Inflammatory Bone Marrow Mesenchymal Stem Cells in Multiple Myeloma: Transcriptional Signature and In Vitro Modeling
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Expression Datasets
2.2. Data Analysis
3. Results
3.1. Transcriptional Signature of Primary MSCs from MM Patients
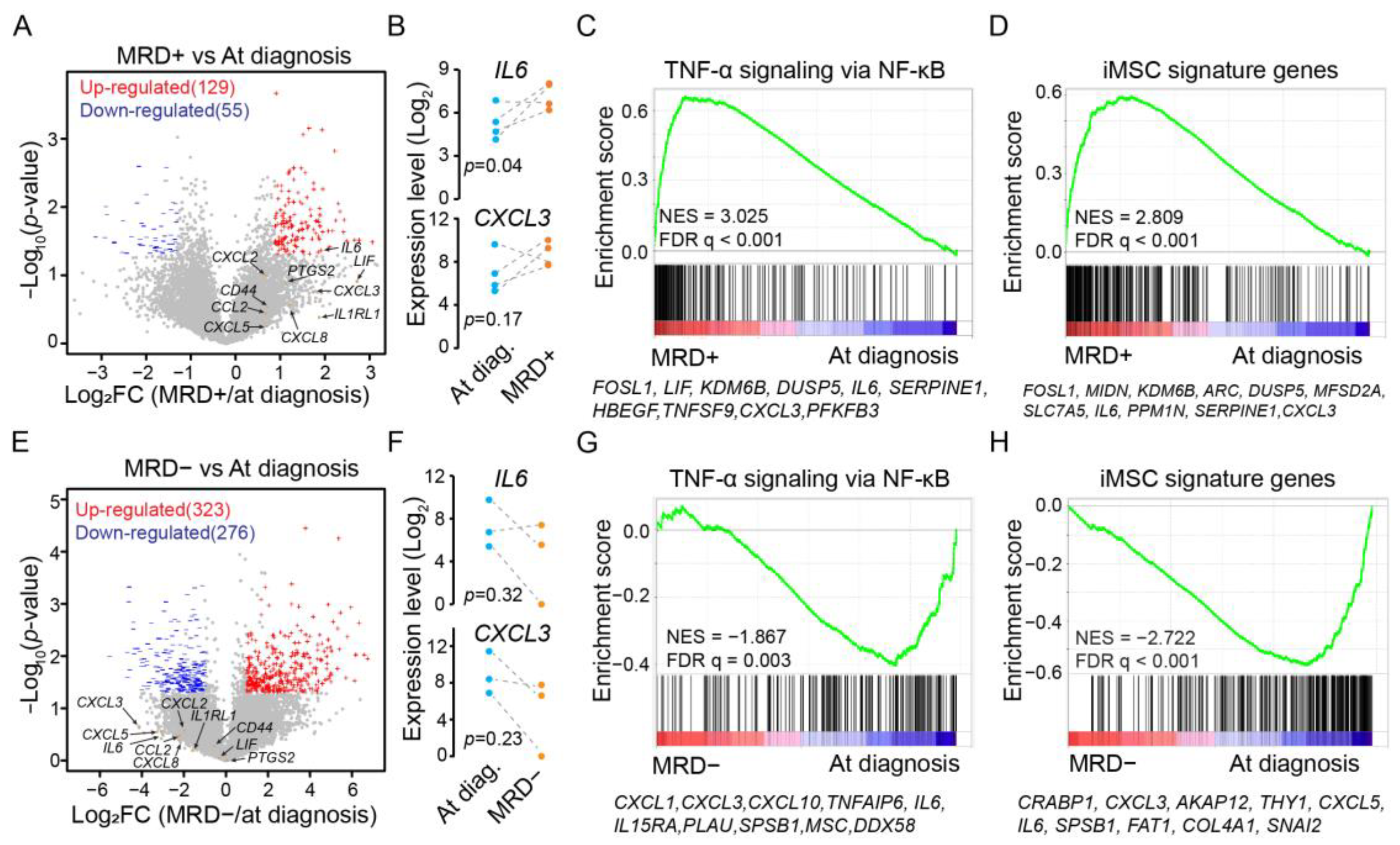
3.2. Expression of iMSC Signature Genes during Minimal Residual Disease
3.3. Diminished Expression of iMSC Signature Genes during In Vitro Expansion
3.4. Expression Activation of iMSC Signature Genes by Cytokine Stimulation
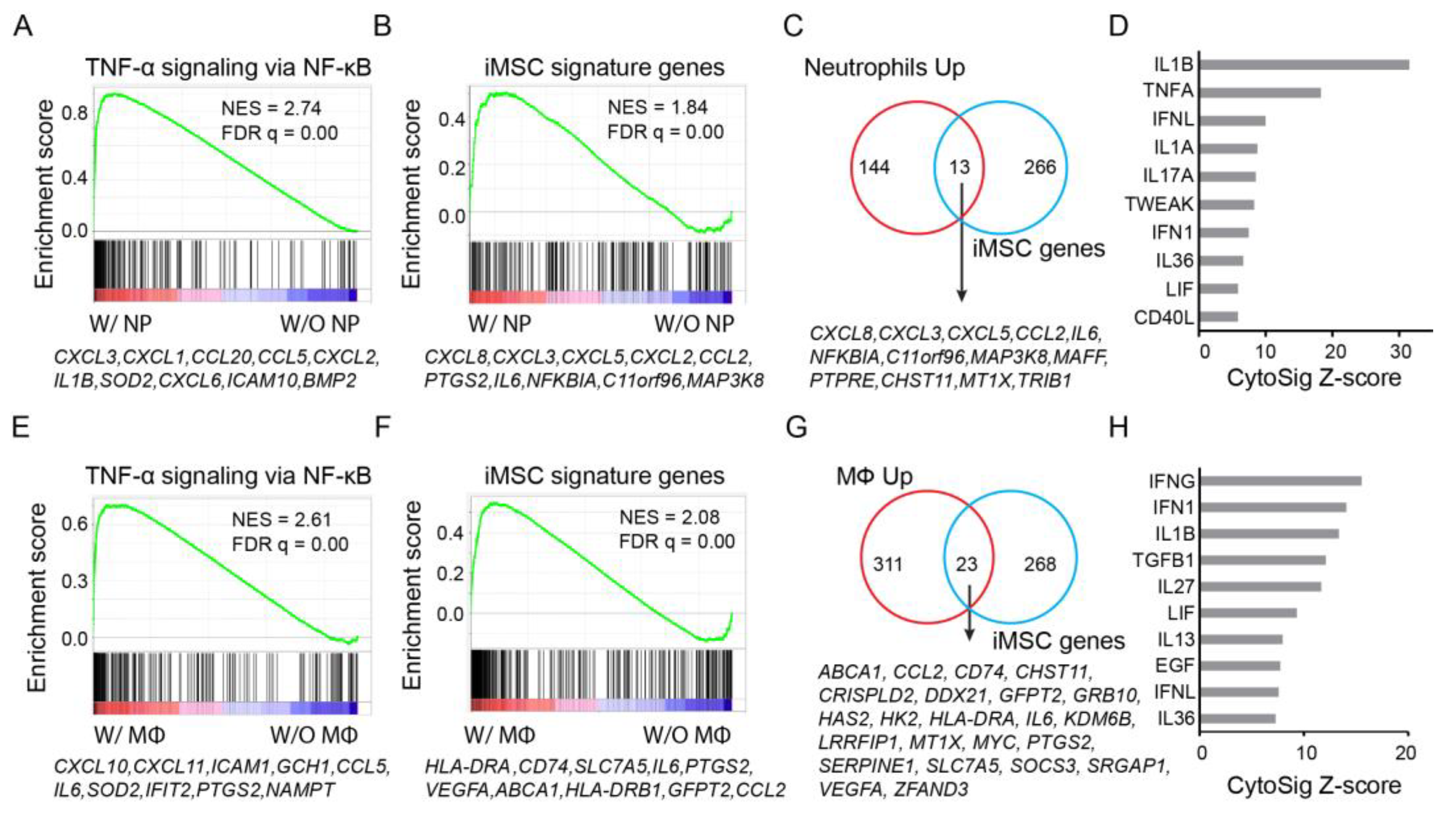
3.5. Expression Activation of iMSC Signature Genes by Immune Cells
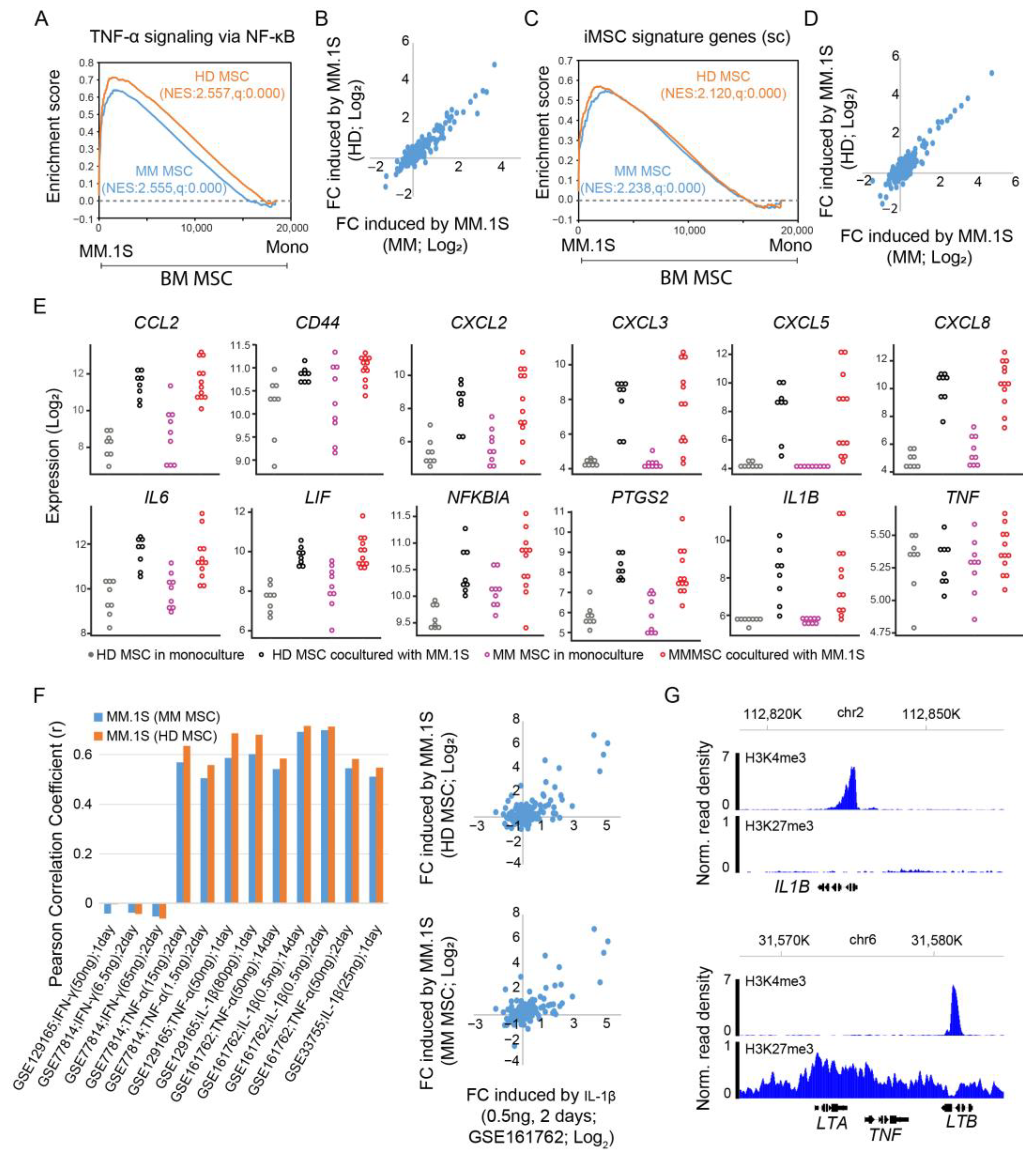
3.6. Expression Activation of iMSC Signature Genes by Multiple Myeloma Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maiso, P.; Mogollon, P.; Ocio, E.M.; Garayoa, M. Bone Marrow Mesenchymal Stromal Cells in Multiple Myeloma: Their Role as Active Contributors to Myeloma Progression. Cancers 2021, 13, 2542. [Google Scholar] [CrossRef]
- Chen, W.C.; Hu, G.; Hazlehurst, L.A. Contribution of the bone marrow stromal cells in mediating drug resistance in hematopoietic tumors. Curr. Opin. Pharmacol. 2020, 54, 36–43. [Google Scholar] [CrossRef]
- Diakos, C.I.; Charles, K.A.; McMillan, D.C.; Clarke, S.J. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014, 15, e493–e503. [Google Scholar] [CrossRef]
- Pinto, V.; Bergantim, R.; Caires, H.R.; Seca, H.; Guimaraes, J.E.; Vasconcelos, M.H. Multiple Myeloma: Available Therapies and Causes of Drug Resistance. Cancers 2020, 12, 407. [Google Scholar] [CrossRef]
- Kumar, S.; Baizer, L.; Callander, N.S.; Giralt, S.A.; Hillengass, J.; Freidlin, B.; Hoering, A.; Richardson, P.G.; Schwartz, E.I.; Reiman, A.; et al. Gaps and opportunities in the treatment of relapsed-refractory multiple myeloma: Consensus recommendations of the NCI Multiple Myeloma Steering Committee. Blood Cancer J. 2022, 12, 98. [Google Scholar] [CrossRef]
- Filippi, I.; Saltarella, I.; Aldinucci, C.; Carraro, F.; Ria, R.; Vacca, A.; Naldini, A. Different Adaptive Responses to Hypoxia in Normal and Multiple Myeloma Endothelial Cells. Cell. Physiol. Biochem. 2018, 46, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Reagan, M.R.; Ghobrial, I.M. Multiple myeloma mesenchymal stem cells: Characterization, origin, and tumor-promoting effects. Clin. Cancer Res. 2012, 18, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Russell, K.C.; Phinney, D.G.; Lacey, M.R.; Barrilleaux, B.L.; Meyertholen, K.E.; O’Connor, K.C. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells 2010, 28, 788–798. [Google Scholar] [CrossRef]
- Banfi, A.; Muraglia, A.; Dozin, B.; Mastrogiacomo, M.; Cancedda, R.; Quarto, R. Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: Implications for their use in cell therapy. Exp. Hematol. 2000, 28, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J.; Oh, J.Y. Mesenchymal Stem/Stromal Cells (MSCs): Role as Guardians of Inflammation. Mol. Ther. 2012, 20, 14–20. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.M.E.; Kellermayer, Z.; Papazian, N.; Tahri, S.; Hofste Op Bruinink, D.; Hoogenboezem, R.; Sanders, M.A.; van de Woestijne, P.C.; Bos, P.K.; Khandanpour, C.; et al. The multiple myeloma microenvironment is defined by an inflammatory stromal cell landscape. Nat. Immunol. 2021, 22, 769–780. [Google Scholar] [CrossRef]
- Alameda, D.; Saez, B.; Lara-Astiaso, D.; Sarvide, S.; Lasa, M.; Alignani, D.; Rodriguez, I.; Garate, S.; Vilas, A.; Paiva, B.; et al. Characterization of freshly isolated bone marrow mesenchymal stromal cells from healthy donors and patients with multiple myeloma: Transcriptional modulation of the microenvironment. Haematologica 2020, 105, e470–e473. [Google Scholar] [CrossRef]
- Sklavenitis-Pistofidis, R.; Haradhvala, N.J.; Getz, G.; Ghobrial, I.M. Inflammatory stromal cells in the myeloma microenvironment. Nat. Immunol. 2021, 22, 677–678. [Google Scholar] [CrossRef] [PubMed]
- Sarsenova, M.; Kim, Y.; Raziyeva, K.; Kazybay, B.; Ogay, V.; Saparov, A. Recent advances to enhance the immunomodulatory potential of mesenchymal stem cells. Front. Immunol. 2022, 13, 1010399. [Google Scholar] [CrossRef]
- Fernando, R.C.; Mazzotti, D.R.; Azevedo, H.; Sandes, A.F.; Rizzatti, E.G.; de Oliveira, M.B.; Alves, V.L.F.; Eugenio, A.I.P.; de Carvalho, F.; Dalboni, M.A.; et al. Transcriptome Analysis of Mesenchymal Stem Cells from Multiple Myeloma Patients Reveals Downregulation of Genes Involved in Cell Cycle Progression, Immune Response, and Bone Metabolism. Sci. Rep. 2019, 9, 1056. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gomez, A.; Li, T.L.; de la Calle-Fabregat, C.; Rodriguez-Ubreva, J.; Ciudad, L.; Catala-Moll, F.; Godoy-Tena, G.; Martin-Sanchez, M.; San-Segundo, L.; Muntion, S.; et al. Targeting aberrant DNA methylation in mesenchymal stromal cells as a treatment for myeloma bone disease. Nat. Commun. 2021, 12, 421. [Google Scholar] [CrossRef]
- Lemaitre, L.; DoSouto Ferreira, L.; Joubert, M.V.; Avet-Loiseau, H.; Martinet, L.; Corre, J.; Couderc, B. Imprinting of Mesenchymal Stromal Cell Transcriptome Persists even after Treatment in Patients with Multiple Myeloma. Int. J. Mol. Sci. 2020, 21, 3854. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gomez, A.; De Las Rivas, J.; Ocio, E.M.; Diaz-Rodriguez, E.; Montero, J.C.; Martin, M.; Blanco, J.F.; Sanchez-Guijo, F.M.; Pandiella, A.; San Miguel, J.F.; et al. Transcriptomic profile induced in bone marrow mesenchymal stromal cells after interaction with multiple myeloma cells: Implications in myeloma progression and myeloma bone disease. Oncotarget 2014, 5, 8284–8305. [Google Scholar] [CrossRef]
- Umezu, T.; Imanishi, S.; Azuma, K.; Kobayashi, C.; Yoshizawa, S.; Ohyashiki, K.; Ohyashiki, J.H. Replenishing exosomes from older bone marrow stromal cells with miR-340 inhibits myeloma-related angiogenesis. Blood Adv. 2017, 1, 812–823. [Google Scholar] [CrossRef]
- Andre, T.; Meuleman, N.; Stamatopoulos, B.; De Bruyn, C.; Pieters, K.; Bron, D.; Lagneaux, L. Evidences of early senescence in multiple myeloma bone marrow mesenchymal stromal cells. PLoS ONE 2013, 8, e59756. [Google Scholar] [CrossRef]
- McNee, G.; Eales, K.L.; Wei, W.; Williams, D.S.; Barkhuizen, A.; Bartlett, D.B.; Essex, S.; Anandram, S.; Filer, A.; Moss, P.A.; et al. Citrullination of histone H3 drives IL-6 production by bone marrow mesenchymal stem cells in MGUS and multiple myeloma. Leukemia 2017, 31, 373–381. [Google Scholar] [CrossRef]
- Heinemann, L.; Mollers, K.M.; Ahmed, H.M.M.; Wei, L.; Sun, K.; Nimmagadda, S.C.; Frank, D.; Baumann, A.; Poos, A.M.; Dugas, M.; et al. Inhibiting PI3K-AKT-mTOR Signaling in Multiple Myeloma-Associated Mesenchymal Stem Cells Impedes the Proliferation of Multiple Myeloma Cells. Front. Oncol. 2022, 12, 874325. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xu, H.; Han, H.; Song, S.; Zhang, X.; Ouyang, L.; Qian, C.; Hong, Y.; Qiu, Y.; Zhou, W.; et al. Exosome-mediated transfer of lncRUNX2-AS1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene 2018, 37, 5508–5519. [Google Scholar] [CrossRef] [PubMed]
- Wiese, D.M.; Wood, C.A.; Ford, B.N.; Braid, L.R. Cytokine Activation Reveals Tissue-Imprinted Gene Profiles of Mesenchymal Stromal Cells. Front. Immunol. 2022, 13, 917790. [Google Scholar] [CrossRef]
- Rubinstein-Achiasaf, L.; Morein, D.; Ben-Yaakov, H.; Liubomirski, Y.; Meshel, T.; Elbaz, E.; Dorot, O.; Pichinuk, E.; Gershovits, M.; Weil, M.; et al. Persistent Inflammatory Stimulation Drives the Conversion of MSCs to Inflammatory CAFs That Promote Pro-Metastatic Characteristics in Breast Cancer Cells. Cancers 2021, 13, 1472. [Google Scholar] [CrossRef]
- Carrero, R.; Cerrada, I.; Lledo, E.; Dopazo, J.; Garcia-Garcia, F.; Rubio, M.P.; Trigueros, C.; Dorronsoro, A.; Ruiz-Sauri, A.; Montero, J.A.; et al. IL1beta induces mesenchymal stem cells migration and leucocyte chemotaxis through NF-kappaB. Stem Cell Rev. Rep. 2012, 8, 905–916. [Google Scholar] [CrossRef]
- Guilloton, F.; Caron, G.; Menard, C.; Pangault, C.; Ame-Thomas, P.; Dulong, J.; De Vos, J.; Rossille, D.; Henry, C.; Lamy, T.; et al. Mesenchymal stromal cells orchestrate follicular lymphoma cell niche through the CCL2-dependent recruitment and polarization of monocytes. Blood 2012, 119, 2556–2567. [Google Scholar] [CrossRef]
- Jin, P.; Zhao, Y.; Liu, H.; Chen, J.; Ren, J.; Jin, J.; Bedognetti, D.; Liu, S.; Wang, E.; Marincola, F.; et al. Interferon-gamma and Tumor Necrosis Factor-alpha Polarize Bone Marrow Stromal Cells Uniformly to a Th1 Phenotype. Sci. Rep. 2016, 6, 26345. [Google Scholar] [CrossRef] [PubMed]
- Lerrer, S.; Liubomirski, Y.; Bott, A.; Abnaof, K.; Oren, N.; Yousaf, A.; Korner, C.; Meshel, T.; Wiemann, S.; Ben-Baruch, A. Co-Inflammatory Roles of TGFbeta1 in the Presence of TNFalpha Drive a Pro-inflammatory Fate in Mesenchymal Stem Cells. Front. Immunol. 2017, 8, 479. [Google Scholar] [CrossRef]
- Gregoire, M.; Guilloton, F.; Pangault, C.; Mourcin, F.; Sok, P.; Latour, M.; Ame-Thomas, P.; Flecher, E.; Fest, T.; Tarte, K. Neutrophils trigger a NF-kappaB dependent polarization of tumor-supportive stromal cells in germinal center B-cell lymphomas. Oncotarget 2015, 6, 16471–16487. [Google Scholar] [CrossRef]
- Espagnolle, N.; Balguerie, A.; Arnaud, E.; Sensebe, L.; Varin, A. CD54-Mediated Interaction with Pro-inflammatory Macrophages Increases the Immunosuppressive Function of Human Mesenchymal Stromal Cells. Stem Cell Rep. 2017, 8, 961–976. [Google Scholar] [CrossRef] [PubMed]
- Dotterweich, J.; Schlegelmilch, K.; Keller, A.; Geyer, B.; Schneider, D.; Zeck, S.; Tower, R.J.; Ebert, R.; Jakob, F.; Schutze, N. Contact of myeloma cells induces a characteristic transcriptome signature in skeletal precursor cells -Implications for myeloma bone disease. Bone 2016, 93, 155–166. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Dziadowicz, S.; Wang, L.; Akhter, H.; Aesoph, D.; Sharma, T.; Adjeroh, D.; Hazlehurst, L.; Hu, G. Bone Marrow Stroma-induced Transcriptome and Regulome Signatures of Multiple Myeloma. Cancers 2022, 14, 927. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Zhang, Y.; Ru, B.B.; Yang, Y.; Vu, T.; Paul, R.; Mirza, A.; Altan-Bonnet, G.; Liu, L.R.; Ruppin, E.; et al. Systematic investigation of cytokine signaling activity at the tissue and single-cell levels. Nat. Methods 2021, 18, 1181–1191. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Choi, H.; Kim, Y.; Kang, D.; Kwon, A.; Kim, J.; Kim, J.M.; Park, S.S.; Kim, Y.J.; Min, C.K.; Kim, M. Common and different alterations of bone marrow mesenchymal stromal cells in myelodysplastic syndrome and multiple myeloma. Cell Proliferat. 2020, 53, e12819. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Yang, J.; Gong, Y.; Zhang, H.; Qiu, X.; Liu, Y.; Zhou, C.; Chen, Y.; Greenbaum, J.; et al. Single-cell RNA sequencing deconvolutes the in vivo heterogeneity of human bone marrow-derived mesenchymal stem cells. Int. J. Biol. Sci. 2021, 17, 4192–4206. [Google Scholar] [CrossRef]
- Vijay, J.; Gauthier, M.F.; Biswell, R.L.; Louiselle, D.A.; Johnston, J.J.; Cheung, W.A.; Belden, B.; Pramatarova, A.; Biertho, L.; Gibson, M.; et al. Single-cell analysis of human adipose tissue identifies depot- and disease-specific cell types. Nat. Metab. 2020, 2, 97–109. [Google Scholar] [CrossRef]
- Lara-Castro, C.; Fu, Y.; Chung, B.H.; Garvey, W.T. Adiponectin and the metabolic syndrome: Mechanisms mediating risk for metabolic and cardiovascular disease. Curr. Opin. Lipidol. 2007, 18, 263–270. [Google Scholar] [CrossRef]
- Li, N.; Zhou, L.; Xie, W.L.; Zeng, D.Y.; Cai, D.Q.; Wang, H.Y.; Zhou, C.R.; Wang, J.; Li, L.H. Alkaline phosphatase enzyme-induced biomineralization of chitosan scaffolds with enhanced osteogenesis for bone tissue engineering. Chem. Eng. J. 2019, 371, 618–630. [Google Scholar] [CrossRef]
- Zhytnik, L.; Maasalu, K.; Pashenko, A.; Khmyzov, S.; Reimann, E.; Prans, E.; Koks, S.; Martson, A. COL1A1/2 Pathogenic Variants and Phenotype Characteristics in Ukrainian Osteogenesis Imperfecta Patients. Front. Genet. 2019, 10, 722. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Kim, J.E.; Nakashima, K.; Balmes, G.; Iwai, N.; Deng, J.M.; Zhang, Z.; Martin, J.F.; Behringer, R.R.; Nakamura, T.; et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. USA 2005, 102, 14665–14670. [Google Scholar] [CrossRef]
- Liu, C.F.; Lefebvre, V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015, 43, 8183–8203. [Google Scholar] [CrossRef]
- Ohlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef]
- Kieffer, Y.; Hocine, H.R.; Gentric, G.; Pelon, F.; Bernard, C.; Bourachot, B.; Lameiras, S.; Albergante, L.; Bonneau, C.; Guyard, A.; et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 2020, 10, 1330–1351. [Google Scholar] [CrossRef]
- Nicolas, A.M.; Pesic, M.; Engel, E.; Ziegler, P.K.; Diefenhardt, M.; Kennel, K.B.; Buettner, F.; Conche, C.; Petrocelli, V.; Elwakeel, E.; et al. Inflammatory fibroblasts mediate resistance to neoadjuvant therapy in rectal cancer. Cancer Cell 2022, 40, 168–184.e113. [Google Scholar] [CrossRef]
- Kumar, S.; Paiva, B.; Anderson, K.C.; Durie, B.; Landgren, O.; Moreau, P.; Munshi, N.; Lonial, S.; Blade, J.; Mateos, M.V.; et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016, 17, e328–e346. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Xu, J.; Lin, Z.; Huang, J.; Wang, F.; Yang, Y.; Cui, Y.; Luo, H.; Gao, Y.; Zhai, X.; et al. Minimal residual disease in multiple myeloma: Current status. Biomark. Res. 2021, 9, 75. [Google Scholar] [CrossRef]
- Munshi, N.C.; Avet-Loiseau, H.; Anderson, K.C.; Neri, P.; Paiva, B.; Samur, M.; Dimopoulos, M.; Kulakova, M.; Lam, A.; Hashim, M.; et al. A large meta-analysis establishes the role of MRD negativity in long-term survival outcomes in patients with multiple myeloma. Blood Adv. 2020, 4, 5988–5999. [Google Scholar] [CrossRef]
- Corre, J.; Mahtouk, K.; Attal, M.; Gadelorge, M.; Huynh, A.; Fleury-Cappellesso, S.; Danho, C.; Laharrague, P.; Klein, B.; Reme, T.; et al. Bone marrow mesenchymal stem cells are abnormal in multiple myeloma. Leukemia 2007, 21, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Todoerti, K.; Lisignoli, G.; Storti, P.; Agnelli, L.; Novara, F.; Manferdini, C.; Codeluppi, K.; Colla, S.; Crugnola, M.; Abeltino, M.; et al. Distinct transcriptional profiles characterize bone microenvironment mesenchymal cells rather than osteoblasts in relationship with multiple myeloma bone disease. Exp. Hematol. 2010, 38, 141–153. [Google Scholar] [CrossRef]
- Garderet, L.; Mazurier, C.; Chapel, A.; Ernou, I.; Boutin, L.; Holy, X.; Gorin, N.C.; Lopez, M.; Doucet, C.; Lataillade, J.J. Mesenchymal stem cell abnormalities in patients with multiple myeloma. Leuk. Lymphoma 2007, 48, 2032–2041. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, L.; Hamaidia, M.; Descamps, J.G.; Do Souto Ferreira, L.; Joubert, M.V.; Gadelorge, M.; Avet-Loiseau, H.; Justo, A.; Reina, N.; Deschaseaux, F.; et al. Toll-like receptor 4 selective inhibition in medullar microenvironment alters multiple myeloma cell growth. Blood Adv. 2022, 6, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, H.; Miyawaki, T.; Nomura, H.; Kuno, K.; Murakami, S.; Matsushima, K.; Mukaida, N. Expression of Both Types of Human Interleukin-8 Receptors on Mature Neutrophils, Monocytes, and Natural-Killer-Cells. J. Leukocyte Biol. 1995, 57, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Baumgart, S.J.; Najafova, Z.; Hossan, T.; Xie, W.; Nagarajan, S.; Kari, V.; Ditzel, N.; Kassem, M.; Johnsen, S.A. CHD1 regulates cell fate determination by activation of differentiation-induced genes. Nucleic Acids Res. 2017, 45, 7722–7735. [Google Scholar] [CrossRef]
- Wong, A.H.; Shin, E.M.; Tergaonkar, V.; Chng, W.J. Targeting NF-kappaB Signaling for Multiple Myeloma. Cancers 2020, 12, 2203. [Google Scholar] [CrossRef]
- De Jong, M.M.E.; Fokkema, C.; Papazian, N.; van Heusden, T.; Vermeulen, M.; Tahri, S.; Hoogenboezem, R.; van Duin, M.; van de Woestijne, P.; Langerak, A.; et al. Stromal Cell-Activated Bone Marrow Neutrophils Provide BAFF in Newly Diagnosed and Treated Multiple Myeloma. Blood 2022, 140, 4181–4182. [Google Scholar] [CrossRef]
- Meads, M.B.; Hazlehurst, L.A.; Dalton, W.S. The bone marrow microenvironment as a tumor sanctuary and contributor to drug resistance. Clin. Cancer Res. 2008, 14, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.; Fokkema, C.; Papazian, N.; van Heusden, T.; Vermeulen, M.; Hoogenboezem, R.; van Beek, G.; Tahri, S.; Sanders, M.A.; van de Woestijne, P.; et al. An IL-1β driven neutrophil-stromal cell axis fosters a BAFF-rich microenvironment in multiple myeloma. bioRxiv 2023. [Google Scholar] [CrossRef]
- Holthof, L.C.; van der Schans, J.J.; Katsarou, A.; Poels, R.; Gelderloos, A.T.; Drent, E.; Van Hal-van Veen, S.E.; Li, F.Z.; Zweegman, S.; van de Donk, N.W.C.J.; et al. Bone Marrow Mesenchymal Stromal Cells Can Render Multiple Myeloma Cells Resistant to Cytotoxic Machinery of CAR T Cells through Inhibition of Apoptosis. Clin. Cancer Res. 2021, 27, 3793–3803. [Google Scholar] [CrossRef] [PubMed]
- Dhodapkar, K.M.; Cohen, A.D.; Kaushal, A.; Garfall, A.L.; Manalo, R.J.; Carr, A.R.; McCachren, S.S.; Stadtmauer, E.A.; Lacey, S.F.; Melenhorst, J.J.; et al. Changes in Bone Marrow Tumor and Immune Cells Correlate with Durability of Remissions Following BCMA CAR T Therapy in Myeloma. Blood Cancer Discov. 2022, 3, 490–501. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Yi, W.; Ma, L.; Lecea, E.; Hazlehurst, L.A.; Adjeroh, D.A.; Hu, G. Inflammatory Bone Marrow Mesenchymal Stem Cells in Multiple Myeloma: Transcriptional Signature and In Vitro Modeling. Cancers 2023, 15, 5148. https://doi.org/10.3390/cancers15215148
Wang L, Yi W, Ma L, Lecea E, Hazlehurst LA, Adjeroh DA, Hu G. Inflammatory Bone Marrow Mesenchymal Stem Cells in Multiple Myeloma: Transcriptional Signature and In Vitro Modeling. Cancers. 2023; 15(21):5148. https://doi.org/10.3390/cancers15215148
Chicago/Turabian StyleWang, Lei, Weijun Yi, Li Ma, Emily Lecea, Lori A. Hazlehurst, Donald A. Adjeroh, and Gangqing Hu. 2023. "Inflammatory Bone Marrow Mesenchymal Stem Cells in Multiple Myeloma: Transcriptional Signature and In Vitro Modeling" Cancers 15, no. 21: 5148. https://doi.org/10.3390/cancers15215148
APA StyleWang, L., Yi, W., Ma, L., Lecea, E., Hazlehurst, L. A., Adjeroh, D. A., & Hu, G. (2023). Inflammatory Bone Marrow Mesenchymal Stem Cells in Multiple Myeloma: Transcriptional Signature and In Vitro Modeling. Cancers, 15(21), 5148. https://doi.org/10.3390/cancers15215148








