Simple Summary
This systematic review evaluates the potential of magnetic resonance imaging (MRI) to predict tumor biology in primary squamous cell carcinoma of the head and neck (HNSCC). Fifty-eight articles were analyzed, examining the relationship between MRI parameters and biological features. Most studies focused on HPV status associations, revealing that HPV-positive tumors consistently exhibited lower diffusion-weighted metrics. Moreover, lower diffusion values were also with a high Ki-67 proliferation index, indicating high cellularity. Several perfusion parameters describing the vascularity were significantly associated with HIF-1α. Analysis results of other biological factors (VEGF, EGFR, tumor cell count, p53, and MVD) were inconclusive. Larger datasets are needed to develop and validate radiomic-based prediction models, which already show promising results in capturing diverse tumor biology features. Overall, MRI holds potential for non-invasive and rapid tumor biology characterization, enhancing future clinical outcome predictions and personalized patient management for HNSCC.
Abstract
Magnetic resonance imaging (MRI) is an indispensable, routine technique that provides morphological and functional imaging sequences. MRI can potentially capture tumor biology and allow for longitudinal evaluation of head and neck squamous cell carcinoma (HNSCC). This systematic review and meta-analysis evaluates the ability of MRI to predict tumor biology in primary HNSCC. Studies were screened, selected, and assessed for quality using appropriate tools according to the PRISMA criteria. Fifty-eight articles were analyzed, examining the relationship between (functional) MRI parameters and biological features and genetics. Most studies focused on HPV status associations, revealing that HPV-positive tumors consistently exhibited lower ADCmean (SMD: 0.82; p < 0.001) and ADCminimum (SMD: 0.56; p < 0.001) values. On average, lower ADCmean values are associated with high Ki-67 levels, linking this diffusion restriction to high cellularity. Several perfusion parameters of the vascular compartment were significantly associated with HIF-1α. Analysis of other biological factors (VEGF, EGFR, tumor cell count, p53, and MVD) yielded inconclusive results. Larger datasets with homogenous acquisition are required to develop and test radiomic-based prediction models capable of capturing different aspects of the underlying tumor biology. Overall, our study shows that rapid and non-invasive characterization of tumor biology via MRI is feasible and could enhance clinical outcome predictions and personalized patient management for HNSCC.
1. Introduction
Over the years, magnetic resonance imaging (MRI) has steadily improved in rapid acquisition, image quality, and functional imaging capabilities, rendering it indispensable for the diagnostics and follow-up of the anatomically complex and biologically heterogenous head and neck squamous cell carcinomas (HNSCCs) [1,2]. As a non-invasive and routinely used imaging technique with additional functional sequences, MRI presents a valuable modality to capture this biological heterogeneity and enable longitudinal evaluation of HNSCC [1].
In HNSCC research, biological tumor characteristics have been a frequent topic, with a notable emphasis on subtype entities based on human papillomavirus (HPV) status and tumor components like vascularization and proliferation.
HPV-positive (HPV+) tumors are predominantly found in the oropharynx and are caused by the HPV-16 subtype. They are characterized by the presence of the viral proteinases E6 and E7, which ultimately leads to the degradation of p53 (tumor suppressor protein 53) and retinoblastoma 1 protein (Rb) [3,4]. In contrast, nearly all HPV-negative (HPV−) tumors deactivate the p53 tumor suppressor pathway via TP53 gene mutations [4]. HPV-negative tumors are associated with worse recurrence rates, metastasis, and overall worse clinical outcomes than HPV-positive tumors [3,4].
Providing vascular support for tumor growth and angiogenesis is another critical factor that also influences the efficacy of radiotherapy and chemotherapy [5]. Vascular endothelial growth factor (VEGF), microvessel density (MVD), and hypoxia-inducible factor-1 alpha (HIF-1α) are histopathological markers that are used to estimate the extent of angiogenesis and to deduct the presence of hypoxia [6,7]. Furthermore, tumor cell proliferation, measured using the Ki-67 proliferation index and frequently driven by the epidermal growth factor receptor (EGFR), links those markers to tumor growth [8]. Higher tumor growth, proliferation, and angiogenesis rates are associated with increased metastatic potential, and larger tumors show worse outcomes [5,9]. On a genetic level, signatures such as epithelial-to-mesenchymal transition (EMT), tumor lymphocyte infiltration, and DNA repair deficits, are also known to affect outcomes [10]. To date, complex immunohistochemical (IHC) assays, gene expression analysis, DNA sequencing, and/or transcriptional profiling are required to identify such biological characteristics for outcome prediction or treatment optimization.
MRI offers the means to monitor various biological processes within tumors. Notably, functional techniques like diffusion-weighted imaging (DWI) and dynamic contrast-enhanced (DCE) imaging excel in unveiling tissue microstructures, perfusion patterns, and vascular attributes [1,11]. However, morphological sequences like T1-weighted (T1W) or T2-weighted (T2W) imaging remain indispensable as these functional techniques may distort or suppress the extraction of the exact anatomical location [1].
Multi-sequence data can be combined for quantitative image analysis in the emerging field of radiomics. Radiomics may be deployed to predict important outcomes by defining biological or genomic factors in studies that are often referred to as radiogenomics [12,13]. Radiogenomics has been increasingly investigated, primarily using computed tomography (CT) images [14]. In HNSCC, however, the superior soft tissue depiction of MRI in combination with functional imaging is expected to provide a better representation.
This systematic review and meta-analysis aims to assess the current level of evidence of biological feature prediction using quantitative MRI analysis. For this purpose, it evaluates the diagnostic accuracy of both stand-alone functional (diffusion and perfusion) and anatomical (T1W, T2W) MRI parameters, along with the utility of multi-feature radiomics models.
2. Materials and Methods
This systematic review (PROSPERO registration: 392509) was performed following the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) criteria [15].
2.1. Search Strategy
A comprehensive search was conducted in PubMed (MEDLINE), Embase, and Scopus for original articles published until 28 July 2023. The search query consisted of “Head and neck squamous cell carcinoma” combined with “MRI”, along with either “Genes”, “HNSCC tumor Biomarkers”, or “radiogenomics”, along with their synonyms or adjacent terms.
The literature search is explained in Supplementary Material S1, Appendix S1, and the full search for the different databases is provided in Supplementary Material S1, Appendix S2. Two researchers (BLINDED and BLINDED) independently screened the results for relevant publications using the Rayyan web application [16]. The reviewers discussed and resolved discrepancies during multiple consensus meetings following the screening of the same subset of items.
2.2. Study Selection
The following selection criteria were applied by the reviewers: (1) human subjects were examined; (2) only original research was considered; (3) the research included MRI parameters of the primary tumor; (4) the study compared quantitative MRI parameters with genetics/histopathology markers; (5) the study solely included HNSCCs at any age, gender, and stage; (6) the study was not a case report; (7) and it was written in the English language.
2.3. Quality Assessment
An optimized version of the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool [17] was used to assess the “applicability concerns” and “risk of bias” in the remaining eligible articles (see Supplementary Material S1, Appendix S3). The various biological features were assessed separately to take into account the varying measurement characteristics, often necessitating multiple independent assessments of the same article.
Articles reporting radiomic models were additionally scored using the radiomics quality score (RQS) to evaluate the quality, design, and generalizability of the model [18].
2.4. Data Extraction and Analyses
Data were extracted using a custom-built standardized extraction form by one reviewer (BLINDED). Collected data included study characteristics (e.g., study design), patient characteristics (e.g., gender and cancer subsite), methods used for biological feature evaluation (e.g., DNA polymerase chain reaction (PCR), immunohistochemistry (IHC), and the number of observers for IHC methods), techniques and its properties used for imaging evaluation (e.g., MRI field strength, use of diffusion-weighted and/or perfusion-weighted imaging, and delineation procedure and characteristics), and statistical methods adopted to compare the MRI features with the biological endpoints.
The results from biological factors analyzed in a reasonable number of studies, determined as four or more studies, are compared and summarized in the main text of this review. Survey results of those with fewer studies are listed in the Supplementary Materials.
When comparable data were available for a considerable number of studies, a set of four or more studies, a meta-analysis was conducted. For this reason, either the standardized mean difference (SMD, Cohen’s d [19]) or the correlation [20] with their 95% confidence intervals (95%CI) were calculated for each study and visualized in a forest plot. If a mean value of the imaging parameter was not provided but a median value was reported, we estimated the mean value using the formulas from Lou et al. and Wan et al. after confirming that the data were not too skewed [21,22,23,24]. Supplementary Material S1, Appendix S4, outlines all the statistical formulas used in this study. If data were unsuitable for meta-analyses, a narrative summarization of the available data was provided. In all analysis, p-values < 0.05 were considered as statically significant.
3. Results
3.1. Literature Search
A literature search was conducted to find studies reporting biological and MRI feature associations and produced a total of 5396 unique entries. Most studies were excluded based on title (n = 5173, 95.5%) or abstract (n = 173, 3.2%) evaluation. Full-text evaluation was performed for 70 entries (1.2%), of which 12 were excluded (17% of 70). Figure 1 provides a flow diagram of the study selection and exclusion process, detailing the reasons for exclusion. Finally, a total of 58 studies [25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82] satisfied all inclusion criteria.
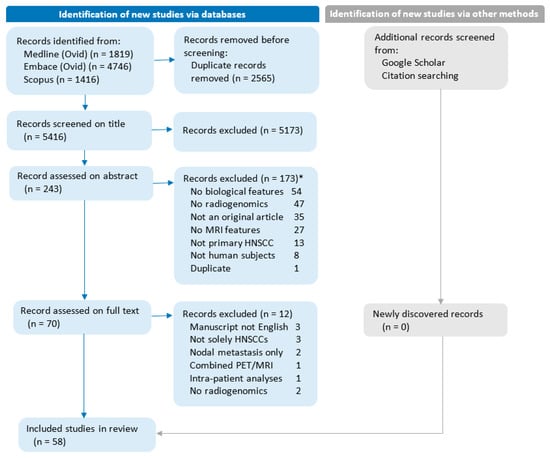
Figure 1.
Flow diagram of the study selection and exclusion process. No radiogenomics: Studies without analyses of biological features compared with MRI features. Combined PET/MRI: Studies focusing on combined PET/MRI parameters rather than stand-alone MRI parameters. Intra-patient analyses: Studies are limited to analyzing multiple biopsies within the same patient. * Studies can be excluded for more than one reason.
Two articles compared genetics and radiomic features, aligning with the research objectives [76,77]. Regarding individual biological endpoints, HPV status, Ki-67, tumor cell count, HIF-1α, VEGF, EGFR, p53, and MVD were each covered by at least four original articles, which together constituted a total of 51 articles. These data provided sufficient material for data analysis, as defined in Section 2, and are evaluated in detail below. The five remaining articles focused on the Epstein–Barr virus (EBV) [78], proliferating cell nuclear antigen (PCNA) [79], carbonic anhydrase 9 (CAIX) [80], tumor–stroma ratio [81,82], or tumor-infiltrating lymphocytes [82]. Supplementary Material S2, Appendix S4, provides an overview of all identified biological endpoints along with the amount of significant correlations or differences reported in these studies.
3.2. Quality Assessment
Quality assessment and applicability analyses were performed using the QUADAS-2 tool [17] and are shown in Supplementary Material S2, Appendices S1 and S2. Across all studies, “reference standard” applicability concerns were most frequently recorded, while “patient selection” and “index test” were acceptable in most studies. Applicability concerns derived from mismatched research questions (i.e., studies including nodal metastasis, radiogenomics not being the main research question, or use of cut-off values for biological factor analyses). Concerns for bias were related to “flow and timing” issues, with discrepancies in MRI-to-biopsy timelines or varied MRI protocols applied. In the “reference standard” category, potential bias could arise from single IHC reviewer use or whether a continuous or cut-off value was used when reporting a biological factor. Table 1 and Table 4 display the results from the RQS evaluation [18] for all radiomic model studies, revealing deficiencies in open science practices, validation approaches, and clinical utility.

Table 1.
Characteristics of studies focused on HPV status associations with shape, stand-alone histogram T1W and T2W parameters, or radiomic features.
3.3. Study Outcome Assessment
The reported MRI and biological feature association study results were structured into HNSCC tumor biology categories. Meta-analyses were performed where applicable. The majority of the MRI-based radiomic studies in HNSCC were focused on links with HPV status, allowing for more extensive cross-study comparisons. Analysis of associations with proliferation, vasculature, and perfusion-related markers was the focus of the assessment of the remainder of the studies.
3.3.1. Human Papilloma Virus (HPV)
HPV status plays a pivotal role in HNSCC staging and outcome prediction. Twenty-nine studies [25,26,27,28,29,30,34,35,36,37,38,43,44,45,46,47,48,49,52,53,54,55,56,57,58,59,69,70,74] involving 2122 patients tested whether HPV status can be deducted using quantitative imaging analysis.
HPV: Independent T1W and T2W Texture Parameters and Radiomic Models
As a standalone parameter, tumor volume was analyzed for associations with HPV status in five studies [35,38,45,56,69] (Table 1). None of these studies observed significant differences in tumor volumes between HPV+ and HPV− tumors (Supplementary Material S2, Appendix S3 (A3.1)).
Only one (Giannitto et al. [37]) out of four studies [37,43,49,55] reported a significant association between T1W and T2W MRI stand-alone histogram features and HPV status. Analyzing 1286 radiomics features, Giannitto et al. [37] suggested that HPV− tumors had a more varied texture than HPV+ oropharyngeal HNSCC, providing the basis for the observed association with stand-alone histogram parameters [37].
MRI radiomic-based prediction models for HPV status were built in eight studies [26,27,28,46,53,58,59,74] (Table 1). These studies based their analysis exclusively on oropharyngeal SCC of different stages. Marzi et al. [46] constructed a model with three DWI or intravoxel incoherent motion (IVIM) features out of the 157 extracted features to predict HPV. However, its diagnostic performance decreased after internal validation (AUC 0.91 to 0.66) [46]. A model starting with 498 native 2D T1W features [26] and trained on images from 249 patients achieved a 5- and 10-fold cross-validation AUC of 0.79 in its training set, yet no validation was conducted. Three studies [27,28,53] employed 3D T1W imaging with gadolinium-based contrast, yielding in-hospital validation AUCs of 0.76 to 0.83 in their respective test sets. In both Sohn et al. and Li et al.’s studies, feature selection of the six most relevant contrast-enhanced T1W and T2W features yielded models with an AUC of 0.74 in their respective test sets [58,74]. Additionally, another multi-sequence model, incorporating potentially native T1W, contrast-enhanced T1W, T2W, and apparent diffusion coefficient (ADC) features, resulted in similar AUC values of 0.76 [59]. Among the combined features, six ADC histogram features and one native T1 feature were identified as the best combination within these analyzed data [59]. None of the models were tested in independent validation or external data sets.
Forest plots of standard mean differences (SMDs) in mean ADC values of HPV-positive and HPV-negative tumors were grouped according to HPV determination methodology. Conventional HPV diagnostics using p16 IHC combined with PCR were used in seven studies. One single assay was used in the remainder (p16 IHC, HC2 High-risk HPV DNA test, or hybrid capture assay kits). In studies that provided median values, standard mean differences were calculated using estimated means as indicated (and as described in Section 2 and Supplementary Material S1, Appendix S4).
HPV: Diffusion-Weighted Imaging
DWI features in relation to HPV status were analyzed in sixteen studies [29,34,35,36,38,43,44,45,46,47,52,54,55,57,69,70], encompassing a total of 897 patients (range n: 20–105), predominantly with oropharyngeal SCC (78.4%) and with a similar average age of 63. Twelve of the articles reported on retrospectively collected data. ADC mapping was acquired using b0 and b1000 in eight of the sixteen studies. Table 2 highlights the key characteristics of the included studies.

Table 2.
Characteristics of MRI diffusion and perfusion parameter association studies with HPV status.
From all analyzed DWI parameters, and with fifteen and seven studies, respectively, ADCmean and ADCmininum were the most reported. ADCmean and ADCmininum standardized mean differences between HPV+ and HPV− as reported or as deduced from estimated mean values (Supplementary Material S1, Appendix S4) are displayed in Forest plots in Figure 2 and Figure 3. Standard mean difference (SMD) analyses are grouped according to HPV determination accuracy (Table 2) as HPV detection accuracy increases when applying combined p16-IHC and HPV16 and HPV18 DNA or E6/7 RNA PCR tests [83].
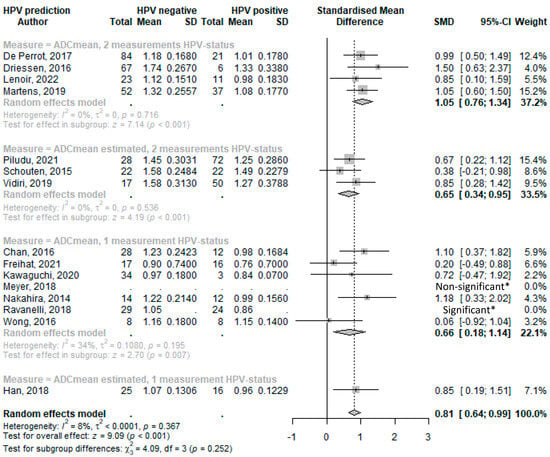
Figure 2.
Forest plot of the mean ADC values with HPV status. * Limited data available and as reported in reference. Abbreviations: SD = standard deviation; SMD = standard mean difference; and CI = confidence interval [34,35,44,45,54,57,69,29,36,43,47,52,55,70,38].
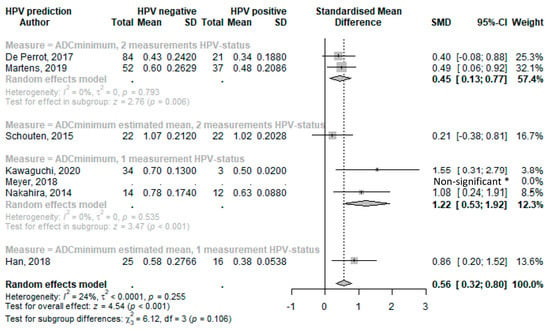
Figure 3.
Forest plot of the minimal ADC values with HPV status. * Limited data available and as reported in reference. Abbreviations: SD = standard deviation; SMD = standard mean difference; CI = confidence interval [34,45,57,43,47,52,38].
The Forest plot analyses (Figure 2) revealed that HPV+ HNSCC has a lower average ADCmean value compared with HPV− subtypes, with an overall SMD of 0.81 (95%CI 0.64–0.99; p < 0.001). Notably, and consistent with an increased specificity, this difference was more pronounced when HPV status was ascertained by using two different HPV determination methods (SMD: 1.05, 95%CI 0.76–1.34; p < 0.001). However, the difference and association were weaker when relying on a single HPV detection method (SMD: 0.66, 95%CI 0.18–1.14; p = 0.007) or on estimated means that generally show smaller SMDs.
Forest plots of standard mean differences in minimal ADC values of HPV-positive and HPV-negative tumors were produced and grouped according to HPV determination methodology. Conventional HPV diagnostics using p16 IHC combined with PCR were used in three studies. One single assay was used in the remainder (either p16 IHC or hybrid capture assay kits). In studies that provided median values, standard mean differences were calculated using estimated means as indicated (and as described in Section 2 and Supplementary Material S1, Appendix S4).
The overall random effect model of all studies that used ADCminimum to predict HPV status resulted in a statistically significant SMD of 0.56 (95%CI 0.32–0.80; p < 0.001) (Figure 3). The ADCminimum SMD values for HPV determined with two different HPV determination methods showed a lower effect compared with the overall values, with an SMD of 0.45 (95%CI 0.13–0.77; p = 0.006) for studies that provided means, and an SMD of 0.21 (95%CI −0.38–0.81) for the study that provided medians (of which we estimated means). ADCminimum with only one measurement of HPV showed a higher SMD of 1.22 (95%CI 0.53–1.92; p < 0.001) for the study that provided a mean, and an SMD of 0.86 (95%CI 0.20–1.52) for studies with an estimated mean.
Three studies [46,54,69] that also evaluated IVIM parameters found significantly lower true diffusion coefficient (Dt) values in HPV+ patients compared with HPV− patients when using higher b-values (b300–b800). This association exhibited overall lower p-values than their reported ADCmean analyses (p <0.001–0.001).
Among the other frequently reported ADC metrics, the histogram-based features of skewness and kurtosis were analyzed in five studies [34,44,45,47,55]. Only two studies [34,44] reported significant differences and higher skewness and kurtosis values for HPV+ tumors. These and other associations can be found in Supplementary Material S2, Appendix S3 (A3.2).
HPV: Perfusion-Based Imaging
The mean values of the DCE parameters, Ktrans, Kep, and Ve were reported by four studies [30,38,48,54]. However, the findings comparing HPV status between groups were inconsistent across these studies. Significant differences in these parameter values were reported in only one study by Choi et al. [30], who reported a significantly higher mean Ktrans for HPV+ tumors compared with HPV−. This finding, however, contrasts with the other studies [38,54] that show (non-significantly) lower Ktrans values in HPV+ tumors.
In addition to mean DCE parameter values, Choi et al. and Meyer et al. also analyzed histogram parameters of Ktrans, Kep, and Ve as 25th, 50th, and 75th percentiles (P), skewness, and kurtosis [30,48]. Significantly higher values for the P25, P50, and P75 of the Ktrans and the P25 of the Kep were found for p16 IHC-positive compared with p16 IHC-negative tumors in Choi et al. These findings were not replicated by Meyer et al. Additional reported associations can be found in Supplementary Material S2, Appendix S3 (A3.3).
3.3.2. Tumor Cell Proliferation and Cellularity Markers: Ki-67, EGFR, Tumor Cell Count, and p53
MRI parameter associations with tumor cell proliferation or density-related markers were discussed in 19 studies, including Ki-67, EGFR, tumor cell count, and p53.
Ki-67 Proliferation Index
The Ki-67 proliferation marker is a reliable IHC method for determining tumor cell proliferation. Associations between the Ki-67 proliferation marker and MRI parameters were assessed in fifteen studies (Table 3).

Table 3.
Characteristics of MRI parameter association studies with other biological endpoints.
The associations between the Ki-67 proliferation index and the DWI-metric ADCmean were examined in six studies [60,61,63,65,71,72]. Four studies [60,61,63,65] directly investigated the correlation and found a significant inverse association (partial correlation r = −0.253 to r = −0.728; p < 0.001–0.024). In contrast, Wu et al. [71] reported a positive but non-significant correlation between ADCmean and Ki-67 (r = 0.238; p = 0.163). An overall effect of COR (−0.37, 95%CI −0.65–0.00; p = 0.051) was calculated; see Figure 4 for the forest plot. Additionally, Shima et al. [72] reported lower ADCmean values for a higher Ki-67 index (p = 0.012).
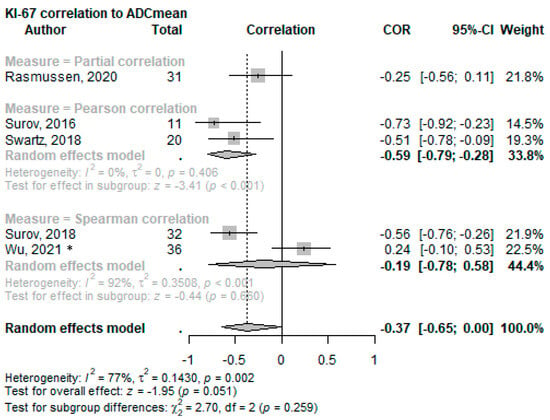
Figure 4.
Forest plot of the correlation of mean ADC values with the Ki-67 proliferation index split using the correlation test applied. * This cohort consists solely of nasopharyngeal squamous cell carcinoma [60,61,65,63,71].
ADCmaximum values also significantly correlated with Ki-67 (ρ = −0.46 to ρ = −0.640; p = 0.0079 to 0.036) in two studies [61,63], while the correlation with ADCminimum was significant in just one [63] of the three [61,63,71] reporting articles (ρ = −0.58; p = 0.0005). Two studies utilized the diffusion kurtosis imaging (DKI) technique, reporting different outcome parameters, but both highlighted the potential value of DKI in evaluating Ki-67 expression [72,73]. Further details on these associations can be found in Supplementary Material S2, Appendix S3 (A3.3).
Out of the five articles [40,41,60,62,64] that analyzed the correlation of Ki-67 with the DCE parameter mean Ktrans, only one study by Surov et al. [62] found significant correlation with Ki-67 (r = −0.62, p = 0.041). The four reporting articles found no significant correlation with Ki-67 for the mean Kep and mean Ve [40,41,62,64]. IVIM parameter analysis also did not reveal significant associations with the Ki-67 proliferation index [71].
Epidermal Growth Factor Receptor (EGFR)
Cellular signaling from the epidermal growth factor receptor (EGFR) induces cell proliferation and is frequently overexpressed or mutated in HNSCC [84]. Seven articles [30,40,47,48,49,60,67] assessed associations between histopathologically determined EGFR expression and MRI parameters, but no consistent results were found (See Table 3 for study characteristics).
No significant relationship was found between the EGFR and T1W or T2W imaging [49], ADCmean [47,60,67], or any other histogram parameter of the ADC map [47].
Mean Ktrans results were inconsistent in the four publications [30,40,48,60] examining associations between EGFR and DCE parameters. One study [40] reported a significantly higher mean Ktrans value in the EGFR overexpressing tumor group (p < 0.0001), while another study [30] found this to be significantly lower (p = 0.047). The remaining two papers [48,60] found no significant association between mean Ktrans and EGFR overexpression status. Similar inconsistencies were seen for Kep; Choi et al. [30] reported a negative association with significantly lower Kep values for EGFR overexpression (p = 0.004), while Huang et al. [40] reported non-significant but higher Kep values for high EGFR expression.
Tumor Cell Count
Six studies [49,60,61,62,63,64] investigated potential associations between pathologically assessed tumor cell count and MRI parameters with moderate success. MRI perfusion studies did not reveal any significant correlations between mean Ktrans or mean Kep and Ve and tumor cell counts [60,62,64]. Only one [63] of the three MR diffusion studies [60,61,63] showed a significant moderate inverse correlation between ADCmean and tumor cell count (ρ = −0.56; p = 0.0009). ADCminimum also exhibited a significant positive correlation with cell count in the same study (ρ = −0.60; p = 0.0003) [63]. See Supplementary Material S2, Appendix S3, for all non-significant tested associations.
Tumor Suppressor Protein p53
Tumor suppressor protein p53 regulates cell cycle progression, and its expression is either affected by HPV in HPV+ HNSCC or by TP53 mutations exclusively found in HPV− HNSCC [3,4].
Five studies [32,47,48,49,60] attempted to link MRI parameters with p53 status in HNSCC. Dang et al. [32] used DWI and contrast-enhanced T1W and T2W features of the oropharyngeal SCC (OPSCC) and classified p53-positive (p53+) and p53-negative (p53−) HNSCC with 81.3% accuracy. Rasmussen et al. analyzed perfusion (mean Ktrans) and diffusion (ADCmean) parameters in a mixed HNSCC group that included 14.3% OPSCC and 24% lymph nodes and observed positive correlations between the p53 percentage values in both Ktrans (partial correlation: 0.193; p = 0.015) and ADCmean (partial correlation r = 0.190; p = 0.010) [60]. However, these findings for Ktrans and ADCmean were not replicated by Meyer et al. [47,82]. Considering HPV status, as determined using p16 IHC, Meyer et al. published three articles [47,49,82] on p53 status prediction, each using a different MRI modality: T1W or T2W [49], DWI [47], and DCE [48]. In the p53− and thus the HPV−-enriched subgroup of Meyer et al., significant negative correlations were observed between lower ADCmaximum, ADC-P75, ADC-P90, ADC-SD [47], Vemax [82] and a larger positively stained p53 expression (ρ = −0.827; p = 0.002, ρ = −0.763; p = 0.01, ρ = −0.836; p = 0.001, ρ = −0.70; p = 0.016, ρ = −0.80; and p = 0.009, respectively). Additionally, T2Wmax (ρ = 0.736; p = 0.015), T2W-P90 (ρ = 0.68; p = 0.028), and T2W-SD (ρ = 0.760; p = 0.011) [49] positively correlated with the stained p53 expression. Within the p16 positive and thus likely HPV-positive oropharyngeal subgroup, higher T2W-mean, T2W-P25, T2W-P75, T2W-median, and T2W-mode [49] were positively associated with higher p53 staining (ρ = 0.569; p = 0.007, ρ = 0.508; p = 0.019, ρ = 0.479; p = 0.028, ρ = 0.555; p = 0.009, ρ = 0.468; p = 0.033, respectively). Lower ADC-kurtosis (ρ = −0.446; p = 0.029) [47] and T1W-entropy (ρ = −0.648; p = 0.001) [49] demonstrated a negative association with highly stained p53 expression.
None of the included studies counted the complete absence of p53 expression as a mutation.
3.3.3. Tumor Vasculature: HIF-1α, VEGF, and MVD
Biological features that impact or reflect tumor vasculature, such as the microvessel density (MVD), hypoxia markers such as HIF-1α, or angiogenesis-promoting vascular endothelial growth factor (VEGF), are bound to affect diffusion and perfusion parameters as determined using functional MRI and have, therefore, been the subject of “radiogenomic” studies [5,6,7].
Hypoxia-Inducible Factor (HIF)-1α
The association of HIF-1α with MRI parameters was addressed in eight publications [31,39,40,42,47,48,49,65] (See Table 3 for all study characteristics).
One study by Chen et al. determined localized higher expression of HIF-1α in tumor cells surrounding MRI-determined necrotic areas. A radiologist identified these areas using gadolinium contrast-enhanced T1W and T2W MRI [31]. Meyer et al. [49] observed a significant negative correlation between several standalone T2W imaging histogram parameters and HIF-1α in p16-negative HNSCC (Supplementary Material S2, Appendix S3 (3A.1)).
Three studies [39,40,42] analyzed the volume and speed of the blood running through the tissue using different metrics: fractional plasma volume (Vp) (r = 0.173; p = 0.327) [39]; tumor blood flow (TBF) (p < 0.001); fluid flow velocity (|ⴎ|) (p < 0.001) [40]; and maximum enhancement (ME) (p = 0.001), relative enhancement (RE) (p = 0.027), and maximum relative enhancement (MRE) (p = 0.017) of the time–signal intensity curve [42]. Most of these parameters showed either a positive correlation or a significantly higher value for higher HIF-1α expression, except for Vp as measured by Hu et al. Yet other DCE parameters describing the extravascular extracellular space (EES) (e.g., Ve) or the transfer between the two compartments (Ktrans, Kep) did not have significant correlations [39,40,48].
Vascular Endothelial Growth Factor (VEGF)
VEGF was analyzed in eight original articles [33,39,47,48,49,60,66,67].
Positive correlations were reported for the T1W-P10 histogram parameter [49] (ρ = 0.371, p = 0.04) and vascular compartment parameter Vp (r = 0.339; p = 0.05) in [39]. A similar blood volume (Vb) parameter measured through a two-compartment model applied by Donaldson et al. [33] did not replicate this. Donaldson et al. found a significant strong inverse relationship between Fb (whole-blood perfusion) and VEGF expression using PCR-derived VEGF mRNA measures (r = −0.82; p = 0.023) [33]. For more details on the non-significant associations tested, refer to Supplementary Material S2, Appendix S3.
Microvessel Density (MVD)
The variability of molecular markers used for microvessel identification and assessment, such as CD105 [41,48,51], CD34 [39,41,68], and CD31 [62,66], hampered comparisons across the seven studies that investigated correlations between MVD and MRI parameters (Table 3).
DCE parameters (mean Ktrans, Kep, and Ve) were analyzed in four studies [39,41,50,62]. One paper by Meyer et al. [50] revealed a significant positive correlation between high mean Kep and a higher number of CD105-derived microvessels. At the same time, another article by Karabay et al. [41] indicated a positive correlation between mean Ktrans and the number of CD34-derived microvessels (r = 0.346; p = 0.049) but not for the CD105-derived count (r = 0.307; p = 0.08). No other discernable trends or correlations were observed in the other studies [39,62]. Unetsubu et al. and Tekiki et al. examined correlations between CD34 and CD31 MVD, respectively, and contrast index (CI) parameters. Both studies observed a significant positive correlation between MVD and CI-gain, representing the maximum gradient during the upslope phase of the enhancement curve (r = 0.46 to r = 0.49; p = 0.00821 to p = 0.037). No significant correlation was found between MVD and CI-max, representing the maximum amplitude of contrast enhancement [66,68].
When investigating the stained vessel area as calculated using either CD105 [50] or CD31 [62], only Surov et al. uncovered a significant positive correlation with mean Kep (r = 0.67; p = 0.02) [62], while Meyer et al. did find other correlations with Kep histogram parameters [50] (see Supplementary Material S2, Appendix S3 (A3.3)).
3.3.4. Radiomics and Genomics Linkage Studies
Two articles [76,77] included broader genomic analyses. Both articles focused on developing radiomic-based treatment outcome prediction models using datasets as outlined in Table 4. Separate, much smaller cohorts, consisting of 9 [76] to 16 [77] patients with gene expression data, were used to explore potential relationships with such biological endpoints. Please note that these separate analyses strongly lack statistical power.

Table 4.
Radiomic prediction model study design characteristics with their radiomics quality score.
Table 4.
Radiomic prediction model study design characteristics with their radiomics quality score.
| Radiomic Models for Biological Signature | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study, Year | Location Inclusion Center | Train (N) | Test (N) | Age (mean) | Male (%) | Tumor Subside | Tumor Stage | Modality | #Features | Total RQS | Domains: IM/ FR/VA/PI/LE/OS |
| Gao, 2021 [76] | Hunan, CHN | 237 | 79 | 47.9 | 69.9 | NA | All | T1+c | 530 | 16 | 8/5/6/3/6/0 |
| Zhang, 2020 [77] | Zuhai, CHN | 220 | 44 + 44 * | 47.4 † | 72.7 | NA | All | T1(c), T2 | 2364 | 19 | 8/6/6/5/7/0 |
The table lists study design characteristics, patient numbers, and total and individual domain quality scores of radiomics-based prediction model studies that were designed to align with biological features. Abbreviations: No. = number of selected patients; Train = number of patients analyzed in the training cohort; Test = number of patients analyzed in the test cohort; #Features = number of features collected; NA = nasopharynx; RQS = radiomics quality score; RSQ domains = IM: image protocol and feature reproducibility, FR: feature reduction and validation, VA: biologic/clinical validation and utility, PI: performance index, LE: high level of evidence, OS: open science. Note: † Median value; * 44 patients of an internal validation cohort and 44 in a separate external validation cohort.
Gao et al. [76] used their newly developed radiomic signature based on T1W with gadolinium features to predict progression-free survival (PFS) risk in a genetic data-equipped subcohort of nine patients. The genes of patients with lower PFS risk were compared with those with higher PFS risk. Furthermore, all genes were correlated to the RAD score. In a similar approach, Zhang et al. [77] initially developed a radiomic model using T1W with and without gadolinium-based agents and T2W imaging to predict failure-free survival (FFS) that was then reduced to twelve key radiomic features. This model was tested against gene mutation data in 508 genes in 16 patients. While the authors report that some texture features were associated with the chromatin remodeling pathway and higher mutational burden, the study size was too small to appropriately adjust for multiple testing, false positive rates, or to conduct multivariate analysis with other (clinical) factors. In contrast to MRI, patient outcome is defined by multiple biologic factors which likely affect the association studies in both reports.
4. Discussion
MRI, with its superior soft tissue definition and functional imaging capabilities, has become a routine and indispensable tool in staging HNSCC [1]. This systematic review evaluated the current investigation status and evidence for the applicability of quantitative MRI techniques to assess the biological characteristics of primary HNSCC. Additionally, it delved into current research examining radiomic models in this context. Key findings include significantly lower ADCmean (SMD: 0.82; p < 0.001) and ADCminimum (SMD: 0.56; p < 0.001) values for HPV+ HNSCC compared with HPV− tumors as described in multiple studies. Moreover, we repeatedly reported correlations between low ADCmean values and high levels of the proliferation marker Ki-67 (COR: −0.37; p = 0.051). Furthermore, functional MRI perfusion parameters that depict increased blood plasma volume and flow showed significant associations with higher HIF-1α. Figure 5 provides a visual overview of the level of significance of all MRI parameters sorted by biological factor.
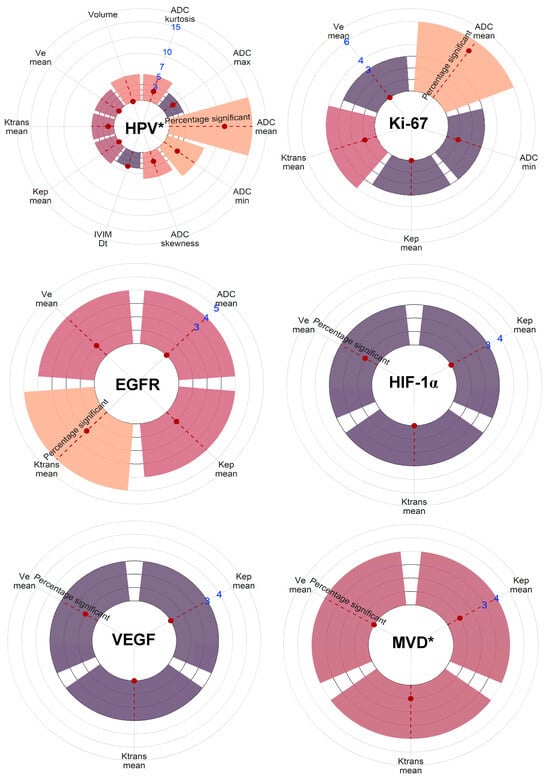
Figure 5.
Visual representation of the number of reporting studies and the level of significance of all MRI parameters sorted by biological factor. MRI parameters are included when three or more studies have reported on associations with the specific metric and the biological factor. The count of studies is indicated by the blue numbers. The statistical significance as reported in the listed studies is indicated by the red dot on the red-striped line within the bars. * As grouped for all biological measurements defining the factor. Note: p53 lacked enough comparable studies.
4.1. Human Papilloma Virus (HPV)
The HPV status of the tumor emerged as the most frequently investigated endpoint, given its significant impact on prognosis and treatment decisions. Consistent with Payabvash et al.’s meta-analysis [85], the comprehensive analyses of the data in this review confirm a link between DWI parameters and HPV status. DWI-derived mean and minimum ADC values seem to be significantly lower in HPV+ tumors compared with HPV− tumors. DWI is a functional MR technique that can assess the random movement of water molecules in the tissue microstructure, with the ADC being a derived metric used to evaluate it [47,65,86]. Hypercellular tissue is in general characterized by a low ADC value due to the restrictions on water movement imposed by the cell walls. This results locally in low water diffusivity. Conversely, local hypo-cellular tissue (e.g., tumor areas with necrosis) has a (slightly) higher ADC value due to fewer cells per volume, allowing more water movement [86,87].
Histopathologically, HPV+ OPSCCs are characterized by immature, ellipsoidal nuclei with more frequent mitosis, a high nuclear-to-cytoplasmic ratio, and decreased keratinization around the tumor periphery, possibly explaining the lower ADC values due to increased cellularity with decreased water diffusivity. HPV− OPSCC typically displays better-keratinized cells with distinct cell borders and a larger amount of cytoplasm [2,3,88,89].
In contrast to this observation, five studies [36,43,47,57,70] reported a higher mean ADC value for HPV+ tumors but this did not reach significance. Possible reasons include limiting the cohort to only the biologically dissimilar nasopharyngeal SCCs (NSCCs) [43], a relatively small sample size [70], a different DWI-acquisition technique (PROPELLER), and higher b-values for the ADC calculation applied [57]. Positive p16 IHC combined with PCR detection of HPV-DNA or RNA increases specificity compared with stand-alone detection methods like p16 staining [90,91]. Incorrectly classified HPV-negative patients in studies that solely use p16 IHC in an already mixed cohort [47] may have also strongly contributed to this discrepancy. Notably, we found that reported significance values may have been erroneous in one study [36], likely based on the misreporting of some values (standard deviation did not match the reported range or p-value); Freihat et al.’s Student’s t-test results, which determined significant difference in ADCmean between HPV+ and HPV− tumors, could not be replicated by us.
In the current set of available studies, MRI perfusion parameters did not correlate with HPV status in a reproducible and consistent manner [30,38,48,54], implying that differences in the DCE parameters for HPV status are less likely to exist, thus suggesting similar vascular biology. More extensive, radiomic methods to distinguish HPV status appear promising, especially when combined with clinical parameters as they may better reflect the diverse biological differences between tumor classes. Importantly, however, as for now, none of the researched radiomic models have been externally validated. Some studies either showed a significant drop-off in AUC after an internal validation on test sets [46] or solely relied on cross-validation values instead of internal validation using independent test sets [26], raising concerns about the robustness of the models. Large, multicenter databases will be needed to build and test definitive models.
In summary, when comparing HPV+ and HPV− OPSCCs, HPV+ tumors show similar perfusion MRI parameters to HPV− but dissimilar diffusion MRI parameters with consistently lower ADC values in DWI.
4.2. Tumor Cell Proliferation and Cellularity Markers: Ki-67, Tumor Cell Count, and EGFR
Cancer development and progression rely significantly on cellular proliferation. Cytotoxic drugs used in cancer therapy target highly proliferative and regenerating cells. Tumor cell proliferation markers, such as Ki-67, tumor cell count, and the EGFR, therefore play critical roles in cancer research, either as potential treatment effect monitors or therapeutic targets [92,93,94,95,96,97,98].
The fraction of Ki-67 positive cells, as determined using IHC, is a frequently used proliferation marker [96]. High Ki-67 expression has been correlated with poor prognosis and increased risk of lymph node metastasis [92]. Being able to monitor tumor cell proliferation through medical imaging techniques can aid in choosing and, when needed, intensifying treatments for high-proliferating tumors.
As described in this review, the DWI parameter ADCmean is inversely correlated with the proliferation index Ki-67 and, to a lesser extent, with tumor cell count, likely due to reduced extracellular space [87]. However, a contradictory study by Wu et al. [71] reported a higher ADCmean for higher Ki-67 values, albeit insignificantly. A positive correlation between ADCmean and Ki-67 is unlikely in any tumor type, suggesting that these results may represent an outlier [99]. Yet if assuming this deviation is significant, this discrepancy could potentially be attributed to the different clinical behavior, epidemiology, histopathological characteristics, and biology of nasopharyngeal tumors compared with other HNSCCs [100]. The link with the Epstein–Barr virus, similar to HPV in OPSCC, could also conceivably impact ADC values. Rasmussen et al. identified a significant correlation using the partial correlation metric and incorporating a random offset; our forest plot analyses did not yield the same level of significance as we could not correct for the offset [60]. For tumor cell count, only one of three reporting articles described significant correlations [60,61,63]. Yet a connection between cellularity and ADC metrics aligns with DWI characteristics. As data are limited, more extensive research is needed to understand these findings.
The EGFR plays a role in endothelial cell proliferation and can be targeted with Cetuximab. It has gained interest due to its link to high mutational burden [8,93,97]. However, no associations have been found between EGFR and DWI parameters such as ADCmean. There have been, however, associations observed between EGFR and DCE parameters, although with contradicting results. It was proposed that such links could possibly be based on the involvement of the EGFR in the promotion of angiogenesis through the stimulation of the production of angiogenic cytokines such as VEGF [84]. Often linked to increased tumor vascularization and aggressiveness, Ktrans represents the constant transfer from blood plasma to extravascular extracellular space (EES), while Kep measures the backflux exchange rate of EES to blood plasma with gadolinium contrast [101]. The presence of necrotic areas may result in lower Ktrans and Kep values [11] and different necrotic profiles and locations could potentially explain the divergent reported results. Higher Ktrans and Kep with higher EGFR values were seen in the less necrotic NSCC by Huang et al. [40], while Choi et al. [30] reported lower Ktrans and Kep values in high-expressing EGFR but large OPSCC that are more likely to have more necrotic areas. Studies focusing on mixed HNSCC groups [48,60] showed no significant effect, calling for further research to be categorized based on anatomical subsites.
In summary, low ADC values reflected high proliferation markers Ki-67 and, to some extent, tumor cell count; this result may have clinical implications if it could accurately guide clinicians toward more appropriate and, if required, aggressive treatment regimens. No consistent results, however, were found for associations between imaging features and the EGFR.
The p53 Pathway
The TP53 gene product plays a crucial role in regulating the cell cycle and trig-gering apoptosis in response to irreparable DNA damage, making it an attractive tar-get for non-invasive patient selection and treatment monitoring [98]. It should be not-ed that virtually all pharyngeal and laryngeal HNSCC tumors are affected in their p53 pathway, either through HPV E7/8 expression or in HPV-negative tumors due to TP53 gene mutations [94]. Given its degradation by HPV, TP53 mutations are absent or not biologically significant in HPV+. In addition, p53 pathway status classification (i.e., in HPV−) can differ across the different studies. Both high percentages of p53 expression and a complete absence of p53 expression in tumor tissue have been found to be asso-ciated with p53 pathway aberrations and TP53 mutations [98]. Together this stresses the need for an HPV and p53 status subgroup analysis that is, however, lacking or in-sufficiently statistically powered within the included studies. With this in mind, some of the included articles elude to a correlation between ADC parameters like ADCmaximum and ADCmean and p53. However, this correlation probably arises from HPV-induced cellular variations observed within the mixed cohorts [32,47,60].
4.3. Tumor Angiogenesis Markers: HIF-1α, VEGF, and MVD
High HIF-1α expression is induced in response to tissue hypoxia [102]. While the HIF1 pathway can have multiple cellular effects that also promote angiogenesis, most studies have not been able to support a simple and unifactorial relationship between increased hypoxia, HIF1 alpha expression, and angiogenesis or tumor growth [103]. Nonetheless, Chen et al. reported a link between the presence of MRI-determined necrotic areas in tumors and a high HIF-1α value [31]. As within hypoxic areas, the effectivity of systematic and radiotherapeutic treatment decreases; therefore, MRI monitoring of hypoxia can have a prominent role in treatment evaluation and follow-up [104]. DCE-imaging can be very useful in depicting the vascularization of tissue as Ktrans has notably been associated with hypoxic fractions in several other tissue types [11,105]. The lack of significant associations between the DCE parameters obtained from pharmacokinetic models and HIF-1α in the studies of this review may be attributable to the focus on nasopharyngeal SCC in two of the articles [39,40]. Tumors at this subsite differ biologically and may induce HIF-1α through alternative pathways unrelated to hypoxia [40].
High tumor blood flow (TBF) and blood velocity (|ⴎ|) perfusion parameters were significantly associated with high IHC HIF-1α values [40]. Similarly, increased values of time–signal intensity curve perfusion parameters (RE, ME, and MRE) also showed a significant correlation with high HIF-1α expression [42]. While hypoxia is a consequence of limited tissue blood flow and permeability, the authors of these studies speculated this correlation to be based on potential angiogenetic effects of HIF-1 in other areas. However, due to the use of different measurement methods in the articles, it is challenging to gauge the true effect.
Similar to HIF-1α, VEGF can be induced in response to tumor hypoxia through the HIF pathway or directly secreted by tumor cells [102]. Higher VEGF expression is expected in situations of low perfusion (hypoxia) or high blood flow (angiogenic tumor growth) [33,106]. This contrast in terms of MRI-based vascular profile may hamper the DCE analyses and align with the overall lack of correlations with mean values of Ktrans, Kep, and Ve [33,39,60]. A higher blood plasma volume could be linked to more angiogenic tumor growth as described by Hu et al. and Donaldson et al. However, differences in models and VEGF assessment methods (IHC [39] and PCR [33]) make direct comparisons challenging [33,39].
The analysis of MVD was complicated by the use of different MVD determination methods and the lack of consistency among studies using identical MVD parameters, potentially obscuring the results. Only the contrast index (CI) perfusion parameter for the maximum gradient of the upslope phase of the enhancement curve (CI gain max) was significantly positively correlated to MVD in both reporting studies using either CD31 [66] or CD34 [68]. This suggests that the simple contrast index curve, which considers only the speed of uptake, may provide a better depiction of the number of microvessels compared with the pharmacokinetic DCE parameters.
In summary, this review presents mixed results with respect to VEGF but reveals associations between HIF-1α and several increased tissue blood flow and permeability perfusion parameters.
4.4. Tumor Heterogeneity and Radiogenomics
As a non-invasive, 3D visualizing modality, medical imaging has some considerable advantages over biopsy-sampling-based techniques. Spatial and temporal intratumor heterogeneity has been shown to be extensive in multi-regional sampling studies, and has been identified as a major cause of treatment resistance and might be related to DNA repair deficits [107,108]. Unlike biopsy-based subtyping, imaging may better capture this heterogeneity, advancing future targeted treatment regimens and driving interest in such advances. In this review, both articles, however, were small in case numbers and primarily developed radiomics-based patient outcome prediction models. Thus, strongly outcome-biased, the articles subsequently investigated the biological differences. While this can help to find MRI feature/biology links, such approaches require an independent assessment of the potentially present associations with MRI features [76,77].
4.5. Limitations
While comprehensive, this review of MRI parameter associations with biological features is limited by the large variety of methods used in the included articles for calculating biological and MRI parameters. Some studies report on histogram parameters based just on the lowest or highest voxel (e.g., ADCminimum or ADCmaximum), posing challenges to reproducibility. Additionally, variations in segmentation methods for tumor delineation (e.g., inclusion/exclusion of necrotic and cystic areas) impact overall measurements. Furthermore, only a modest amount of data are available for certain biological factors such as p53 and MVD, making it difficult to draw firm conclusions and highlighting the need for further research.
It is also crucial to note that biological processes do not occur in isolation. Cell proliferation and angiogenesis are intertwined through multiple cross-linked processes. Similarly, in MRI there are interconnections between parameters such as DCE, DWI, and even T1W and T2W parameters in visualizing the tissue. Understanding these complex relationships can offer more comprehensive insights into the important tumor-specific differences, paving the way for improved clinical outcome prediction and personalized patient management in HNSCC.
5. Conclusions
In this comprehensive review, we analyzed the relationship between biological factors and MRI in HNSCC. Across all studies, we predominantly found that HPV+ tumors showed lower ADCmean (SMD: 0.82; p < 0.001) and ADCminimum (SMD: 0.56; p < 0.001) values than HPV− tumors. Lower ADC values correlated with elevated Ki-67 index levels in most studies (COR: −0.37; p = 0.051). Perfusion parameters that depict increased blood plasma volume and flow showed some associations with HIF-1α. There is potential for radiomic models to capture biological differences in tumors. However, diverse methodologies and limited reports on certain investigated biological factors necessitate further research and larger datasets. Understanding these connections can improve clinical outcome prediction and facilitate personalized patient management in HNSCC.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15205077/s1. Supplementary Material S1: Material and methods; Supplementary Material S2: Results.
Author Contributions
Conceptualization, H.J.v.d.H., R.W.J., J.A.C. and M.W.M.v.d.B.; methodology, H.J.v.d.H. and R.W.J.; formal analysis, H.J.v.d.H.; investigation, H.J.v.d.H., R.W.J. and J.A.C.; data curation, H.J.v.d.H.; writing—original draft preparation, H.J.v.d.H., R.W.J., C.V. and J.A.C.; writing—review and editing, all authors; visualization, H.J.v.d.H.; and supervision, J.A.C., M.W.M.v.d.B. and R.G.H.B.-T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Becker, M.; Zaidi, H. Imaging in head and neck squamous cell carcinoma: The potential role of PET/MRI. Br. J. Radiol. 2014, 87, 20130677. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Canning, M.; Guo, G.; Yu, M.; Myint, C.; Groves, M.W.; Byrd, J.K.; Cui, Y. Heterogeneity of the Head and Neck Squamous Cell Carcinoma Immune Landscape and Its Impact on Immunotherapy. Front. Cell Dev. Biol. 2019, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Nishida, N.; Yano, H.; Nishida, T.; Kamura, T.; Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar] [CrossRef]
- Kyzas, P.A.; Stefanou, D.; Batistatou, A.; Agnantis, N.J. Prognostic significance of VEGF immunohistochemical expression and tumor angiogenesis in head and neck squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2005, 131, 624–630. [Google Scholar] [CrossRef]
- Li, J.; Li, S.X.; Gao, X.H.; Zhao, L.F.; Du, J.; Wang, T.Y.; Wang, L.; Zhang, J.; Wang, H.Y.; Dong, R.; et al. HIF1A and VEGF regulate each other by competing endogenous RNA mechanism and involve in the pathogenesis of peritoneal fibrosis. Pathol. Res. Pract. 2019, 215, 644–652. [Google Scholar] [CrossRef]
- Rude Voldborg, B.; Damstrup, L.; Spang-Thomsen, M.; Skovgaard Poulsen, H. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann. Oncol. 1997, 8, 1197–1206. [Google Scholar] [CrossRef]
- Knegjens, J.L.; Hauptmann, M.; Pameijer, F.A.; Balm, A.J.; Hoebers, F.J.; de Bois, J.A.; Kaanders, J.H.; van Herpen, C.M.; Verhoef, C.G.; Wijers, O.B.; et al. Tumor volume as prognostic factor in chemoradiation for advanced head and neck cancer. Head Neck 2011, 33, 375–382. [Google Scholar] [CrossRef]
- van der Heijden, M.; Essers, P.B.M.; de Jong, M.C.; de Roest, R.H.; Sanduleanu, S.; Verhagen, C.V.M.; Hamming-Vrieze, O.; Hoebers, F.; Lambin, P.; Bartelink, H.; et al. Biological Determinants of Chemo-Radiotherapy Response in HPV-Negative Head and Neck Cancer: A Multicentric External Validation. Front. Oncol. 2019, 9, 1470. [Google Scholar] [CrossRef]
- Gaustad, J.V.; Hauge, A.; Wegner, C.S.; Simonsen, T.G.; Lund, K.V.; Hansem, L.M.K.; Rofstad, E.K. DCE-MRI of Tumor Hypoxia and Hypoxia-Associated Aggressiveness. Cancers 2020, 12, 1979. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.W.; van Amstel, P.; Martens, R.M.; Kooi, I.E.; Wesseling, P.; de Langen, A.J.; Menke-Van der Houven van Oordt, C.W.; Jansen, B.H.E.; Moll, A.C.; Dorsman, J.C.; et al. Non-invasive tumor genotyping using radiogenomic biomarkers, a systematic review and oncology-wide pathway analysis. Oncotarget 2018, 9, 20134–20155. [Google Scholar] [CrossRef] [PubMed]
- Park, A.Y.; Han, M.-R.; Seo, B.K.; Ju, H.-Y.; Son, G.S.; Lee, H.Y.; Chang, Y.W.; Choi, J.; Cho, K.R.; Song, S.E.; et al. MRI-based breast cancer radiogenomics using RNA profiling: Association with subtypes in a single-center prospective study. Breast Cancer Res. 2023, 25, 79. [Google Scholar] [CrossRef] [PubMed]
- Rabasco Meneghetti, A.; Zwanenburg, A.; Linge, A.; Lohaus, F.; Grosser, M.; Baretton, G.B.; Kalinauskaite, G.; Tinhofer, I.; Guberina, M.; Stuschke, M.; et al. Integrated radiogenomics analyses allow for subtype classification and improved outcome prognosis of patients with locally advanced HNSCC. Sci. Rep. 2022, 12, 16755. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Faraone, S.V. Interpreting estimates of treatment effects: Implications for managed care. Pharm. Ther. 2008, 33, 700–711. [Google Scholar]
- Balduzzi, S.; Rücker, G.; Schwarzer, G. How to perform a meta-analysis with R: A practical tutorial. Evid. Based Ment. Health 2019, 22, 153–160. [Google Scholar] [CrossRef]
- Luo, D.; Wan, X.; Liu, J.; Tong, T. Optimally estimating the sample mean from the sample size, median, mid-range, and/or mid-quartile range. Stat. Methods Med. Res. 2018, 27, 1785–1805. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Wang, W.; Liu, J.; Tong, T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med. Res. Methodol. 2014, 14, 135. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Luo, D.; Wan, X.; Liu, Y.; Liu, J.; Bian, Z.; Tong, T. Detecting the skewness of data from the five-number summary and its application in meta-analysis. Stat. Methods Med. Res. 2023, 32, 1338–1360. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Luo, D.; Wan, X.; Liu, Y.; Liu, J.; Bian, Z.; Tong, T. Detecting the skewness of data from the sample size and the five-number summary. arXiv 2020, arXiv:201005749. [Google Scholar]
- Ahn, Y.; Choi, Y.J.; Sung, Y.S.; Pfeuffer, J.; Suh, C.H.; Chung, S.R.; Baek, J.H.; Lee, J.H. Histogram analysis of arterial spin labeling perfusion data to determine the human papillomavirus status of oropharyngeal squamous cell carcinomas. Neuroradiology 2021, 63, 1345–1352. [Google Scholar] [CrossRef]
- Boot, P.A.; Mes, S.W.; de Bloeme, C.M.; Martens, R.M.; Leemans, C.R.; Boellaard, R.; van de Wiel, M.A.; de Graaf, P. Magnetic resonance imaging based radiomics prediction of Human Papillomavirus infection status and overall survival in oropharyngeal squamous cell carcinoma. Oral Oncol. 2023, 137, 106307. [Google Scholar] [CrossRef]
- Bos, P.; van den Brekel, M.W.M.; Gouw, Z.A.R.; Al-Mamgani, A.; Waktola, S.; Aerts, H.; Beets-Tan, R.G.H.; Castelijns, J.A.; Jasperse, B. Clinical variables and magnetic resonance imaging-based radiomics predict human papillomavirus status of oropharyngeal cancer. Head Neck 2021, 43, 485–495. [Google Scholar] [CrossRef]
- Bos, P.; van den Brekel, M.W.M.; Taghavi, M.; Gouw, Z.A.R.; Al-Mamgani, A.; Waktola, S.; Aerts, H.J.W.L.; Beets-Tan, R.G.H.; Castelijns, J.A.; Jasperse, B. Largest diameter delineations can substitute 3D tumor volume delineations for radiomics prediction of human papillomavirus status on MRI’s of oropharyngeal cancer. Phys. Medica 2022, 101, 36–43. [Google Scholar] [CrossRef]
- Chan, M.W.; Higgins, K.; Enepekides, D.; Poon, I.; Symons, S.P.; Moineddin, R.; Weinreb, I.; Shearkhani, O.; Chen, A.; Beelen, J.; et al. Radiologic Differences between Human Papillomavirus-Related and Human Papillomavirus-Unrelated Oropharyngeal Carcinoma on Diffusion-Weighted Imaging. ORL J. Oto-Rhino-Laryngol. Its Relat. Spec. 2016, 78, 344–352. [Google Scholar] [CrossRef]
- Choi, Y.S.; Park, M.; Kwon, H.J.; Koh, Y.W.; Lee, S.-K.; Kim, J. Human Papillomavirus and Epidermal Growth Factor Receptor in Oral Cavity and Oropharyngeal Squamous Cell Carcinoma: Correlation with Dynamic Contrast-Enhanced MRI Parameters. AJR Am. J. Roentgenol. 2016, 206, 408–413. [Google Scholar] [CrossRef]
- Chen, T.C.; Wu, C.T.; Wang, C.P.; Hsu, W.L.; Yang, T.L.; Lou, P.J.; Ko, J.Y.; Chang, Y.L. Associations among pretreatment tumor necrosis and the expression of HIF-1α and PD-L1 in advanced oral squamous cell carcinoma and the prognostic impact thereof. Oral Oncol. 2015, 51, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Dang, M.; Lysack, J.T.; Wu, T.; Matthews, T.W.; Chandarana, S.P.; Brockton, N.T.; Bose, P.; Bansal, G.; Cheng, H.; Mitchell, J.R.; et al. MRI texture analysis predicts p53 status in head and neck squamous cell carcinoma. AJNR Am. J. Neuroradiol. 2015, 36, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, S.B.; Betts, G.; Bonington, S.C.; Homer, J.J.; Slevin, N.J.; Kershaw, L.E.; Valentine, H.; West, C.M.L.; Buckley, D.L. Perfusion estimated with rapid dynamic contrast-enhanced magnetic resonance imaging correlates inversely with vascular endothelial growth factor expression and pimonidazole staining in head-and-neck cancer: A pilot study. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, 1176–1183. [Google Scholar] [CrossRef] [PubMed]
- de Perrot, T.; Lenoir, V.; Domingo Ayllón, M.; Dulguerov, N.; Pusztaszeri, M.; Becker, M. Apparent Diffusion Coefficient Histograms of Human Papillomavirus-Positive and Human Papillomavirus-Negative Head and Neck Squamous Cell Carcinoma: Assessment of Tumor Heterogeneity and Comparison with Histopathology. AJNR Am. J. Neuroradiol. 2017, 38, 2153–2160. [Google Scholar] [CrossRef] [PubMed]
- Driessen, J.P.; van Bemmel, A.J.M.; van Kempen, P.M.W.; Janssen, L.M.; Terhaard, C.H.J.; Pameijer, F.A.; Willems, S.M.; Stegeman, I.; Grolman, W.; Philippens, M.E.P. Correlation of human papillomavirus status with apparent diffusion coefficient of diffusion-weighted MRI in head and neck squamous cell carcinomas. Head Neck 2016, 38, E613–E618. [Google Scholar] [CrossRef] [PubMed]
- Freihat, O.; Tóth, Z.; Pintér, T.; Kedves, A.; Sipos, D.; Cselik, Z.; Lippai, N.; Repa, I.; Kovács, Á. Pre-treatment PET/MRI based FDG and DWI imaging parameters for predicting HPV status and tumor response to chemoradiotherapy in primary oropharyngeal squamous cell carcinoma (OPSCC). Oral Oncol. 2021, 116, 105239. [Google Scholar] [CrossRef]
- Giannitto, C.; Marvaso, G.; Botta, F.; Raimondi, S.; Alterio, D.; Ciardo, D.; Volpe, S.; De Piano, F.; Ancona, E.; Tagliabue, M.; et al. Association of quantitative MRI-based radiomic features with prognostic factors and recurrence rate in oropharyngeal squamous cell carcinoma. Neoplasma 2020, 67, 1437–1446. [Google Scholar] [CrossRef]
- Han, M.; Lee, S.J.; Lee, D.; Kim, S.Y.; Choi, J.W. Correlation of human papilloma virus status with quantitative perfusion/diffusion/metabolic imaging parameters in the oral cavity and oropharyngeal squamous cell carcinoma: Comparison of primary tumour sites and metastatic lymph nodes. Clin. Radiol. 2018, 73, 757.e721–757.e727. [Google Scholar] [CrossRef]
- Hu, Y.; E, H.; Yu, X.; Li, F.; Zeng, L.; Lu, Q.; Xi, X.; Shen, L. Correlation of quantitative parameters of magnetic resonance perfusion-weighted imaging with vascular endothelial growth factor, microvessel density and hypoxia-inducible factor-1alpha in nasopharyngeal carcinoma: Evaluation on radiosensitivity study. Clin. Otolaryngol. 2018, 43, 425–433. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, Q.; Wu, G.; Chen, P.P.; Li, J.; McCabe Gillen, K.; Spincemaille, P.; Chiang, G.C.; Gupta, A.; Wang, Y.; et al. DCE-MRI quantitative transport mapping for noninvasively detecting hypoxia inducible factor-1alpha, epidermal growth factor receptor overexpression, and Ki-67 in nasopharyngeal carcinoma patients. Radiother. Oncol. 2021, 164, 146–154. [Google Scholar] [CrossRef]
- Karabay, N.; Bülbül, H.; Doğan, E.; İkiz, A.; Bülbül, G.; Sarıoğlu, S. The correlations between dynamic contrast enhanced magnetic resonance imaging and immunohistochemical data in head and neck squamous cell carcinomas. Turk. J. Med. Sci. 2022, 52, 1950–1957. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Hu, L.; Zeng, Q.; Peng, D.; Chen, Z.; Huang, C.; Liu, Z.; Wen, Q.; Zou, F.; Yan, L. Dynamic contrast-enhanced MRI of nasopharyngeal carcinoma: Correlation of quantitative dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) parameters with hypoxia-inducible factor 1alpha expression and tumor grade/stage. Ann. Palliat. Med. 2021, 10, 2238–2253. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Kato, H.; Tomita, H.; Hara, A.; Suzui, N.; Miyazaki, T.; Matsuo, M. Comparison of Imaging Findings between Human Papillomavirus-positive and -Negative Squamous Cell Carcinomas of the Maxillary Sinus. J. Clin. Imaging Sci. 2020, 10, 59. [Google Scholar] [CrossRef]
- Lenoir, V.; Delattre, B.M.A.; M’Ra, D.Y.; De Vito, C.; de Perrot, T.; Becker, M. Diffusion-Weighted Imaging to Assess HPV-Positive versus HPV-Negative Oropharyngeal Squamous Cell Carcinoma: The Importance of b-Values. Am. J. Neuroradiol. 2022, 43, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Martens, R.M.; Noij, D.P.; Koopman, T.; Zwezerijnen, B.; Heymans, M.; de Jong, M.C.; Hoekstra, O.S.; Vergeer, M.R.; de Bree, R.; Leemans, C.R.; et al. Predictive value of quantitative diffusion-weighted imaging and 18-F-FDG-PET in head and neck squamous cell carcinoma treated by (chemo)radiotherapy. Eur. J. Radiol. 2019, 113, 39–50. [Google Scholar] [CrossRef]
- Marzi, S.; Piludu, F.; Avanzolini, I.; Muneroni, V.; Sanguineti, G.; Farneti, A.; D’Urso, P.; Benevolo, M.; Rollo, F.; Covello, R.; et al. Multifactorial Model Based on DWI-Radiomics to Determine HPV Status in Oropharyngeal Squamous Cell Carcinoma. Appl. Sci. 2022, 12, 7244. [Google Scholar] [CrossRef]
- Meyer, H.J.; Leifels, L.; Hamerla, G.; Höhn, A.K.; Surov, A. ADC-histogram analysis in head and neck squamous cell carcinoma. Associations with different histopathological features including expression of EGFR, VEGF, HIF-1 alpha, Her 2 and p53. A preliminary study. Magn. Reson. Imaging 2018, 54, 214–217. [Google Scholar] [CrossRef]
- Meyer, H.J.; Leifels, L.; Hamerla, G.; Höhn, A.K.; Surov, A. Associations between Histogram Analysis Parameters Derived from DCE-MRI and Histopathological Features including Expression of EGFR, p16, VEGF, Hif1-alpha, and p53 in HNSCC. Contrast Media Mol. Imaging 2019, 2019, 5081909. [Google Scholar] [CrossRef]
- Meyer, H.J.; Leifels, L.; Hamerla, G.; Höhn, A.K.; Surov, A. Histogram Analysis Parameters Derived from Conventional T1-and T2-Weighted Images Can Predict Different Histopathological Features Including Expression of Ki67, EGFR, VEGF, HIF-1 alpha, and p53 and Cell Count in Head and Neck Squamous Cell Carcinoma. Mol. Imaging Biol. 2019, 21, 740–746. [Google Scholar] [CrossRef]
- Meyer, H.J.; Hamerla, G.; Leifels, L.; Höhn, A.K.; Surov, A. Histogram analysis parameters derived from DCE-MRI in head and neck squamous cell cancer—Associations with microvessel density. Eur. J. Radiol. 2019, 120, 108669. [Google Scholar] [CrossRef]
- Meyer, H.J.; Hamerla, G.; Leifels, L.; Höhn, A.K.; Surov, A. Whole-lesion ADC histogram analysis is not able to reflect microvessel density in HNSCC. Medicine 2019, 98, e15520. [Google Scholar] [CrossRef] [PubMed]
- Nakahira, M.; Saito, N.; Yamaguchi, H.; Kuba, K.; Sugasawa, M. Use of quantitative diffusion-weighted magnetic resonance imaging to predict human papilloma virus status in patients with oropharyngeal squamous cell carcinoma. Eur. Arch. Oto-Rhino-Laryngol. 2014, 271, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M.; Lim, J.Y.; Koh, Y.W.; Kim, S.H.; Choi, E.C. Machine learning and magnetic resonance imaging radiomics for predicting human papilloma virus status and prognostic factors in oropharyngeal squamous cell carcinoma. Head Neck 2022, 44, 897–903. [Google Scholar] [CrossRef]
- Piludu, F.; Marzi, S.; Gangemi, E.; Farneti, A.; Marucci, L.; Venuti, A.; Benevolo, M.; Pichi, B.; Pellini, R.; Sperati, F.; et al. Multiparametric MRI Evaluation of Oropharyngeal Squamous Cell Carcinoma. A Mono-Institutional Study. J. Clin. Med. 2021, 10, 3865. [Google Scholar] [CrossRef]
- Ravanelli, M.; Grammatica, A.; Tononcelli, E.; Morello, R.; Leali, M.; Battocchio, S.; Agazzi, G.M.; Bastia, M.; Maroldi, R.; Nicolai, P.; et al. Correlation between Human Papillomavirus Status and Quantitative MR Imaging Parameters including Diffusion-Weighted Imaging and Texture Features in Oropharyngeal Carcinoma. Am. J. Neuroradiol. 2018, 39, 1878–1883. [Google Scholar] [CrossRef] [PubMed]
- Samolyk-Kogaczewska, N.; Sierko, E.; Dziemianczyk-Pakiela, D.; Nowaszewska, K.B.; Lukasik, M.; Reszec, J. Usefulness of Hybrid PET/MRI in Clinical Evaluation of Head and Neck Cancer Patients. Cancers 2020, 12, 511. [Google Scholar] [CrossRef]
- Schouten, C.S.; de Graaf, P.; Bloemena, E.; Witte, B.I.; Braakhuis, B.J.M.; Brakenhoff, R.H.; Leemans, C.R.; Castelijns, J.A.; de Bree, R. Quantitative Diffusion-Weighted MRI Parameters and Human Papillomavirus Status in Oropharyngeal Squamous Cell Carcinoma. Am. J. Neuroradiol. 2015, 36, 763–767. [Google Scholar] [CrossRef]
- Sohn, B.; Choi, Y.S.; Ahn, S.S.; Kim, H.; Han, K.; Lee, S.K.; Kim, J. Machine Learning Based Radiomic HPV Phenotyping of Oropharyngeal SCC: A Feasibility Study Using MRI. Laryngoscope 2020, 131, E851–E856. [Google Scholar] [CrossRef]
- Suh, C.H.; Lee, K.H.; Choi, Y.J.; Chung, S.R.; Baek, J.H.; Lee, J.H.; Yun, J.; Ham, S.; Kim, N. Oropharyngeal squamous cell carcinoma: Radiomic machine-learning classifiers from multiparametric MR images for determination of HPV infection status. Sci. Rep. 2020, 10, 17525. [Google Scholar] [CrossRef]
- Rasmussen, J.H.; Olin, A.; Lelkaitis, G.; Hansen, A.E.; Andersen, F.L.; Johannesen, H.H.; Kjær, A.; Vogelius, I.R.; Specht, L.; Bentzen, S.M.; et al. Does multiparametric imaging with 18F-FDG-PET/MRI capture spatial variation in immunohistochemical cancer biomarkers in head and neck squamous cell carcinoma? Br. J. Cancer 2020, 123, 46–53. [Google Scholar] [CrossRef]
- Surov, A.; Stumpp, P.; Meyer, H.J.; Gawlitza, M.; Hohn, A.-K.; Boehm, A.; Sabri, O.; Kahn, T.; Purz, S. Simultaneous (18)F-FDG-PET/MRI: Associations between diffusion, glucose metabolism and histopathological parameters in patients with head and neck squamous cell carcinoma. Oral Oncol. 2016, 58, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Surov, A.; Meyer, H.J.; Gawlitza, M.; Hohn, A.-K.; Boehm, A.; Kahn, T.; Stumpp, P. Correlations between DCE MRI and Histopathological Parameters in Head and Neck Squamous Cell Carcinoma. Transl. Oncol. 2017, 10, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Surov, A.; Meyer, H.J.; Winter, K.; Richter, C.; Hoehn, A.-K. Histogram analysis parameters of apparent diffusion coefficient reflect tumor cellularity and proliferation activity in head and neck squamous cell carcinoma. Oncotarget 2018, 9, 23599–23607. [Google Scholar] [CrossRef]
- Surov, A.; Meyer, H.J.; Leifels, L.; Hohn, A.-K.; Richter, C.; Winter, K. Histogram analysis parameters of dynamic contrast-enhanced magnetic resonance imaging can predict histopathological findings including proliferation potential, cellularity, and nucleic areas in head and neck squamous cell carcinoma. Oncotarget 2018, 9, 21070–21077. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Swartz, J.E.; Driessen, J.P.; van Kempen, P.M.W.; de Bree, R.; Janssen, L.M.; Pameijer, F.A.; Terhaard, C.H.J.; Philippens, M.E.P.; Willems, S. Influence of tumor and microenvironment characteristics on diffusion-weighted imaging in oropharyngeal carcinoma: A pilot study. Oral Oncol. 2018, 77, 9–15. [Google Scholar] [CrossRef]
- Tekiki, N.; Fujita, M.; Okui, T.; Kawai, H.; Oo, M.W.; Kawazu, T.; Hisatomi, M.; Okada, S.; Takeshita, Y.; Barham, M.; et al. Dynamic contrast-enhanced MRI as a predictor of programmed death ligand-1 expression in patients with oral squamous cell carcinoma. Oncol. Lett. 2021, 22, 778. [Google Scholar] [CrossRef]
- Tse, G.M.; King, A.D.; Yu, A.M.; Lam, C.C.; Chan, A.W.; Chen, G.G.; Yeung, D.K.; Yu, K.H.; Bhatia, K.S.; Chan, A.B. Correlation of biomarkers in head and neck squamous cell carcinoma. Otolaryngol.-Head Neck Surg. 2010, 143, 795–800. [Google Scholar] [CrossRef]
- Unetsubo, T.; Konouchi, H.; Yanagi, Y.; Murakami, J.; Fujii, M.; Matsuzaki, H.; Hisatomi, M.; Nagatsuka, H.; Asaumi, J.-i. Dynamic contrast-enhanced magnetic resonance imaging for estimating tumor proliferation and microvessel density of oral squamous cell carcinomas. Oral Oncol. 2009, 45, 621–626. [Google Scholar] [CrossRef][Green Version]
- Vidiri, A.; Marzi, S.; Gangemi, E.; Benevolo, M.; Rollo, F.; Farneti, A.; Marucci, L.; Spasiano, F.; Sperati, F.; Di Giuliano, F.; et al. Intravoxel incoherent motion diffusion-weighted imaging for oropharyngeal squamous cell carcinoma: Correlation with human papillomavirus Status. Eur. J. Radiol. 2019, 119, 108640. [Google Scholar] [CrossRef]
- Wong, K.H.; Panek, R.; Welsh, L.; McQuaid, D.; Dunlop, A.; Riddell, A.; Murray, I.; Du, Y.; Chua, S.; Koh, D.M.; et al. The Predictive Value of Early Assessment After 1 Cycle of Induction Chemotherapy with 18F-FDG PET/CT and Diffusion-Weighted MRI for Response to Radical Chemoradiotherapy in Head and Neck Squamous Cell Carcinoma. J. Nucl. Med. 2016, 57, 1843–1850. [Google Scholar] [CrossRef]
- Wu, W.; Jiang, G.; Xu, Z.; Wang, R.; Pan, A.; Gao, M.; Yu, T.; Huang, L.; Quan, Q.; Li, J. Three-dimensional pulsed continuous arterial spin labeling and intravoxel incoherent motion imaging of nasopharyngeal carcinoma: Correlations with Ki-67 proliferation status. Quant. Imaging Med. Surg. 2021, 11, 1394–1405. [Google Scholar] [CrossRef] [PubMed]
- Shima, T.; Fujima, N.; Yamano, S.; Kameda, H.; Suzuka, M.; Takeuchi, A.; Kinoshita, Y.; Iwai, N.; Kudo, K.; Minowa, K. Non-Gaussian model-based diffusion-weighted imaging of oral squamous cell carcinoma: Associations with Ki-67 proliferation status. Oral Radiol. 2023, 39, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Huang, L.; Li, B.; Li, H. Correlation between Diagnostic Value of Diffusion Kurtosis Imaging for Nasopharyngeal Carcinoma and Ki-67 Expression. B-Ent 2023, 19, 69–73. [Google Scholar] [CrossRef]
- Li, Q.; Xu, T.; Gong, J.; Xiang, S.; Shen, C.; Zhou, X.; Hu, C.; Wu, B.; Lu, X. Applying multisequence MRI radiomics of the primary tumor and lymph node to predict HPV-related p16 status in patients with oropharyngeal squamous cell carcinoma. Quant. Imaging Med. Surg. 2023, 13, 2234–2247. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dang, H.; Wu, X.; Zhang, Z.; Shi, X.; Zhang, T.; Chen, X.; Zhu, X.; Su, T.; Wang, Y.; et al. Correlation between 18F-FDG PET/MR parameters with the expression level of epidermal growth factor receptor and the diagnostic value of PET/MR in head and neck squamous cell carcinoma. Heliyon 2023, 9, e14822. [Google Scholar] [CrossRef]
- Gao, Y.; Mao, Y.; Lu, S.; Tan, L.; Li, G.; Chen, J.; Huang, D.; Zhang, X.; Qiu, Y.; Liu, Y. Magnetic resonance imaging-based radiogenomics analysis for predicting prognosis and gene expression profile in advanced nasopharyngeal carcinoma. Head Neck 2021, 43, 3730–3742. [Google Scholar] [CrossRef]
- Zhang, F.; Zhong, L.Z.; Zhao, X.; Dong, D.; Yao, J.J.; Wang, S.Y.; Liu, Y.; Zhu, D.; Wang, Y.; Wang, G.J.; et al. A deep-learning-based prognostic nomogram integrating microscopic digital pathology and macroscopic magnetic resonance images in nasopharyngeal carcinoma: A multi-cohort study. Ther. Adv. Med. Oncol. 2020, 12, 1758835920971416. [Google Scholar] [CrossRef]
- Sriyook, A.; Lertbutsayanukul, C.; Jittapiromsak, N. Value of dynamic contrast-enhanced magnetic resonance imaging for determining the plasma Epstein-Barr virus status and staging of nasopharyngeal carcinoma. Clin. Imaging 2021, 72, 1–7. [Google Scholar] [CrossRef]
- Konouchi, H.; Asaumi, J.; Yanagi, Y.; Shigehara, H.; Hisatomi, M.; Matsuzaki, H.; Kishi, K. Evaluation of tumor proliferation using dynamic contrast enhanced-MRI of oral cavity and oropharyngeal squamous cell carcinoma. Oral Oncol. 2003, 39, 290–295. [Google Scholar] [CrossRef]
- Newbold, K.; Castellano, I.; Charles-Edwards, E.; Mears, D.; Sohaib, A.; Leach, M.; Rhys-Evans, P.; Clarke, P.; Fisher, C.; Harrington, K.; et al. An exploratory study into the role of dynamic contrast-enhanced magnetic resonance imaging or perfusion computed tomography for detection of intratumoral hypoxia in head-and-neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 29–37. [Google Scholar] [CrossRef]
- Choi, J.W.; Lee, D.; Hyun, S.H.; Han, M.; Kim, J.H.; Lee, S.J. Intratumoural heterogeneity measured using FDG PET and MRI is associated with tumour-stroma ratio and clinical outcome in head and neck squamous cell carcinoma. Clin. Radiol. 2017, 72, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.J.; Höhn, A.K.; Surov, A. Associations between dynamic-contrast enhanced MRI and tumor infiltrating lymphocytes and tumor-stroma ratio in head and neck squamous cell cancer. Cancer Imaging 2021, 21, 60. [Google Scholar] [CrossRef] [PubMed]
- Galati, L.; Chiocca, S.; Duca, D.; Tagliabue, M.; Simoens, C.; Gheit, T.; Arbyn, M.; Tommasino, M. HPV and head and neck cancers: Towards early diagnosis and prevention. Tumour Virus Res. 2022, 14, 200245. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, J. Targeting the epidermal growth factor receptor for cancer therapy. J. Clin. Oncol. 2002, 20 (Suppl. S18), 1s–13s. [Google Scholar]
- Payabvash, S.; Chan, A.; Jabehdar Maralani, P.; Malhotra, A. Quantitative diffusion magnetic resonance imaging for prediction of human papillomavirus status in head and neck squamous-cell carcinoma: A systematic review and meta-analysis. Neuroradiol. J. 2019, 32, 232–240. [Google Scholar] [CrossRef]
- Vandecaveye, V.; Dirix, P.; De Keyzer, F.; de Beeck, K.O.; Vander Poorten, V.; Roebben, I.; Nuyts, S.; Hermans, R. Predictive value of diffusion-weighted magnetic resonance imaging during chemoradiotherapy for head and neck squamous cell carcinoma. Eur. Radiol. 2010, 20, 1703–1714. [Google Scholar] [CrossRef]
- Bammer, R. Basic principles of diffusion-weighted imaging. Eur. J. Radiol. 2003, 45, 169–184. [Google Scholar] [CrossRef]
- Westra, W.H. The morphologic profile of HPV-related head and neck squamous carcinoma: Implications for diagnosis, prognosis, and clinical management. Head. Neck Pathol. 2012, 6 (Suppl. S1), S48–S54. [Google Scholar] [CrossRef]
- Rahimi, S. HPV-related squamous cell carcinoma of oropharynx: A review. J. Clin. Pathol. 2020, 73, 624–629. [Google Scholar] [CrossRef]
- Prigge, E.-S.; Arbyn, M.; von Knebel Doeberitz, M.; Reuschenbach, M. Diagnostic accuracy of p16INK4a immunohistochemistry in oropharyngeal squamous cell carcinomas: A systematic review and meta-analysis. Int. J. Cancer 2017, 140, 1186–1198. [Google Scholar] [CrossRef]
- Schache, A.G.; Liloglou, T.; Risk, J.M.; Filia, A.; Jones, T.M.; Sheard, J.; Woolgar, J.A.; Helliwell, T.R.; Triantafyllou, A.; Robinson, M.; et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: Sensitivity, specificity, and prognostic discrimination. Clin. Cancer Res. 2011, 17, 6262–6271. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.A.; Jung, M.; Zlobec, I.; Green, E.; Storck, C.; Tornillo, L.; Lugli, A.; Wolfensberger, M.; Terracciano, L.M. Co-overexpression of p21 and Ki-67 in head and neck squamous cell carcinoma relative to a significantly poor prognosis. Head Neck 2011, 33, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S. Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59 (Suppl. S2), 21–26. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, H.; Yasui, T.; Ishikawa-Fujiwara, T.; Morii, E.; Yamamoto, Y.; Yoshii, T.; Takenaka, Y.; Nakahara, S.; Todo, T.; Hongyo, T.; et al. Human papillomavirus and p53 mutations in head and neck squamous cell carcinoma among Japanese population. Cancer Sci. 2014, 105, 409–417. [Google Scholar] [CrossRef]
- Pitolli, C.; Wang, Y.; Mancini, M.; Shi, Y.; Melino, G.; Amelio, I. Do Mutations Turn p53 into an Oncogene? Int. J. Mol. Sci. 2019, 20, 6241. [Google Scholar] [CrossRef]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Rehmani, H.S.; Issaeva, N. EGFR in head and neck squamous cell carcinoma: Exploring possibilities of novel drug combinations. Ann. Transl. Med. 2020, 8, 813. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; O’Grady, S.; Crown, J. Targeting p53 for the treatment of cancer. Semin. Cancer Biol. 2022, 79, 58–67. [Google Scholar] [CrossRef]
- Surov, A.; Meyer, H.J.; Wienke, A. Associations between apparent diffusion coefficient (ADC) and KI 67 in different tumors: A meta-analysis. Part 1: ADC(mean). Oncotarget 2017, 8, 75434–75444. [Google Scholar] [CrossRef]
- Cantù, G. Nasopharyngeal carcinoma. A “different” head and neck tumour. Part A: From histology to staging. Acta Otorhinolaryngol. Ital. 2023, 43, 85–98. [Google Scholar] [CrossRef]
- Sourbron, S.P.; Buckley, D.L. On the scope and interpretation of the Tofts models for DCE-MRI. Magn. Reson. Med. 2011, 66, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, K.M.; Le, A.; Duong, H.; Wu, Y.; Zhang, Q.; Messadi, D.V. Correlation between VEGF and HIF-1α expression in human oral squamous cell carcinoma. Exp. Mol. Pathol. 2004, 76, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Pugh, C.W.; Ratcliffe, P.J. Regulation of angiogenesis by hypoxia: Role of the HIF system. Nat. Med. 2003, 9, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Heukelom, J.; Hamming, O.; Bartelink, H.; Hoebers, F.; Giralt, J.; Herlestam, T.; Verheij, M.; van den Brekel, M.; Vogel, W.; Slevin, N.; et al. Adaptive and innovative Radiation Treatment FOR improving Cancer treatment outcomE (ARTFORCE); a randomized controlled phase II trial for individualized treatment of head and neck cancer. BMC Cancer 2013, 13, 84. [Google Scholar] [CrossRef]
- Xie, Q.; Wu, J.; Du, Z.; Di, N.; Yan, R.; Pang, H.; Jin, T.; Zhang, H.; Wu, Y.; Zhang, Y.; et al. DCE-MRI in Human Gliomas: A Surrogate for Assessment of Invasive Hypoxia Marker HIF-1A Based on MRI-Neuronavigation Stereotactic Biopsies. Acad. Radiol. 2019, 26, 179–187. [Google Scholar] [CrossRef]
- Kut, C.; Mac Gabhann, F.; Popel, A.S. Where is VEGF in the body? A meta-analysis of VEGF distribution in cancer. Br. J. Cancer 2007, 97, 978–985. [Google Scholar] [CrossRef]
- Andor, N.; Graham, T.A.; Jansen, M.; Xia, L.C.; Aktipis, C.A.; Petritsch, C.; Ji, H.P.; Maley, C.C. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016, 22, 105–113. [Google Scholar] [CrossRef]
- Clasen, K.; Leibfarth, S.; Hilke, F.J.; Admard, J.; Winter, R.M.; Welz, S.; Gatidis, S.; Nann, D.; Ossowski, S.; Breuer, T.; et al. PET/MRI and genetic intrapatient heterogeneity in head and neck cancers. Strahlenther. Onkol. 2020, 196, 542–551. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).