Establishment of a Prediction Model Based on Preoperative MRI Radiomics for Diffuse Astrocytic Glioma, IDH-Wildtype, with Molecular Features of Glioblastoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Patients and Methods
2.1. Patients
2.2. Inclusion and Exclusion Criteria
3. MRI Scanning Parameters
4. Pathological Grading and Molecular Diagnosis
5. Data Preprocessing
6. Image Segmentation
7. Feature Extraction and Selection
8. Classifier Evaluation and Statistical Analysis
- GNB = GaussianNB ()
- KNN = KneighborsClassifier ()
- RF = RandomForestClassifier (n_estimators = 100, criterion = ‘entropy’, max_depth = 5)
- AB = AdaBoostClassifier (DecisionTreeClassifier (max_depth = 3), n_estimators = 100,learning_rate = 1, algorithm = “SAMME”)
- SVM = SVC (kernel = ‘linear’, C = 1, decision_function_shape = ‘ovr’, probability = True)
- MLP = MLPClassifier (random_state = 1, max_iter = 500)
9. Results
Baseline Characteristics
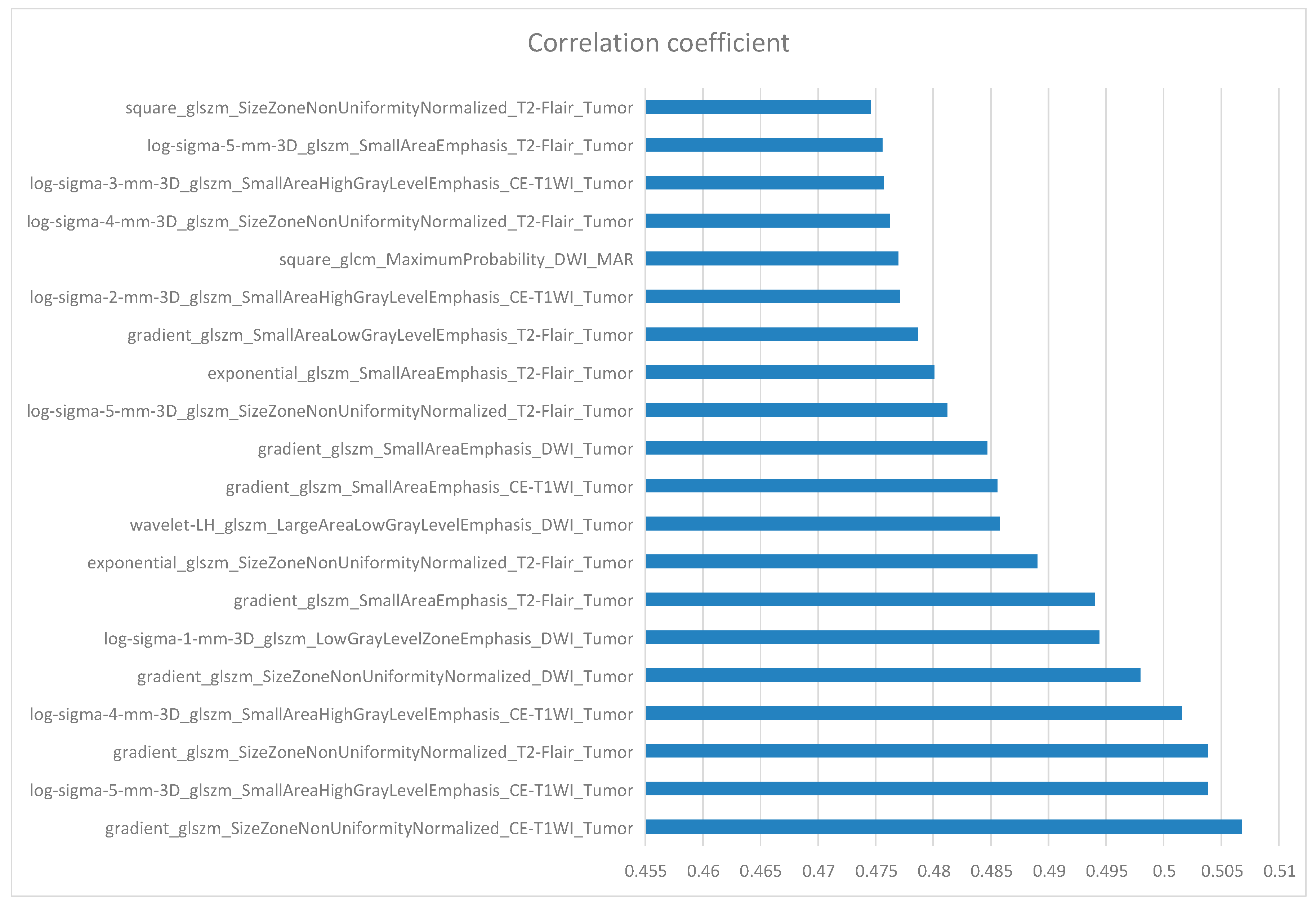
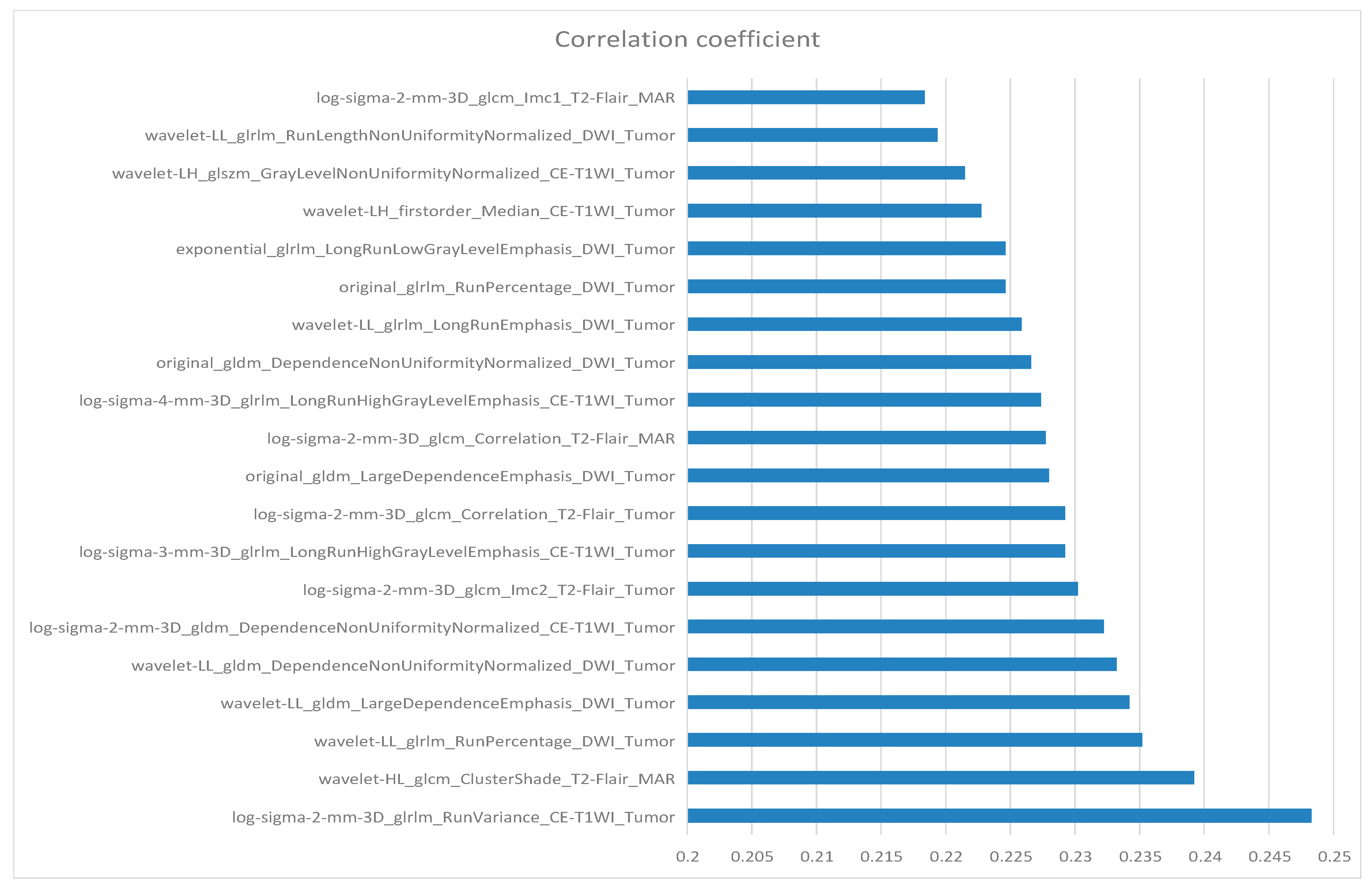
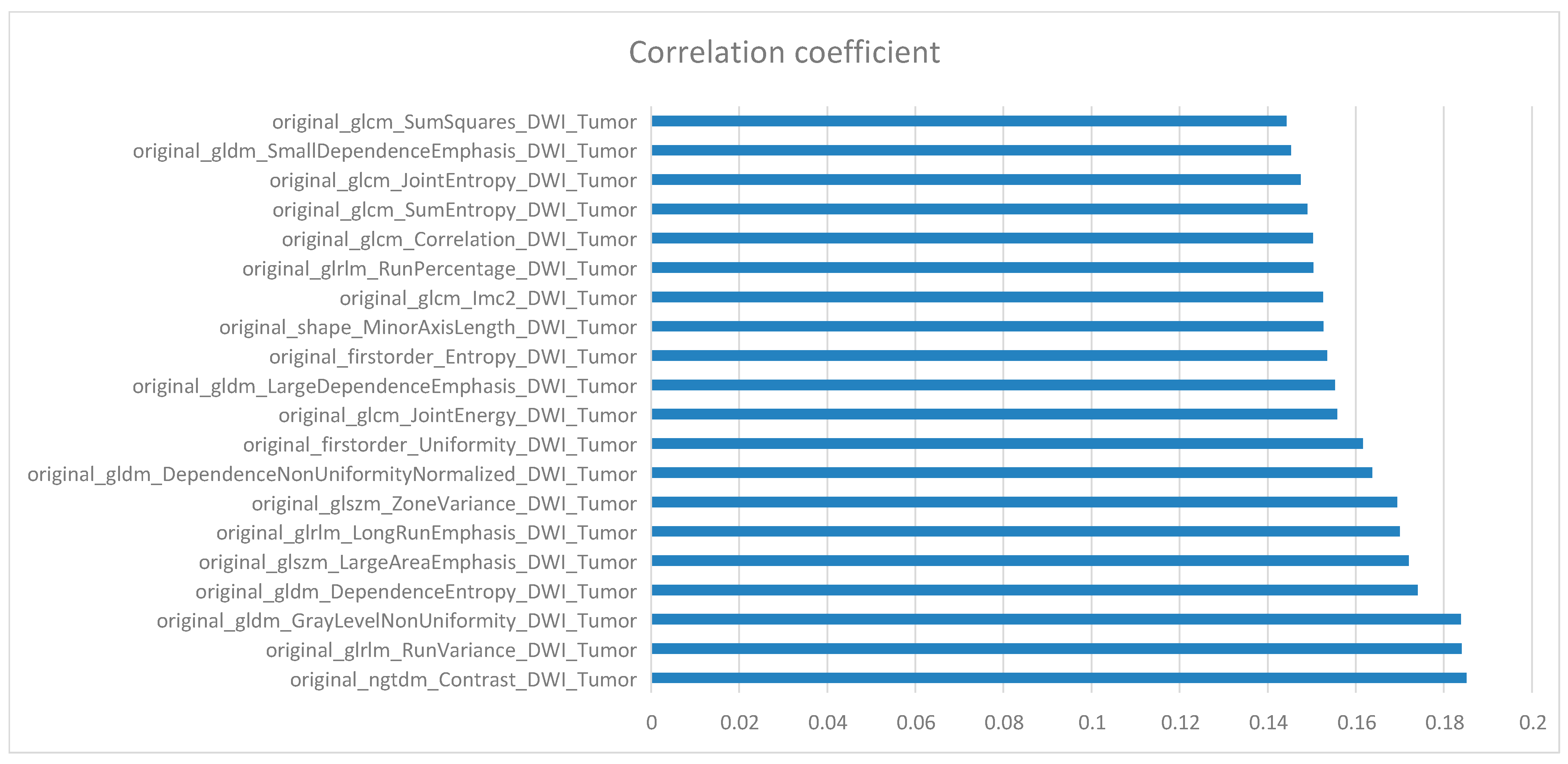
10. Feature Selection
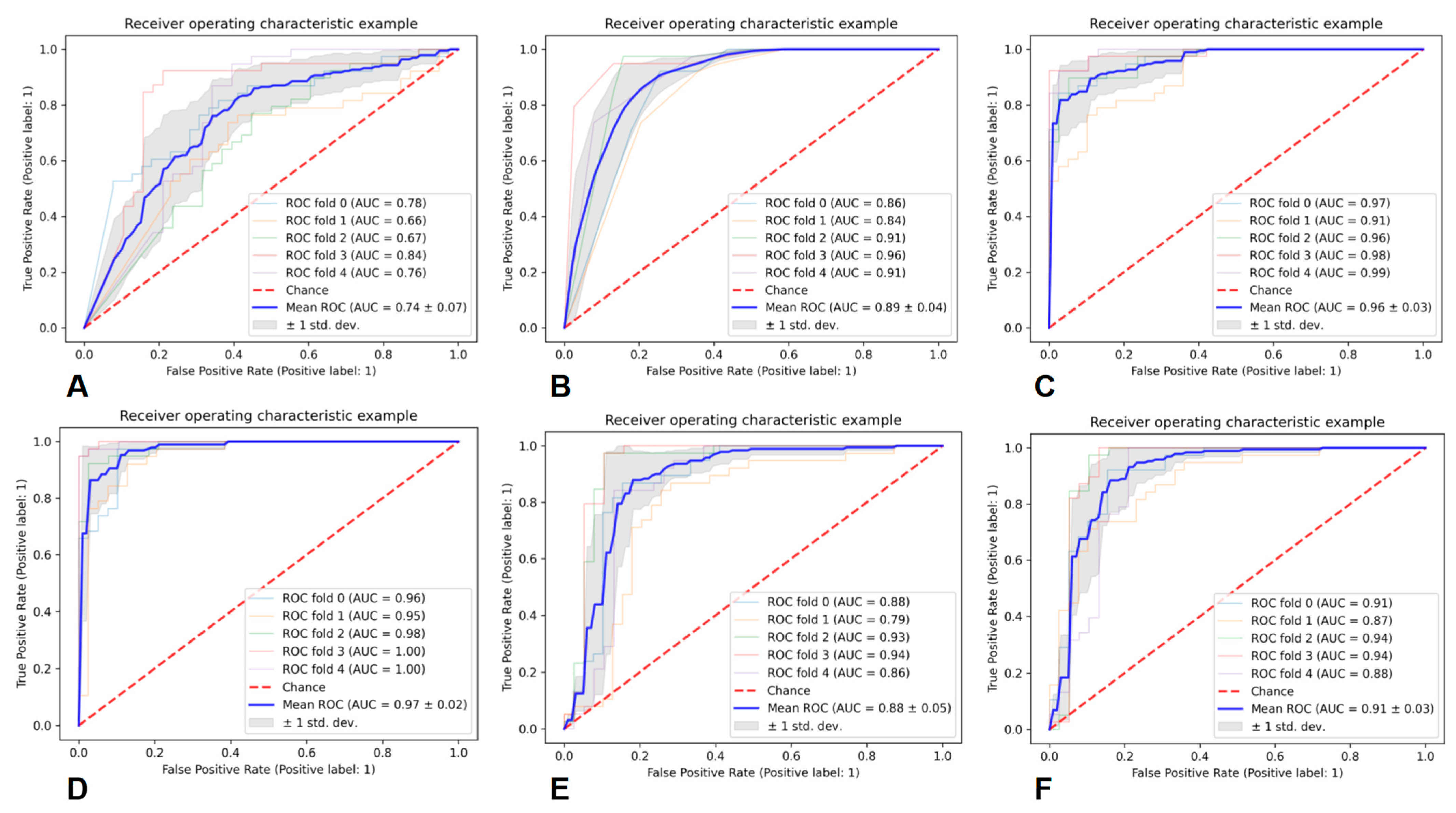
11. Performance of the Prediction Models
12. Discussion
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Horbinski, C.; Berger, T.; Packer, R.J.; Wen, P.Y. Clinical implications of the 2021 edition of the WHO classification of central nervous system tumours. Nat. Rev. Neurol. 2022, 18, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Verdugo, E.; Puerto, I.; Medina, M.Á. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun. 2022, 42, 1083–1111. [Google Scholar] [CrossRef] [PubMed]
- Lopes Abath Neto, O.; Aldape, K. Morphologic and Molecular Aspects of Glioblastomas. Neurosurg. Clin. N. Am. 2021, 32, 149–158. [Google Scholar] [CrossRef]
- Richardson, T.E.; Hatanpaa, K.J.; Walker, J.M. Molecular Characterization of “True” Low-Grade IDH-Wildtype Astrocytomas. J. Neuropathol. Exp. Neurol. 2021, 80, 431–435. [Google Scholar] [CrossRef]
- Zacharaki, E.I.; Wang, S.; Chawla, S.; Soo Yoo, D.; Wolf, R.; Melhem, E.R.; Davatzikos, C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009, 62, 1609–1618. [Google Scholar] [CrossRef]
- Ludwig, K.; Kornblum, H.I. Molecular markers in glioma. J. Neurooncol. 2017, 134, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, K.; Snuderl, M. Molecular Pathology of Gliomas. Surg. Pathol. Clin. 2021, 14, 379–386. [Google Scholar] [CrossRef]
- Reifenberger, G.; Wirsching, H.G.; Knobbe-Thomsen, C.B.; Weller, M. Advances in the molecular genetics of gliomas—Implications for classification and therapy. Nat. Rev. Clin. Oncol. 2017, 14, 434–452. [Google Scholar] [CrossRef]
- Chai, R.; Fang, S.; Pang, B.; Liu, Y.; Wang, Y.; Zhang, W.; Jiang, T. Molecular pathology and clinical implications of diffuse glioma. Chin. Med. J. (Engl.) 2022, 135, 2914–2925. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Aldape, K.; Colman, H.; Holland, E.C.; Louis, D.N.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.K.; Perry, A.; Reifenberger, G.; Stupp, R.; et al. cIMPACT-NOW update 3: Recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018, 136, 805–810. [Google Scholar] [CrossRef]
- Thakkar, J.P.; Dolecek, T.A.; Horbinski, C.; Ostrom, Q.T.; Lightner, D.D.; Barnholtz-Sloan, J.S.; Villano, J.L. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 1985–1996. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Lee, D.; Riestenberg, R.A.; Haskell-Mendoza, A.; Bloch, O. Diffuse astrocytic glioma, IDH-Wildtype, with molecular features of glioblastoma, WHO grade IV: A single-institution case series and review. J. Neurooncol. 2021, 152, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Berzero, G.; Di Stefano, A.L.; Ronchi, S.; Bielle, F.; Villa, C.; Guillerm, E.; Capelle, L.; Mathon, B.; Laurenge, A.; Giry, M.; et al. IDH-wildtype lower-grade diffuse gliomas: The importance of histological grade and molecular assessment for prognostic stratification. Neuro-oncology 2021, 23, 955–966. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, X.; Li, G.; Chang, X.; Li, S.; Chen, J.; Zhao, Z.; Wang, J.; Jiang, T.; Chai, R. Clinical management and survival outcomes of patients with different molecular subtypes of diffuse gliomas in China (2011–2017): A multicenter retrospective study from CGGA. Cancer Biol. Med. 2022, 19, 1460–1476. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.; Bray, D.P.; Cosgrove, M.; Boucher, A.; Erwood, A.; Linder, D.F.; Mendoza, P.; Morales, B.; Pradilla, G.; Nduom, E.K.; et al. Clinical and radiographic characteristics of diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma: A single institution review. J. Neurooncol. 2022, 157, 187–195. [Google Scholar] [CrossRef]
- van Breemen, M.S.; Wilms, E.B.; Vecht, C.J. Epilepsy in patients with brain tumours: Epidemiology, mechanisms, and management. Lancet Neurol. 2007, 6, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Wen, P.Y.; Hurwitz, S.; Black, P.; Kesari, S.; Drappatz, J.; Golby, A.J.; Wells, W.M.; Warfield, S.K.; Kikinis, R.; et al. Morphological characteristics of brain tumors causing seizures. Arch. Neurol. 2010, 67, 336–342. [Google Scholar] [CrossRef]
- Liigant, A.; Haldre, S.; Oun, A.; Linnamägi, U.; Saar, A.; Asser, T.; Kaasik, A.E. Seizure disorders in patients with brain tumors. Eur. Neurol. 2001, 45, 46–51. [Google Scholar] [CrossRef]
- Tesileanu, C.M.S.; Dirven, L.; Wijnenga, M.M.J.; Koekkoek, J.A.F.; Vincent, A.J.P.E.; Dubbink, H.J.; Atmodimedjo, P.N.; Kros, J.M.; van Duinen, S.G.; Smits, M.; et al. Survival of diffuse astrocytic glioma, IDH1/2 wildtype, with molecular features of glioblastoma, WHO grade IV: A confirmation of the cIMPACT-NOW criteria. Neuro-oncology 2020, 22, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Forst, D.A.; Nahed, B.V.; Loeffler, J.S.; Batchelor, T.T. Low-grade gliomas. Oncologist 2014, 19, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Fresnedo, A.; Pullen, M.W.; Perez-Vega, C.; Domingo, R.A.; Akinduro, O.O.; Almeida, J.P.; Suarez-Meade, P.; Marenco-Hillembrand, L.; Jentoft, M.E.; Bendok, B.R.; et al. The survival outcomes of molecular glioblastoma IDH-wildtype: A multicenter study. J. Neurooncol. 2022, 157, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Han, K.; Kim, H.; Ahn, S.S.; Choi, D.; Park, Y.W.; Chang, J.H.; Kim, S.H.; Cha, S.; Lee, S.K. MRI Features May Predict Molecular Features of Glioblastoma in Isocitrate Dehydrogenase Wild-Type Lower-Grade Gliomas. AJNR Am. J. Neuroradiol. 2021, 42, 448–456. [Google Scholar] [CrossRef]
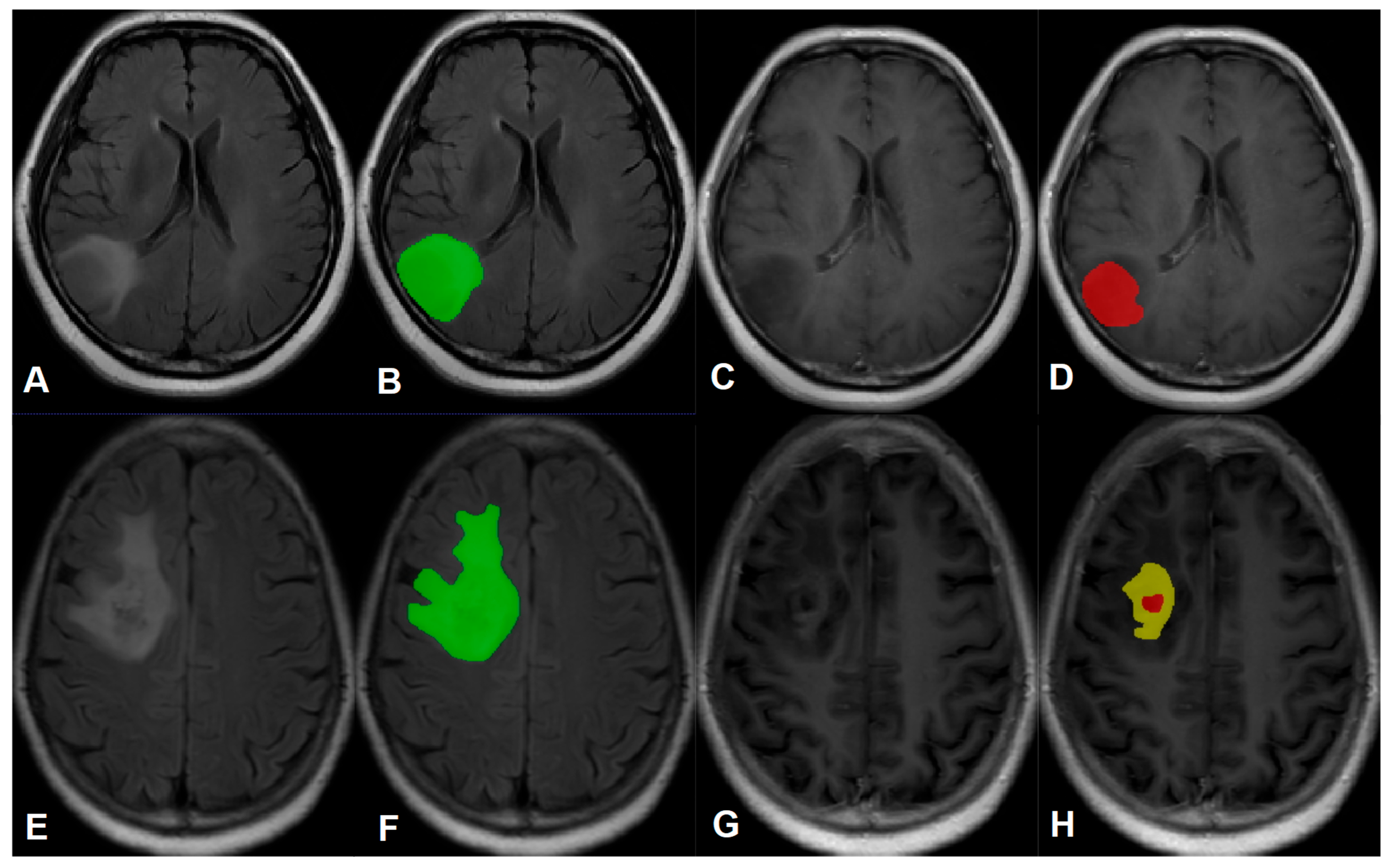
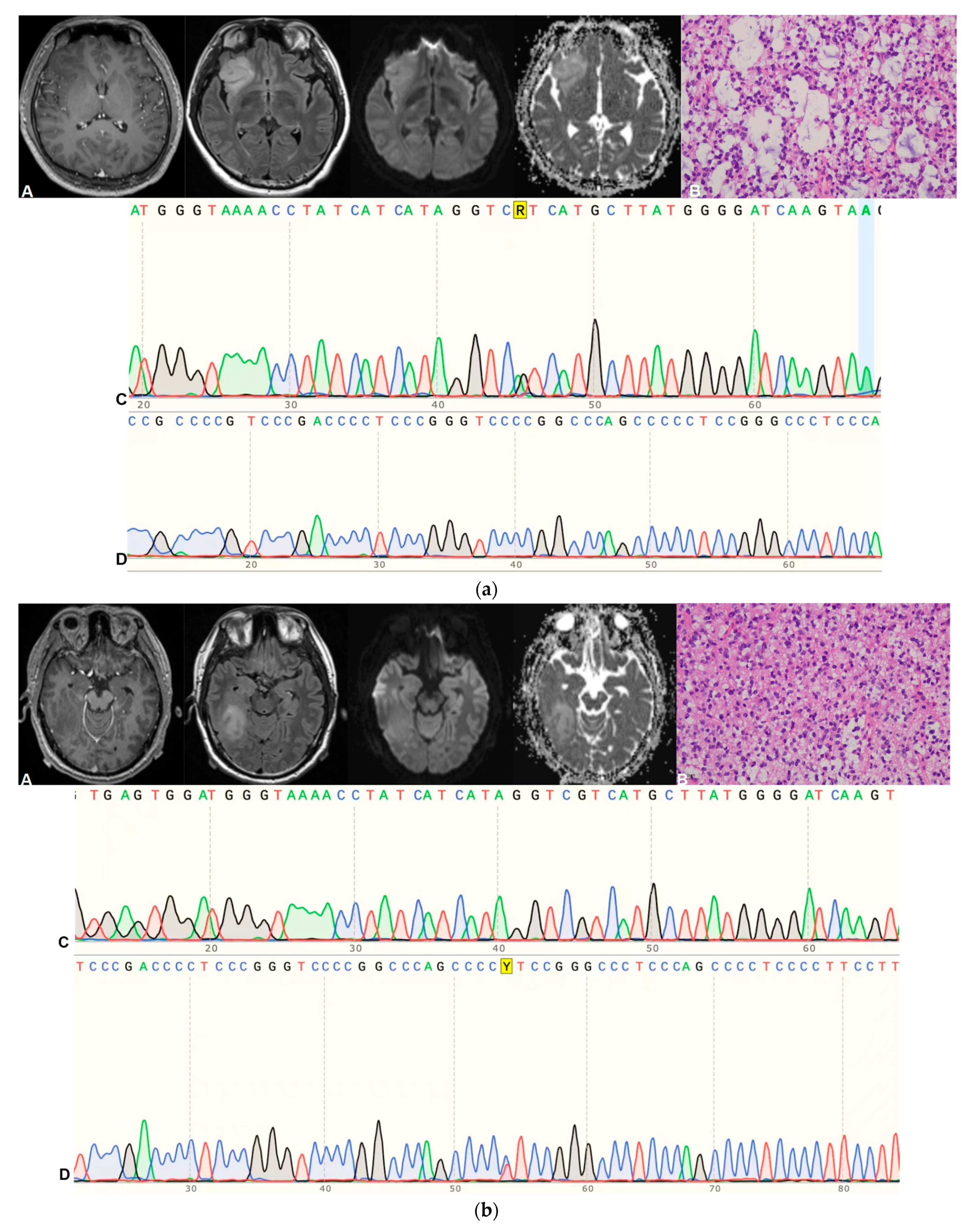
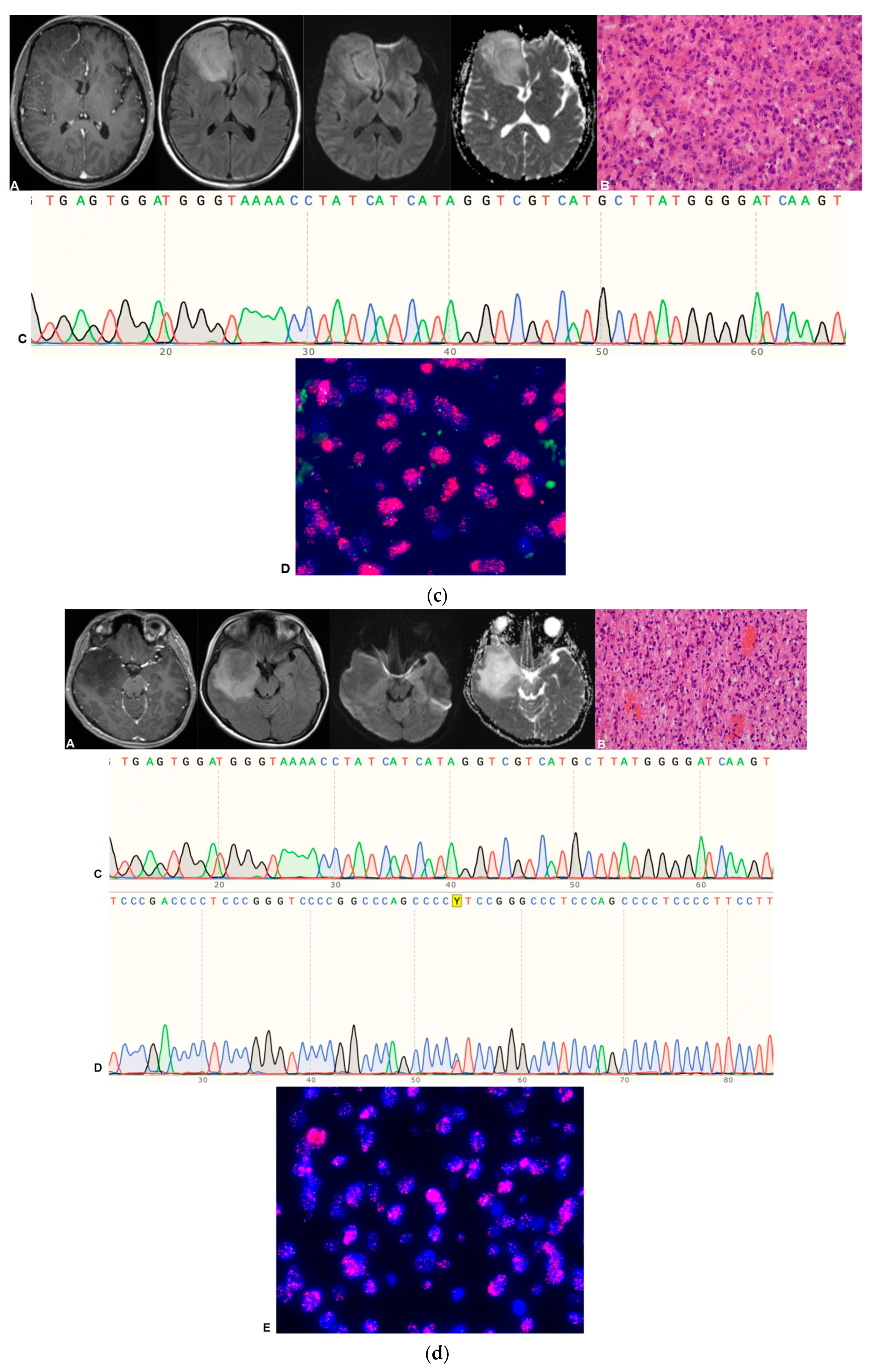
| Sequence | Parameters |
|---|---|
| T1WI | TR = 2992 ms; TI = 869 ms; TE = 24 ms; Matrix = 320 × 320; FOV = 240 × 240 mm2; Thickness = 5 mm; Interval = 1.5 mm |
| T2WI | TR = 4599 ms; TE = 102 ms; Matrix = 320 × 224; FOV = 240 × 240 mm2; Thickness = 5 mm; Interval = 1.5 mm |
| T2-Flair | TR = 8000 ms; TI = 2100 ms; TE = 160 ms; Matrix = 256 × 256; FOV = 240 × 240 mm2; Thickness = 5 mm; Interval = 1.5 mm |
| DWI | TR = 4800 ms; TE = 74 ms; Matrix = 128 × 130; FOV = 240 × 240 mm2; Thickness = 8 mm; Interval = 0.94 mm |
| CE-T1WI | TR = 2992 ms; TI = 869 ms; TE = 24 ms; Matrix = 320 × 320; FOV = 240 × 240 mm2; Thickness = 5 mm; Interval = 1.5 mm |
| Baseline Characteristics | Number of Astrocytoma Patients (Percentage) | Number of DAG-G Patients (Percentage) |
|---|---|---|
| Total | 172 | 44 |
| Gender | ||
| Male | 103 (60%) | 30 (68%) |
| Female | 69 (40%) | 14 (32%) |
| Age (year) | ||
| Mean ± standard deviation | 47.97 ± 13.81 | 46.58 ± 12.55 |
| Pathology grading | ||
| WHO grade 2 | 105 (61%) | 20 (45%) |
| WHO grade 3 | 67 (39%) | 24 (55%) |
| IDH | ||
| Mutant | 85 (49%) | 0 (0%) |
| Wild-type | 87 (51%) | 44 (100%) |
| TERT promoter | ||
| Mutant | 66 (38%) | 31 (70%) |
| Non-mutant | 106 (62%) | 13 (30%) |
| EGFR | ||
| Amplification | 17 (10%) | 14 (32%) |
| Non-amplification | 155 (90%) | 30 (68%) |
| Clinical symptom | ||
| Epilepsy | 88 (51%) | 25 (57%) |
| Headache | 23 (13%) | 7 (16%) |
| Aphasia | 22 (13%) | 5 (11%) |
| Altered mental status | 18 (10%) | 3 (7%) |
| Others | 21 (12%) | 4 (9%) |
| Radiology manifestation | ||
| -Hemisphere | ||
| Left | 79 (46%) | 18 (41%) |
| Right | 74 (43%) | 23 (52%) |
| Both | 19 (11%) | 3 (7%) |
| -Location | ||
| Frontal lobe | 83 (48%) | 22 (50%) |
| Temporal lobe | 34 (20%) | 10 (23%) |
| Parietal lobe | 24 (14%) | 5 (11%) |
| Insula | 20 (12%) | 4 (9%) |
| Others | 11 (6%) | 3 (7%) |
| -Contrast enhancement | ||
| Non | 92 (53%) | 21 (48%) |
| Mild | 35 (20%) | 10 (23%) |
| Moderate | 25 (15%) | 7 (16%) |
| Ring-like | 6 (3%) | 2 (4%) |
| Heterogenous | 14 (8%) | 4 (9%) |
| Classifier | Average AUC | Average PRE | Average REC | Average F1 |
|---|---|---|---|---|
| GNB | 0.74 ± 0.07 | 0.68 ± 0.06 | 0.68 ± 0.08 | 0.67 ± 0.06 |
| KNN | 0.89 ± 0.04 | 0.83 ± 0.08 | 0.79 ± 0.12 | 0.79 ± 0.05 |
| RF | 0.96 ± 0.03 | 0.88 ± 0.06 | 0.87 ± 0.08 | 0.87 ± 0.05 |
| AB | 0.97 ± 0.02 | 0.91 ± 0.06 | 0.90 ± 0.08 | 0.89 ± 0.05 |
| SVM | 0.88 ± 0.05 | 0.85 ± 0.08 | 0.83 ± 0.10 | 0.83 ± 0.07 |
| MLP | 0.91 ± 0.03 | 0.87 ± 0.07 | 0.86 ± 0.08 | 0.86 ± 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, P.; Wu, X.; Liu, X.; Chen, J.; Cao, A.; Geng, D. Establishment of a Prediction Model Based on Preoperative MRI Radiomics for Diffuse Astrocytic Glioma, IDH-Wildtype, with Molecular Features of Glioblastoma. Cancers 2023, 15, 5094. https://doi.org/10.3390/cancers15205094
Du P, Wu X, Liu X, Chen J, Cao A, Geng D. Establishment of a Prediction Model Based on Preoperative MRI Radiomics for Diffuse Astrocytic Glioma, IDH-Wildtype, with Molecular Features of Glioblastoma. Cancers. 2023; 15(20):5094. https://doi.org/10.3390/cancers15205094
Chicago/Turabian StyleDu, Peng, Xuefan Wu, Xiao Liu, Jiawei Chen, Aihong Cao, and Daoying Geng. 2023. "Establishment of a Prediction Model Based on Preoperative MRI Radiomics for Diffuse Astrocytic Glioma, IDH-Wildtype, with Molecular Features of Glioblastoma" Cancers 15, no. 20: 5094. https://doi.org/10.3390/cancers15205094
APA StyleDu, P., Wu, X., Liu, X., Chen, J., Cao, A., & Geng, D. (2023). Establishment of a Prediction Model Based on Preoperative MRI Radiomics for Diffuse Astrocytic Glioma, IDH-Wildtype, with Molecular Features of Glioblastoma. Cancers, 15(20), 5094. https://doi.org/10.3390/cancers15205094





