RNA Editing in Cancer Progression
Simple Summary
Abstract
1. Introduction
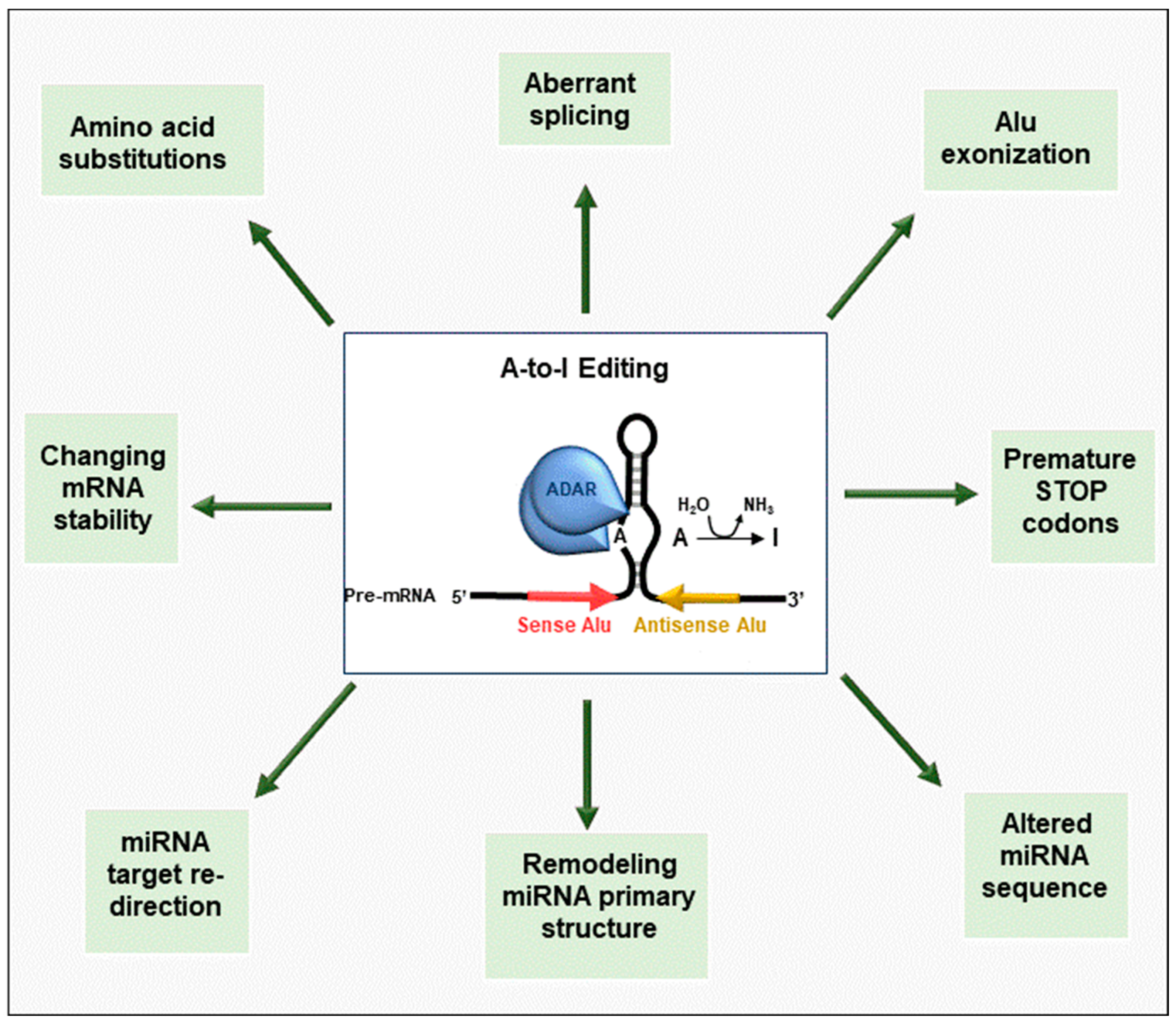
2. An Overview of RNA Editing
3. Dysregulation of RNA Editing in Cancer Progression
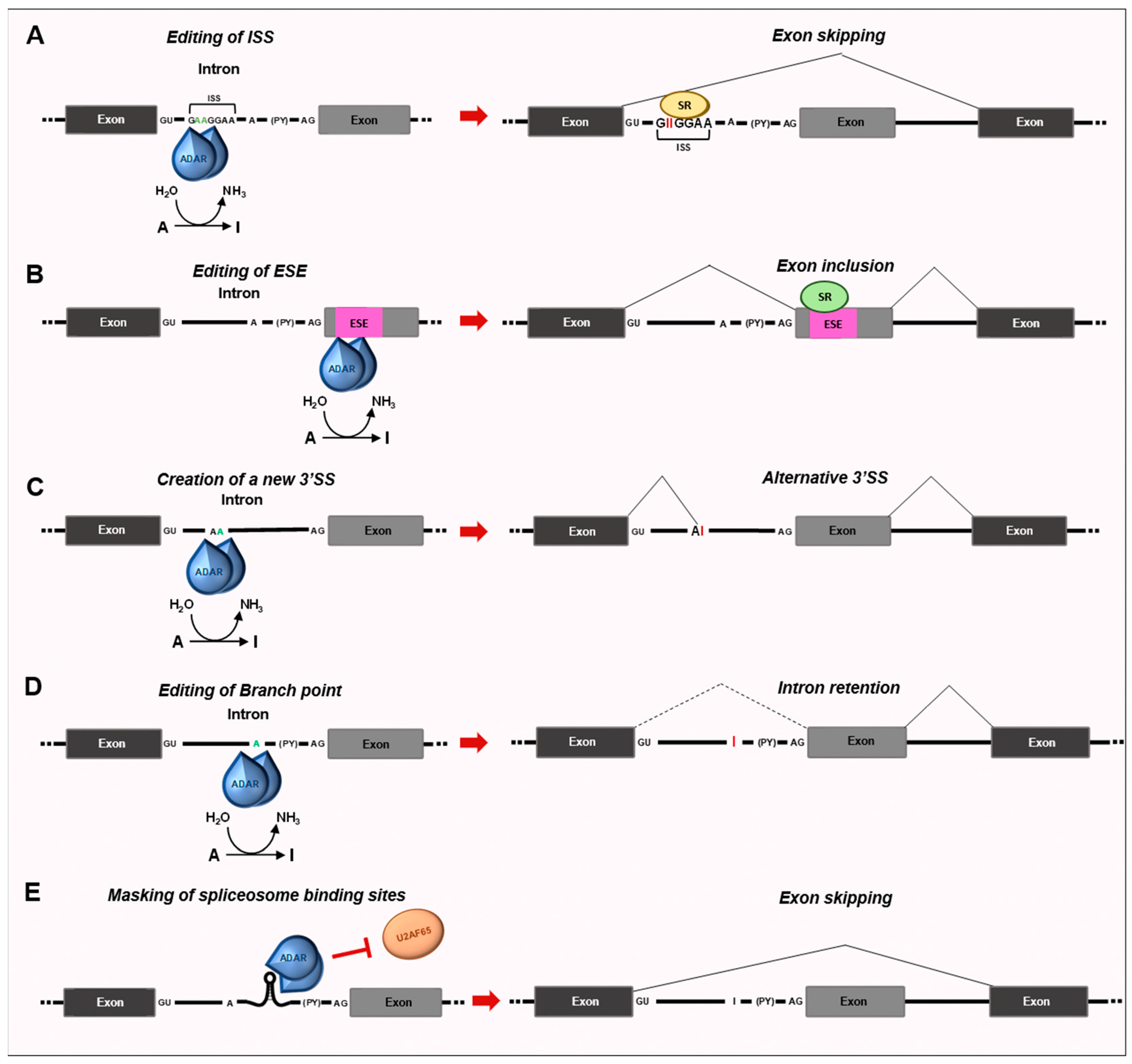
3.1. Impact of RNA Editing on Splicing Decisions
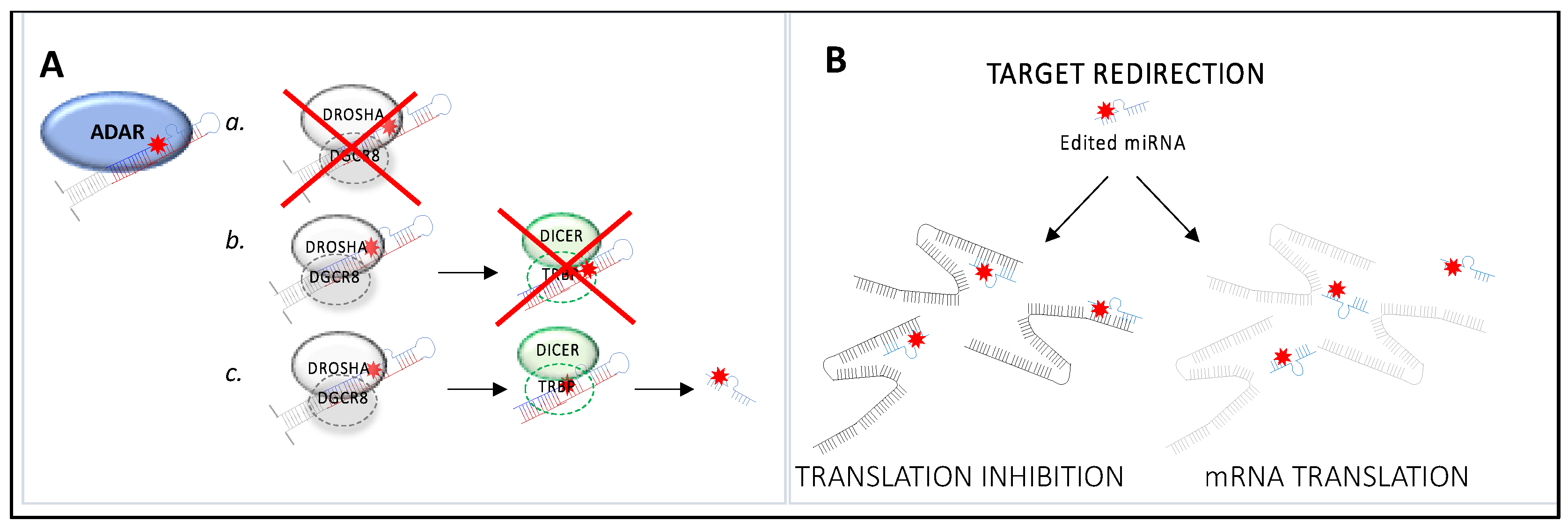
3.2. Impact of RNA Editing on miRNA Function
| Type of Cancer | Enzyme Involved | Type of Activity | miRNA Name | Downstream Effect |
|---|---|---|---|---|
| Thyroid cancer [86] | ADAR1 | Overedited (A-I) | miR-200b | Alteration of miRNA ability to target 3′UTR of ZEB1, promoting cancer aggressiveness |
| Melanoma [87] | ADAR1 | Underedited (A-I) | miR-455-5p | Binding to the 3′UTR of CPEB1, promoting cancer progression |
| [89] | ADAR1 | Overedited (A-I) | miR-378a-3p | Binding to the 3′UTR of PARVA, preventing cancer progression |
| Chronic lymphocytic leukemia [90] | ADARB1 | Overedited | miR-15/16 | Dysregulated miRNA processing, promoting cancer progression |
| [91] | ADAR1 | Overedited | miR-3157 miR-6503 | Shortened Progression-Free Survival |
| Nonsmall cell lung cancer [93] | ADAR | Underedited (A-I) | miR-411-5p | Changes in miRNA targets, promoting cancer |
| Breast cancer [94] | ADAR1 | Overedited (A-I) | miR-30b-3p miR-573 | Alteration of miRNA’s ability to target 3′UTR of ARHGAP26, promoting cancer progression |
| Glioblastoma [88] | ADARB1 | Underedited (A-I) | miR-376a | Binding to the 3′UTR of RAP2A, promoting invasiveness; |
| [94] | ADAR1 | Overedited (A-I) | miR-30b-3p miR-573 | Alteration of miRNAs ability to target 3′UTR of ARHGAP26, promoting cancer progression |
4. ADAR RNA Editing as a Novel Biomarker for Cancer Diagnosis and Treatment
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Herbert, A.; Rich, A. RNA processing and the evolution of eukaryotes. Nat. Genet. 1999, 21, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Alfonzo, J.D.; Thiemann, O.; Simpson, L. The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 1997, 25, 3751–3759. [Google Scholar] [CrossRef] [PubMed]
- Benne, R.; Van den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Feagin, J.E.; Abraham, J.M.; Stuart, K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 1988, 53, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.M.; Feagin, J.E.; Stuart, K.; Simpson, L. Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell 1988, 53, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Eisen, H. RNA editing: Who’s on first? Cell 1988, 53, 331–332. [Google Scholar] [CrossRef]
- Blum, B.; Bakalara, N.; Simpson, L. A model for RNA editing in kinetoplastid mitochondria: RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 1990, 60, 189–198. [Google Scholar] [CrossRef]
- Rusché, L.N.; Cruz-Reyes, J.; Piller, K.J.; Sollner-Webb, B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997, 16, 4069–4081. [Google Scholar] [CrossRef]
- Porath, H.T.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol. 2017, 18, 185. [Google Scholar] [CrossRef]
- Melcher, T.; Maas, S.; Higuchi, M.; Keller, W.; Seeburg, P.H. Editing of α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J. Biol. Chem. 1995, 270, 8566–8570. [Google Scholar] [CrossRef]
- Bass, B.L.; Nishikura, K.; Keller, W.; Seeburg, P.H.; Emeson, R.B.; O’Connell, M.A.; Samuel, C.E.; Herbert, A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA 1997, 3, 947–949. [Google Scholar] [PubMed]
- Snyder, E.; Chukrallah, L.; Seltzer, K.; Goodwin, L.; Braun, R.E. ADAD1 and ADAD2, testis-specific adenosine deaminase domain-containing proteins, are required for male fertility. Sci. Rep. 2020, 10, 11536. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, K.A.; Bass, B.L. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 2000, 39, 12875–12884. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A.; Alfken, J.; Kim, Y.G.; Mian, I.S.; Nishikura, K.; Rich, A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. USA 1997, 94, 8421–8426. [Google Scholar] [CrossRef]
- Athanasiadis, A.; Placido, D.; Maas, S.; Brown, B.A.; Lowenhaupt, K.; Rich, A. The crystal structure of the Zβ domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains. J. Mol. Biol. 2005, 351, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Nakahama, T.; Kawahara, Y. Deciphering the Biological Significance of ADAR1-Z-RNA Interactions. Int. J. Mol. Sci. 2021, 22, 11435. [Google Scholar] [CrossRef]
- Sansam, C.L.; Wells, K.S.; Emeson, R.B. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. USA 2003, 100, 14018–14023. [Google Scholar] [CrossRef]
- Liu, J.; Wang, F.; Zhang, Y.; Liu, J.; Zhao, B. ADAR1-Mediated RNA Editing and Its Role in Cancer. Front. Cell. Dev. Biol. 2022, 10, 956649. [Google Scholar] [CrossRef]
- Polson, A.G.; Bass, B.L. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994, 13, 5701–5711. [Google Scholar] [CrossRef]
- Lehmann, K.A.; Bass, B.L. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 1999, 291, 1–13. [Google Scholar] [CrossRef]
- Ensterö, M.; Akerborg, O.; Lundin, D.; Wang, B.; Furey, T.S.; Ohman, M.; Lagergren, J. A computational screen for site selective A-to-I editing detects novel sites in neuron specific Hu proteins. BMC Bioinform. 2010, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Daniel, C.; Widmark, A.; Rigardt, D.; Öhman, M. Editing inducer elements increases A-to-I editing efficiency in the mammalian transcriptome. Genome Biol. 2017, 18, 195. [Google Scholar] [CrossRef]
- Carmi, S.; Borukhov, I.; Levanon, E.Y. Identification of widespread ultra-edited human RNAs. PLoS Genet. 2011, 7, e1002317. [Google Scholar] [CrossRef] [PubMed]
- Porath, H.T.; Carmi, S.; Levanon, E.Y. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat. Commun. 2014, 5, 4726. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004, 2, e391. [Google Scholar] [CrossRef] [PubMed]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef]
- Blow, M.; Futreal, P.A.; Wooster, R.; Stratton, M.R. A survey of RNA editing in human brain. Genome Res. 2004, 14, 2379–2387. [Google Scholar] [CrossRef]
- Kim, D.D.; Kim, T.T.; Walsh, T.; Kobayashi, Y.; Matise, T.C.; Buyske, S.; Gabriel, A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004, 14, 1719–1725. [Google Scholar] [CrossRef]
- Powell, L.M.; Wallis, S.C.; Pease, R.J.; Edwards, Y.H.; Knott, T.J.; Scott, J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell 1987, 50, 831–840. [Google Scholar] [CrossRef]
- Chen, S.H.; Habib, G.; Yang, C.Y.; Gu, Z.W.; Lee, B.R.; Weng, S.A.; Silberman, S.R.; Cai, S.J.; Deslypere, J.P.; Rosseneu, M.; et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science 1987, 238, 363–366. [Google Scholar] [CrossRef]
- Chester, A.; Scott, J.; Anant, S.; Navaratnam, N. RNA editing: Cytidine to uridine conversion in apolipoprotein B mRNA. Biochim. Biophys. Acta 2000, 1494, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Greeve, J.; Altkemper, I.; Dieterich, J.H.; Greten, H.; Windler, E. Apolipoprotein B mRNA editing in 12 different mammalian species: Hepatic expression is reflected in low concentrations of apoB-containing plasma lipoproteins. J. Lipid Res. 1993, 34, 1367–1383. [Google Scholar] [CrossRef] [PubMed]
- Boström, K.; Garcia, Z.; Poksay, K.S.; Johnson, D.F.; Lusis, A.J.; Innerarity, T.L. Apolipoprotein B mRNA editing. Direct determination of the edited base and occurrence in non-apolipoprotein B-producing cell lines. J. Biol. Chem. 1990, 265, 22446–22452. [Google Scholar] [CrossRef] [PubMed]
- Navaratnam, N.; Morrison, J.R.; Bhattacharya, S.; Patel, D.; Funahashi, T.; Giannoni, F.; Teng, B.B.; Davidson, N.O.; Scott, J. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 1993, 268, 20709–20712. [Google Scholar] [CrossRef]
- Anant, S.; MacGinnitie, A.J.; Davidson, N.O. The binding of apobec-1 to mammalian apo B RNA is stabilized by the presence of complementation factors which are required for post-transcriptional editing. Nucleic Acids Symp. Ser. 1995, 33, 99–102. [Google Scholar]
- Mehta, A.; Kinter, M.T.; Sherman, N.E.; Driscoll, D.M. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 2000, 20, 1846–1854. [Google Scholar] [CrossRef]
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef]
- Deffit, S.N.; Hundley, H.A. To edit or not to edit: Regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA 2016, 7, 113–127. [Google Scholar] [CrossRef]
- Daniel, C.; Silberberg, G.; Behm, M.; Öhman, M. Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol. 2014, 15, R28. [Google Scholar] [CrossRef]
- Kung, C.P.; Maggi, L.B., Jr.; Weber, J.D. The Role of RNA Editing in Cancer Development and Metabolic Disorders. Front. Endocrinol. 2018, 9, 762. [Google Scholar] [CrossRef]
- Behroozi, J.; Shahbazi, S.; Bakhtiarizadeh, M.R.; Mahmoodzadeh, H. ADAR expression and copy number variation in patients with advanced gastric cancer. BMC Gastroenterol. 2020, 20, 152. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, E.; Mondala, P.K.; Santos, N.D.; Miller, A.C.; Pineda, G.; Jiang, Q.; Leu, H.; Ali, S.A.; Ganesan, A.P.; Wu, C.N.; et al. Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma. Nat. Commun. 2017, 8, 1922. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, D.; Gacquer, D.; Rothé, F.; Lefort, A.; Libert, F.; Brown, D.; Kheddoumi, N.; Shlien, A.; Konopka, T.; Salgado, R.; et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell Rep. 2015, 13, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Cenci, C.; Barzotti, R.; Galeano, F.; Corbelli, S.; Rota, R.; Massimi, L.; Di Rocco, C.; O’Connell, M.A.; Gallo, A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 2008, 283, 7251–7260. [Google Scholar] [CrossRef] [PubMed]
- Mendez Ruiz, S.; Chalk, A.M.; Goradia, A.; Heraud-Farlow, J.; Walkley, C.R. Over-expression of ADAR1 in mice does not initiate or accelerate cancer formation in vivo. NAR Cancer 2023, 5, zcad023. [Google Scholar] [CrossRef]
- Amin, E.M.; Liu, Y.; Deng, S.; Tan, K.S.; Chudgar, N.; Mayo, M.W.; Sanchez-Vega, F.; Adusumilli, P.S.; Schultz, N.; Jones, D.R. The RNA-editing enzyme ADAR promotes lung adenocarcinoma migration and invasion by stabilizing FAK. Sci. Signal 2017, 10, aah3941. [Google Scholar] [CrossRef]
- Fu, L.; Qin, Y.R.; Ming, X.Y.; Zuo, X.B.; Diao, Y.W.; Zhang, L.Y.; Ai, J.; Liu, B.L.; Huang, T.X.; Cao, T.T.; et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc. Natl. Acad. Sci USA 2017, 114, E4631–E4640. [Google Scholar] [CrossRef]
- Sagredo, E.A.; Blanco, A.; Sagredo, A.I.; Pérez, P.; Sepúlveda-Hermosilla, G.; Morales, F.; Müller, B.; Verdugo, R.; Marcelain, K.; Harismendy, O.; et al. ADAR1-mediated RNA-editing of 3′UTRs in breast cancer. Biol. Res. 2018, 51, 36. [Google Scholar] [CrossRef]
- Galeano, F.; Rossetti, C.; Tomaselli, S.; Cifaldi, L.; Lezzerini, M.; Pezzullo, M.; Boldrini, R.; Massimi, L.; Di Rocco, C.M.; Locatelli, F.; et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene 2013, 32, 998–1009. [Google Scholar] [CrossRef]
- Maas, S.; Patt, S.; Schrey, M.; Rich, A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. USA 2001, 98, 14687–14692. [Google Scholar] [CrossRef]
- Ishiuchi, S.; Tsuzuki, K.; Yoshida, Y.; Yamada, N.; Hagimura, N.; Okado, H.; Miwa, A.; Kurihara, H.; Nakazato, Y.; Tamura, M.; et al. Blockage of Ca(2+)-permeable AMPA receptors suppresses migration and induces apoptosis in human glioblastoma cells. Nat. Med. 2002, 8, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Galeano, F.; Leroy, A.; Rossetti, C.; Gromova, I.; Gautier, P.; Keegan, L.P.; Massimi, L.; Di Rocco, C.; O’Connell, M.A.; Gallo, A. Human BLCAP transcript: New editing events in normal and cancerous tissues. Int. J. Cancer 2010, 127, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; He, W.; Cai, H.; Hu, B.; Zheng, C.; Ke, X.; Xie, L.; Zheng, Z.; Wu, X.; Wang, H. A-to-I RNA editing of BLCAP lost the inhibition to STAT3 activation in cervical cancer. Oncotarget 2017, 8, 39417–39429. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Y.; Lin, C.H.; Chan, T.H.; Chow, R.K.; Song, Y.; Liu, M.; Yuan, Y.F.; Fu, L.; Kong, K.L.; et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013, 19, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.H.; Lin, C.H.; Qi, L.; Fei, J.; Li, Y.; Yong, K.J.; Liu, M.; Song, Y.; Chow, R.K.; Ng, V.H.; et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut 2014, 63, 832–843. [Google Scholar] [CrossRef]
- Qin, Y.R.; Qiao, J.J.; Chan, T.H.; Zhu, Y.H.; Li, F.F.; Liu, H.; Fei, J.; Li, Y.; Guan, X.Y.; Chen, L. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014, 74, 840–851. [Google Scholar] [CrossRef]
- HHu, X.; Chen, J.; Shi, X.; Feng, F.; Lau, K.W.; Chen, Y.; Chen, Y.; Jiang, L.; Cui, F.; Zhang, Y.; et al. RNA editing of AZIN1 induces the malignant progression of non-small-cell lung cancers. Tumor Biol. 2017, 39, 1010428317700001. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Shigeyasu, K.; Yamamoto, A.; Shigemori, T.; Yin, C.; Ichikawa, T.; Yasuda, H.; Fujikawa, H.; Yoshiyama, S.; et al. Enhanced AZIN1 RNA editing and overexpression of its regulatory enzyme ADAR1 are important prognostic biomarkers in gastric cancer. J. Transl. Med. 2018, 16, 366. [Google Scholar] [CrossRef]
- Chan, T.H.; Qamra, A.; Tan, K.T.; Guo, J.; Yang, H.; Qi, L.; Lin, J.S.; Ng, V.H.; Song, Y.; Hong, H.; et al. ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer. Gastroenterology 2016, 151, 637–650.e610. [Google Scholar] [CrossRef]
- Shigeyasu, K.; Okugawa, Y.; Toden, S.; Miyoshi, J.; Toiyama, Y.; Nagasaka, T.; Takahashi, N.; Kusunoki, M.; Takayama, T.; Yamada, Y.; et al. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight 2018, 3, e99976. [Google Scholar] [CrossRef]
- Takeda, S.; Shigeyasu, K.; Okugawa, Y.; Yoshida, K.; Mori, Y.; Yano, S.; Noma, K.; Umeda, Y.; Kondo, Y.; Kishimoto, H.; et al. Activation of AZIN1 RNA editing is a novel mechanism that promotes invasive potential of cancer-associated fibroblasts in colorectal cancer. Cancer Lett. 2019, 444, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Han, S.W.; Kim, H.P.; Shin, J.Y.; Jeong, E.G.; Lee, W.C.; Kim, K.Y.; Park, S.Y.; Lee, D.W.; Won, J.K.; Jeong, S.Y.; et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J. Exp. Med. 2014, 211, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, T.; Rahman, M.F.; Tostar, U.; Sonkoly, E.; Ståhle, M.; Pivarcsi, A.; Palaniswamy, R.; Zaphiropoulos, P.G. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol. 2013, 10, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Gumireddy, K.; Li, A.; Kossenkov, A.V.; Sakurai, M.; Yan, J.; Li, Y.; Xu, H.; Wang, J.; Zhang, P.J.; Zhang, L.; et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat. Commun. 2016, 7, 10715. [Google Scholar] [CrossRef] [PubMed]
- Skuse, G.R.; Cappione, A.J.; Sowden, M.; Metheny, L.J.; Smith, H.C. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res. 1996, 24, 478–485. [Google Scholar] [CrossRef]
- Mukhopadhyay, D.; Anant, S.; Lee, R.M.; Kennedy, S.; Viskochil, D.; Davidson, N.O. C→U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am. J. Hum. Genet. 2002, 70, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Law, E.K.; Sieuwerts, A.M.; LaPara, K.; Leonard, B.; Starrett, G.J.; Molan, A.M.; Temiz, N.A.; Vogel, R.I.; Meijer-van Gelder, M.E.; Sweep, F.C.; et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2016, 2, e1601737. [Google Scholar] [CrossRef]
- Niavarani, A.; Currie, E.; Reyal, Y.; Anjos-Afonso, F.; Horswell, S.; Griessinger, E.; Luis Sardina, J.; Bonnet, D. APOBEC3A is implicated in a novel class of G-to-A mRNA editing in WT1 transcripts. PLoS ONE 2015, 10, e0120089. [Google Scholar] [CrossRef]
- Hsiao, Y.E.; Bahn, J.H.; Yang, Y.; Lin, X.; Tran, S.; Yang, E.W.; Quinones-Valdez, G.; Xiao, X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018, 28, 812–823. [Google Scholar] [CrossRef]
- Laurencikiene, J.; Källman, A.M.; Fong, N.; Bentley, D.L.; Ohman, M. RNA editing and alternative splicing: The importance of co-transcriptional coordination. EMBO Rep. 2006, 7, 303–307. [Google Scholar] [CrossRef]
- Ryman, K.; Fong, N.; Bratt, E.; Bentley, D.L.; Ohman, M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA 2007, 13, 1071–1078. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rueter, S.M.; Dawson, T.R.; Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature 1999, 399, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, Z.; Du, C.; Qi, B.; Zhao, X.; Wang, L.; Bi, L.; Wang, G.; Zhang, X.; Su, X.; et al. Abnormal expression of an ADAR2 alternative splicing variant in gliomas downregulates adenosine-to-inosine RNA editing. Acta Neurochir. 2014, 156, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Chang, I.Y.; Liu, H.; Ma, C.P.; Kuo, Y.P.; Shih, C.T.; Shih, Y.H.; Kang, L.; Tan, B.C. Tumor-associated intronic editing of HNRPLL generates a novel splicing variant linked to cell proliferation. J. Biol. Chem. 2018, 293, 10158–10171. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.J.; Shen, H.; An, O.; Hong, H.; Li, J.; Song, Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Bellido Molias, F.; et al. Cis- and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development. Nat. Commun. 2020, 11, 799. [Google Scholar] [CrossRef]
- Beghini, A.; Ripamonti, C.B.; Peterlongo, P.; Roversi, G.; Cairoli, R.; Morra, E.; Larizza, L. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum. Mol. Genet. 2000, 9, 2297–2304. [Google Scholar] [CrossRef]
- Eisenberg, E.; Nemzer, S.; Kinar, Y.; Sorek, R.; Rechavi, G.; Levanon, E.Y. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005, 21, 77–81. [Google Scholar] [CrossRef]
- Wallace, M.R.; Andersen, L.B.; Saulino, A.M.; Gregory, P.E.; Glover, T.W.; Collins, F.S. A de novo Alu insertion results in neurofibromatosis type 1. Nature 1991, 353, 864–866. [Google Scholar] [CrossRef]
- Lev-Maor, G.; Sorek, R.; Shomron, N.; Ast, G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 2003, 300, 1288–1291. [Google Scholar] [CrossRef]
- Lev-Maor, G.; Sorek, R.; Levanon, E.Y.; Paz, N.; Eisenberg, E.; Ast, G. RNA-editing-mediated exon evolution. Genome Biol. 2007, 8, R29. [Google Scholar] [CrossRef]
- Sakurai, M.; Yano, T.; Kawabata, H.; Ueda, H.; Suzuki, T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 2010, 6, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Chan, T.W.; Bahn, J.H.; Kim, T.H.; Rowat, A.C.; Xiao, X. Multifaceted role of RNA editing in promoting loss-of-function of PODXL in cancer. iScience 2022, 25, 104836. [Google Scholar] [CrossRef] [PubMed]
- Marceca, G.P.; Distefano, R.; Tomasello, L.; Lagana, A.; Russo, F.; Calore, F.; Romano, G.; Bagnoli, M.; Gasparini, P.; Ferro, A.; et al. MiREDiBase, a manually curated database of validated and putative editing events in microRNAs. Sci. Data 2021, 8, 199. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Marceca, G.P.; Tomasello, L.; Distefano, R.; Acunzo, M.; Croce, C.M.; Nigita, G. Detecting and Characterizing A-To-I microRNA Editing in Cancer. Cancers 2021, 13, 1699. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Moya, J.; Baker, A.R.; Slack, F.J.; Santisteban, P. ADAR1-mediated RNA editing is a novel oncogenic process in thyroid cancer and regulates miR-200 activity. Oncogene 2020, 39, 3738–3753. [Google Scholar] [CrossRef]
- Shoshan, E.; Mobley, A.K.; Braeuer, R.R.; Kamiya, T.; Huang, L.; Vasquez, M.E.; Salameh, A.; Lee, H.J.; Kim, S.J.; Ivan, C.; et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015, 17, 311–321. [Google Scholar] [CrossRef]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012, 122, 4059–4076. [Google Scholar] [CrossRef]
- Velazquez-Torres, G.; Shoshan, E.; Ivan, C.; Huang, L.; Fuentes-Mattei, E.; Paret, H.; Kim, S.J.; Rodriguez-Aguayo, C.; Xie, V.; Brooks, D.; et al. A-to-I miR-378a-3p editing can prevent melanoma progression via regulation of PARVA expression. Nat. Commun. 2018, 9, 461. [Google Scholar] [CrossRef]
- Allegra, D.; Bilan, V.; Garding, A.; Döhner, H.; Stilgenbauer, S.; Kuchenbauer, F.; Mertens, D. Defective DROSHA processing contributes to downregulation of MiR-15/-16 in chronic lymphocytic leukemia. Leukemia 2014, 28, 98–107. [Google Scholar] [CrossRef]
- Gassner, F.J.; Zaborsky, N.; Buchumenski, I.; Levanon, E.Y.; Gatterbauer, M.; Schubert, M.; Rauscher, S.; Hebenstreit, D.; Nadeu, F.; Campo, E.; et al. RNA editing contributes to epitranscriptome diversity in chronic lymphocytic leukemia. Leukemia 2020, 35, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Gassner, F.J.; Zaborsky, N.; Feldbacher, D.; Greil, R.; Geisberger, R. RNA Editing Alters miRNA Function in Chronic Lymphocytic Leukemia. Cancers 2020, 12, 1159. [Google Scholar] [CrossRef] [PubMed]
- Nigita, G.; Distefano, R.; Veneziano, D.; Romano, G.; Rahman, M.; Wang, K.; Pass, H.; Croce, C.M.; Acunzo, M.; Nana-Sinkam, P. Tissue and exosomal miRNA editing in Non-Small Cell Lung Cancer. Sci. Rep. 2018, 8, 10222. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hui, H.; Guo, Z.; Zhang, W.; Hu, Y.; He, T.; Tai, Y.; Peng, P.; Wang, L. ADAR1 regulates ARHGAP26 gene expression through RNA editing by disrupting miR-30b-3p and miR-573 binding. RNA 2013, 19, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Paz, N.; Levanon, E.Y.; Amariglio, N.; Heimberger, A.B.; Ram, Z.; Constantini, S.; Barbash, Z.S.; Adamsky, K.; Safran, M.; Hirschberg, A.; et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007, 17, 1586–1595. [Google Scholar] [CrossRef]
- Paz-Yaacov, N.; Bazak, L.; Buchumenski, I.; Porath, H.T.; Danan-Gotthold, M.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 2015, 13, 267–276. [Google Scholar] [CrossRef]
- Han, L.; Diao, L.; Yu, S.; Xu, X.; Li, J.; Zhang, R.; Yang, Y.; Werner, H.M.J.; Eterovic, A.K.; Yuan, Y.; et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 2015, 28, 515–528. [Google Scholar] [CrossRef]
- Wang, B.; Tian, P.; Sun, Q.; Zhang, H.; Han, L.; Zhu, B. A novel, effective machine learning-based RNA editing profile for predicting the prognosis of lower-grade gliomas. Heliyon 2023, 9, e18075. [Google Scholar] [CrossRef]
- Shah, S.P.; Morin, R.D.; Khattra, J.; Prentice, L.; Pugh, T.; Burleigh, A.; Delaney, A.; Gelmon, K.; Guliany, R.; Senz, J.; et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 2009, 461, 809–813. [Google Scholar] [CrossRef]
- Chalk, A.M.; Taylor, S.; Heraud-Farlow, J.E.; Walkley, C.R. The majority of A-to-I RNA editing is not required for mammalian homeostasis. Genome Biol. 2019, 20, 268. [Google Scholar] [CrossRef]
- Higuchi, M.; Maas, S.; Single, F.N.; Hartner, J.; Rozov, A.; Burnashev, N.; Feldmeyer, D.; Sprengel, R.; Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000, 406, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Hartner, J.C.; Schmittwolf, C.; Kispert, A.; Müller, A.M.; Higuchi, M.; Seeburg, P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004, 279, 4894–4902. [Google Scholar] [CrossRef] [PubMed]
- Spremulli, E.N.; Cummings, F.J.; Crabtree, G.W.; LaBresh, K.; Jordan, M.; Calabresi, P. Hemodynamic effects of potentially useful antineoplastic agents. J. Natl. Cancer Inst. 1983, 70, 499–504. [Google Scholar] [PubMed]
- Steinman, R.A.; Yang, Q.; Gasparetto, M.; Robinson, L.J.; Liu, X.; Lenzner, D.E.; Hou, J.; Smith, C.; Wang, Q. Deletion of the RNA-editing enzyme ADAR1 causes regression of established chronic myelogenous leukemia in mice. Int. J. Cancer 2013, 132, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Gannon, H.S.; Zou, T.; Kiessling, M.K.; Gao, G.F.; Cai, D.; Choi, P.S.; Ivan, A.P.; Buchumenski, I.; Berger, A.C.; Goldstein, J.T.; et al. Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells. Nat. Commun. 2018, 9, 5450. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, J.J.; Manguso, R.T.; Cheruiyot, C.K.; Bi, K.; Panda, A.; Iracheta-Vellve, A.; Miller, B.C.; Du, P.P.; Yates, K.B.; Dubrot, J.; et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 2019, 565, 43–48. [Google Scholar] [CrossRef]
- Liu, X.; Fu, Y.; Huang, J.; Wu, M.; Zhang, Z.; Xu, R.; Zhang, P.; Zhao, S.; Liu, L.; Jiang, H. ADAR1 promotes the epithelial-to-mesenchymal transition and stem-like cell phenotype of oral cancer by facilitating oncogenic microRNA maturation. J. Exp. Clin. Cancer Res. 2019, 38, 315. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, S.; Galeano, F.; Alon, S.; Raho, S.; Galardi, S.; Polito, V.A.; Presutti, C.; Vincenti, S.; Eisenberg, E.; Locatelli, F.; et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015, 16, 5. [Google Scholar] [CrossRef]
- Patterson, J.B.; Samuel, C.E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 1995, 15, 5376–5388. [Google Scholar] [CrossRef]
- Valente, L.; Nishikura, K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 2007, 282, 16054–16061. [Google Scholar] [CrossRef]
- Hong, H.; An, O.; Chan, T.H.M.; Ng, V.H.E.; Kwok, H.S.; Lin, J.S.; Qi, L.; Han, J.; Tay, D.J.T.; Tang, S.J.; et al. Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer. Nucleic Acids Res. 2018, 46, 7953–7969. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Grosse, F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair 2011, 10, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Fidaleo, M.; De Paola, E.; Paronetto, M.P. The RNA helicase A in malignant transformation. Oncotarget 2016, 7, 28711–28723. [Google Scholar] [CrossRef] [PubMed]
- Palombo, R.; Verdile, V.; Paronetto, M.P. Poison-Exon Inclusion in DHX9 Reduces Its Expression and Sensitizes Ewing Sarcoma Cells to Chemotherapeutic Treatment. Cells 2020, 9, 328. [Google Scholar] [CrossRef] [PubMed]
- Chellini, L.; Pieraccioli, M.; Sette, C.; Paronetto, M.P. The DNA/RNA helicase DHX9 contributes to the transcriptional program of the androgen receptor in prostate cancer. J. Exp. Clin. Cancer Res. 2022, 41, 178. [Google Scholar] [CrossRef] [PubMed]
- Solomon, O.; Oren, S.; Safran, M.; Deshet-Unger, N.; Akiva, P.; Jacob-Hirsch, J.; Cesarkas, K.; Kabesa, R.; Amariglio, N.; Unger, R.; et al. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR). RNA 2013, 19, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Licht, K.; Jantsch, M.F. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 2016, 213, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Licht, K.; Kapoor, U.; Mayrhofer, E.; Jantsch, M.F. Adenosine to Inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 2016, 44, 6398–6408. [Google Scholar] [CrossRef]
- Hundley, H.A.; Bass, B.L. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 2010, 35, 377–383. [Google Scholar] [CrossRef]
- Havens, M.A.; Hastings, M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef]
- Singh, N.N.; Howell, M.D.; Androphy, E.J.; Singh, R.N. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017, 24, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhao, X.; Li, Z.; Wei, J.; Tian, Y. Splicing variants of ADAR2 and ADAR2-mediated RNA editing in glioma. Oncol. Lett. 2016, 12, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tian, Y.; Tian, N.; Zhao, X.; Du, C.; Han, L.; Zhang, H. Aberrant alternative splicing pattern of ADAR2 downregulates adenosine-to-inosine editing in glioma. Oncol. Rep. 2015, 33, 2845–2852. [Google Scholar] [CrossRef] [PubMed]
| Type of Cancer | Enzyme Involved | Type of Activity | Downstream Effect |
|---|---|---|---|
| Nonsmall-cell lung cancers [46] | ADAR | Overedited (A-I) | Stabilization and increase of FAK transcript, promoting tumor progression |
| [57] | ADAR | Overedited (A-I) | Alteration of the AZIN1 transcript, resulting in nuclear translocation, promoting the malignant phenotype |
| Esophageal Squamous Cell [47] | ADAR2 | Overedited (A-I) | Decrease of SLC22A3 transcript, promoting tumor progression |
| [56] | ADAR1 | Overedited (A-I) | Alteration of the AZIN1 transcript, promoting tumor progression |
| Breast Cancer [48] | ADAR1 | Overedited (A-I) | Alteration in GINS4 transcript stability, alteration in ATM and POLH transcript expression, promoting tumorigenesis |
| [64] | ADAR1 | Overedited (A-I) | Alteration of the GABRA transcript, causing amino acid substitution and suppressing tumor progression |
| [67] | APOBEC3B | Underedited (C-U) | Not investigated |
| Astrocytoma [49] [44] | ADAR2 ADAR1/2 | Underedited (A-I) Underedited (A-I) | Alteration of the CDC14B pre-mRNA, increasing its expression with consequent reduction of tumorigenicity Not investigated |
| Glyoblastoma [51] | ADAR2 | Underedited (A-I) | Alteration of the GluR-B transcript, causing amino acid substitution and tumor invasiveness |
| Pediatric Astrocytoma [44] | ADAR2 | Underedited (A-I) | Not investigated |
| Cervical Cancer [53] | ADAR1 | Overedited (A-I) | Alteration of the binding motif of BLCAP for STAT3, promoting tumor progression |
| Hepatocellular carcinoma [54] | ADAR1 | Overedited (A-I) | Alteration of the AZIN1 transcript, causing amino acid substitution and promoting tumor progression |
| Gastric Cancer [58] | ADAR1 | Overedited (A-I) | Not investigated |
| [59] | ADAR2 | Overedited (A-I) | Alteration of the PODXL transcript, causing an amino acid substitution, reducing tumorigenicity |
| Colorectal cancer [60] | ADAR1 | Overedited (A-I) | Alteration of the AZIN1 transcript, promoting ODC accumulation and tumor progression |
| [62] | ADAR | Overedited (A-I) | Alteration of the RHOQ transcript, causing an amino acid substitution and promoting tumor progression |
| Multiple Myeloma [63] | ADAR1 | Overedited (A-I) | Alteration of the GLI1 transcript, causing an amino acid substitution and promoting tumor progression |
| [68] | APOBEC3A | Overedited (G-A) | Alteration in WT1 transcript, promoting tumorigenesis |
| Type of Cancer | Enzyme Involved | Type of Activity | Gene Name | Downstream Effect |
|---|---|---|---|---|
| Kidney and bladder [74] | ADAR1/2 | Overedited (A-I) | HNRNPLL | Creation of a binding site for SRSF1 and inclusion of exon 12A, promoting tumorigenesis |
| Esophageal squamous carcinoma [75] | ADAR1/2 | Overedited (A-I) | CCDC15 | Enhancing the binding sites for SRSF7 and repression of exon 9 inclusion, promoting tumorigenesis |
| Acute myeloid leukemia [76] | Not reported | Overedited (A-I) | PTPN6 | Retaining of intron 3, causing a nonsense translation, favoriting leukemogenesis |
| Glyoblastoma [81] | ADAR1 | Overedited (A-I) | SARS | Preventing aberrant exonization of Alu sequence into mature mRNA, protecting by degrading aberrant transcripts |
| Kidney Renal Clear Cell Carcinoma [82] | ADAR2 | Underedited (A-I) | PODXL | Inclusion of an alternative exon and production of a longer isoform, promoting cell migration |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frezza, V.; Chellini, L.; Del Verme, A.; Paronetto, M.P. RNA Editing in Cancer Progression. Cancers 2023, 15, 5277. https://doi.org/10.3390/cancers15215277
Frezza V, Chellini L, Del Verme A, Paronetto MP. RNA Editing in Cancer Progression. Cancers. 2023; 15(21):5277. https://doi.org/10.3390/cancers15215277
Chicago/Turabian StyleFrezza, Valentina, Lidia Chellini, Arianna Del Verme, and Maria Paola Paronetto. 2023. "RNA Editing in Cancer Progression" Cancers 15, no. 21: 5277. https://doi.org/10.3390/cancers15215277
APA StyleFrezza, V., Chellini, L., Del Verme, A., & Paronetto, M. P. (2023). RNA Editing in Cancer Progression. Cancers, 15(21), 5277. https://doi.org/10.3390/cancers15215277






