korsmeyer, S.J.; Heerema, N.A.; Hammond, G.D.; Kersey, J.H. Molecular rearrangements on chromosome 11q23 predominate in infant acute lymphoblastic leukemia and are associated with specific biologic variables and poor outcome. Blood 1993, 81, 2386–2393. [Google Scholar] [CrossRef] [PubMed]
- Mrózek, K.; Heinonen, K.; Lawrence, D.; Carroll, A.J.; Koduru, P.R.; Rao, K.W.; Strout, M.P.; Hutchison, R.E.; Moore, J.O.; Mayer, R.J. Adult patients with de novo acute myeloid leukemia and t (9; 11)(p22; q23) have a superior outcome to patients with other translocations involving band 11q23: A cancer and leukemia group B study. Blood J. Am. Soc. Hematol. 1997, 90, 4532–4538. [Google Scholar]
- Tomizawa, D.; Koh, K.; Sato, T.; Kinukawa, N.; Morimoto, A.; Isoyama, K.; Kosaka, Y.; Oda, T.; Oda, M.; Hayashi, Y. Outcome of risk-based therapy for infant acute lymphoblastic leukemia with or without an MLL gene rearrangement, with emphasis on late effects: A final report of two consecutive studies, MLL96 and MLL98, of the Japan Infant Leukemia Study Group. Leukemia 2007, 21, 2258–2263. [Google Scholar] [CrossRef] [PubMed]
- Hilden, J.M.; Dinndorf, P.A.; Meerbaum, S.O.; Sather, H.; Villaluna, D.; Heerema, N.A.; McGlennen, R.; Smith, F.O.; Woods, W.G.; Salzer, W.L. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: Report on CCG 1953 from the Children’s Oncology Group. Blood 2006, 108, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.-H.; Gaynon, P.S.; Boyett, J.M.; Chessells, J.M.; Baruchel, A.; Kamps, W.; Silverman, L.B.; Biondi, A.; Harms, D.O.; Vilmer, E. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet 2002, 359, 1909–1915. [Google Scholar] [CrossRef]
- Li, X.; Song, Y. Structure, function and inhibition of critical protein–protein interactions involving mixed lineage leukemia 1 and its fusion oncoproteins. J. Hematol. Oncol. 2021, 14, 56. [Google Scholar] [CrossRef]
- Sasaki, K.; Ravandi, F.; Kadia, T.M.; DiNardo, C.D.; Short, N.J.; Borthakur, G.; Jabbour, E.; Kantarjian, H.M. De novo acute myeloid leukemia: A population-based study of outcome in the United States based on the Surveillance, Epidemiology, and End Results (SEER) database, 1980 to 2017. Cancer 2021, 127, 2049–2061. [Google Scholar] [CrossRef] [PubMed]
- Super, H.; McCabe, N.R.; Thirman, M.J.; Larson, R.A.; Le Beau, M.M.; Pedersen-Bjergaard, J.; Philip, P.; Diaz, M.O.; Rowley, J.D. Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood 1993, 82, 3705–3711. [Google Scholar] [CrossRef]
- Felix, C.A. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Et Biophys. Acta (BBA)-Gene Struct. Expr. 1998, 1400, 233–255. [Google Scholar] [CrossRef]
- Britten, O.; Ragusa, D.; Tosi, S.; Mostafa Kamel, Y. MLL-Rearranged Acute Leukemia with t (4; 11)(q21; q23)—Current Treatment Options. Is There a Role for CAR-T Cell Therapy? Cells 2019, 8, 1341. [Google Scholar] [CrossRef]
- Stein, E.M.; Garcia-Manero, G.; Rizzieri, D.A.; Tibes, R.; Berdeja, J.G.; Savona, M.R.; Jongen-Lavrenic, M.; Altman, J.K.; Thomson, B.; Blakemore, S.J. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood J. Am. Soc. Hematol. 2018, 131, 2661–2669. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Burmeister, T.; Gröger, D.; Tsaur, G.; Fechina, L.; Renneville, A.; Sutton, R.; Venn, N.; Emerenciano, M.; Pombo-de-Oliveira, M. The MLL recombinome of acute leukemias in 2017. Leukemia 2018, 32, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Marschalek, R. Systematic classification of mixed-lineage leukemia fusion partners predicts additional cancer pathways. Ann. Lab. Med. 2016, 36, 85. [Google Scholar] [CrossRef]
- Liu, H.; Cheng, E.H.; Hsieh, J.J. MLL fusions: Pathways to leukemia. Cancer Biol. Ther. 2009, 8, 1204–1211. [Google Scholar] [CrossRef]
- Yokoyama, A.; Lin, M.; Naresh, A.; Kitabayashi, I.; Cleary, M.L. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 2010, 17, 198–212. [Google Scholar] [CrossRef]
- Mohan, M.; Herz, H.-M.; Takahashi, Y.-H.; Lin, C.; Lai, K.C.; Zhang, Y.; Washburn, M.P.; Florens, L.; Shilatifard, A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 2010, 24, 574–589. [Google Scholar] [CrossRef]
- Lin, C.; Smith, E.R.; Takahashi, H.; Lai, K.C.; Martin-Brown, S.; Florens, L.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C.; Shilatifard, A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 2010, 37, 429–437. [Google Scholar] [CrossRef]
- De Boer, J.; Walf-Vorderwülbecke, V.; Williams, O. In focus: MLL-rearranged leukemia. Leukemia 2013, 27, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Zeisig, B.B.; Milne, T.; García-Cuéllar, M.-P.; Schreiner, S.; Martin, M.-E.; Fuchs, U.; Borkhardt, A.; Chanda, S.K.; Walker, J.; Soden, R. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol. Cell. Biol. 2004, 24, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yao, Y.; Wu, F.; Song, Y. A proteolysis-targeting chimera molecule selectively degrades ENL and inhibits malignant gene expression and tumor growth. J. Hematol. Oncol. 2022, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Nie, S.; Yao, Y.; Huo, T.; Li, X.; Wu, X.; Zhao, J.; Lin, Y.L.; Zhang, Y.; Mo, Q.; et al. Small-molecule inhibitor of AF9/ENL-DOT1L/AF4/AFF4 interactions suppresses malignant gene expression and tumor growth. Theranostics 2021, 11, 8172–8184. [Google Scholar] [CrossRef]
- Kuntimaddi, A.; Achille, N.J.; Thorpe, J.; Lokken, A.A.; Singh, R.; Hemenway, C.S.; Adli, M.; Zeleznik-Le, N.J.; Bushweller, J.H. Degree of recruitment of DOT1L to MLL-AF9 defines level of H3K79 Di- and tri-methylation on target genes and transformation potential. Cell Rep. 2015, 11, 808–820. [Google Scholar] [CrossRef]
- Leach, B.I.; Kuntimaddi, A.; Schmidt, C.R.; Cierpicki, T.; Johnson, S.A.; Bushweller, J.H. Leukemia fusion target AF9 is an intrinsically disordered transcriptional regulator that recruits multiple partners via coupled folding and binding. Structure 2013, 21, 176–183. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Feng, Z.; Lemieux, M.E.; Faber, J.; Vempati, S.; Sinha, A.U.; Xia, X.; Jesneck, J.; Bracken, A.P.; Silverman, L.B.; et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 2008, 14, 355–368. [Google Scholar] [CrossRef]
- Daigle, S.R.; Olhava, E.J.; Therkelsen, C.A.; Majer, C.R.; Sneeringer, C.J.; Song, J.; Johnston, L.D.; Scott, M.P.; Smith, J.J.; Xiao, Y. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011, 20, 53–65. [Google Scholar] [CrossRef]
- Daigle, S.R.; Olhava, E.J.; Therkelsen, C.A.; Basavapathruni, A.; Jin, L.; Boriack-Sjodin, P.A.; Allain, C.J.; Klaus, C.R.; Raimondi, A.; Scott, M.P. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood J. Am. Soc. Hematol. 2013, 122, 1017–1025. [Google Scholar] [CrossRef]
- Anglin, J.L.; Deng, L.; Yao, Y.; Cai, G.; Liu, Z.; Jiang, H.; Cheng, G.; Chen, P.; Dong, S.; Song, Y. Synthesis and structure-activity relationship investigation of adenosine-containing inhibitors of histone methyltransferase DOT1L. J. Med. Chem. 2012, 55, 8066–8074. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Grigsby, S.M.; Yao, A.; Chang, Y.; Johnson, G.; Sun, H.; Nikolovska-Coleska, Z. Peptidomimetics for targeting protein–protein interactions between DOT1L and MLL oncofusion proteins AF9 and ENL. ACS Med. Chem. Lett. 2018, 9, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Du, L.; Tan, R.; Yu, Y.; Jiang, J.; Yao, A.; Luo, J.; Tang, R.; Xiao, Y.; Sun, H. Design, Synthesis, and Biological Evaluations of DOT1L Peptide Mimetics Targeting the Protein–Protein Interactions between DOT1L and MLL-AF9/MLL-ENL. J. Med. Chem. 2022, 65, 7770–7785. [Google Scholar] [CrossRef] [PubMed]
- Erfurth, F.; Hemenway, C.; De Erkenez, A.; Domer, P. MLL fusion partners AF4 and AF9 interact at subnuclear foci. Leukemia 2004, 18, 92–102. [Google Scholar] [CrossRef]
- Bitoun, E.; Oliver, P.L.; Davies, K.E. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 2007, 16, 92–106. [Google Scholar] [CrossRef]
- Monroe, S.C.; Jo, S.Y.; Sanders, D.S.; Basrur, V.; Elenitoba-Johnson, K.S.; Slany, R.K.; Hess, J.L. MLL-AF9 and MLL-ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia. Exp. Hematol. 2011, 39, 77–86.e75. [Google Scholar] [CrossRef]
- Wan, L.; Wen, H.; Li, Y.; Lyu, J.; Xi, Y.; Hoshii, T.; Joseph, J.K.; Wang, X.; Loh, Y.E.; Erb, M.A.; et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017, 543, 265–269. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Miller, D.M.; Thomas, S.D.; Islam, A.; Muench, D.; Sedoris, K. c-Myc and cancer metabolism. Clin. Cancer. Res. 2012, 18, 5546–5553. [Google Scholar] [CrossRef]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, metabolism, and cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef]
- Nie, S.; Zhao, J.; Wu, X.; Yao, Y.; Wu, F.; Lin, Y.-L.; Li, X.; Kneubehl, A.R.; Vogt, M.B.; Rico-Hesse, R. Synthesis, structure-activity relationship and antiviral activity of indole-containing inhibitors of Flavivirus NS2B-NS3 protease. Eur. J. Med. Chem. 2021, 225, 113767. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).