R-Loops in Genome Instability and Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. R-Loops’ Formation in Physiological and Pathological Conditions
2.1. R-Loops during Transcription
2.2. R-Loops during Replication
2.3. R-Loops in Genome Editing
2.4. R-Loops at DNA Damage Sites of Transcriptionally Active Loci
2.5. Mitochondrial R-Loops
2.6. R-Loops in Telomeres and Sub-Telomeres
2.7. R-Loops in Centromeres
2.8. Cytoplasmic R-Loops and RNA–DNA Hybrids
3. Suppression and Resolution Mechanisms of R-Loops
3.1. RNase H1 and RNase H2
3.2. RNA–DNA Helicases
3.3. Chromatin Remodeling Factors
3.4. DNA Repair Proteins
| Type of Protein | Functions | References |
|---|---|---|
| Topoisomerase | ||
| TOP1 | A class of loci overlapping with efficient early replication origins showed an unexpected loss of R-loops upon Top1 depletion | [143,144] |
| TOP3B | TOP3B works in an epistatic manner with the helicase DDX5 to resolve cellular R-loops | [145] |
| Ribonucleases | ||
| RNaseH1 | Cleaves RNA in RNA–DNA hybrids | [7,98] |
| RNaseH2 | Cleaves RNA in RNA–DNA hybrids; Removes ribonucleotides | [99,146,147] |
| RNA–DNA helicases | ||
| DDX1 | Associates with ATAD5 and suppresses R-loops during DNA replication | [148] |
| DDX2A/ DDX2B | Unwinds the 5′ leader structure as part of the eIF4F complex; reduces affinity of eIF3j for the ribosome to enable the mRNA to access the entry channel | [149] |
| DDX3X | Pre-mRNA splicing; general mRNA export | [150,151] |
| DDX5 | Associates with BRCA2 and suppresses R-loops at DSBs; together with ATAD5, it suppresses R-loops during replication | [140,148,152] |
| DDX19 | Enters the nucleus following DNA damage in an ATR-dependent manner; suppresses R-loops | [153] |
| DDX21 | Unwinds RNA–DNA hybrids in vitro; functions with SIRT7 to suppress R-loops at specific genes; associates with ATAD5; suppresses R-loops during replication | [113] |
| DDX23 | Involved in mRNA processing by regulating of FOXM1 | [154,155] |
| DDX39B(UAP56) | Unwinds RNA–DNA hybrids in vitro; suppresses co-transcriptional R-loops | [156,157,158,159] |
| DDX41 | Enriched in promoter regions in vivo; unwind RNA–DNA hybrids in vitro | [160,161] |
| DDX43 | ATP-dependent; aids piRNA amplification by liberating cleaved RNAs from Ago3-piRISC | [162,163] |
| DHX9 | Binds RNA–DNA hybrids and unwinds R-loops; associates with ATAD5 and suppresses R-loops during replication; prevents R-loop formation by melting RNA–DNA hybrids with a 3′–5′ polarity | [96,117,118,164] |
| DHX29 | Rearrangement of 43S complexes leading to higher processivity in unwinding during scanning | [165,166] |
| DHX36 | Regulates the translation of Gnai2 mRNA by unwinding its 5′ UTR rG4 structures | [167] |
| WRN | Protects the replication fork by preventing unprogrammed R-loops formation | [168] |
| AQR | Putative RNA–DNA helicase; suppresses R-loops in human cells | [109] |
| BLM | Unwinds RNA–DNA hybrids in vitro; suppresses R-loops in human cells | [169,170] |
| SETX | Suppresses R-loops at transcription termination sites and DSBs; binds to replication forks to protect its integrity across RNA-Polymerase-II-transcribed gene and unwinds unnecessary R-loops | [63,171] |
| RECQL4 | Resolves concatenated DNA molecules during HR, at replication forks, and in anaphase | [172,173,174] |
| FANCM | Unwinds telomeric RNA–DNA hybrids; suppresses telomeric R-loops in human cells | [80,120,124] |
| DNA-damage checkpoint proteins | ||
| ATR, CHK1 | May suppress R-loops by preventing collisions between R-loops and replication forks | [153,175] |
| ATM, CHK2 | May suppress R-loops by promoting the repair of DSBs at replication forks collapsed at R-loops | [176,177] |
| Chromatin modulators | ||
| SWI/SNF | ATP-dependent chromatin-remodeling complexes, may suppress R-loops during transcription–replication conflicts | [128,178] |
| PCR1 | Promotes repressive chromatin, suppresses R-loops by decreasing transcription | [179] |
| BRD2 | A reader of histone lysine acetylation; recruits TOP1 | [180] |
| BRD4 | A reader of histone lysine acetylation; suppresses R-loops at specific genes by preventing Pol II pausing | [39,130] |
| FACT | Histone chaperone, may reorganize chromatin at sites of R-loop-replication collisions | [127] |
| KAT8 | A histone acetyltransferase that functions with c and BRD4 to suppress R-loops | [180] |
| SIN3A | Part of a histone deacetylase complex that interacts with the THO complex | [181] |
| INO80 | Part of the ATP-dependent chromatin-remodeling complex, may reorganize chromatin to resolve R-loops | [46] |
| DNA repair proteins | ||
| BRCA1 | Suppresses R-loops at promoter-proximal Pol II pause sites and transcription termination sites; recruits SETX to transcription termination sites | [82,90,182,183] |
| BRCA2 | Suppresses R-loops at promoter-proximal Pol II pause sites; interacts with DDX5 and TREX2 | [139,140,141,142] |
| FA factors(A/I/D2/L) Likely entire pathway | FANCD2–FANCI bind RNA–DNA hybrids and RNA processing factors suppress R-loops to coordinate replication and transcription; the repair of RFs blocked at R-loop-containing sites | [135,136,137] |
| RPA | Recruits, binds, and stimulates RNaseH1 | [138] |
| CtIP | May process R-loops at active genes | [184] |
| APTX/TDP1 | Suppresses R-loops through its function in single-strand break repair | [185] |
| MRE11 | Suppresses R-loops upon collisions of R-loops and replication forks, promotes functions of FA repair proteins | [186] |
| MUS81 | A structure-specific nuclease that cleaves Holliday junctions and stalled or reversed replication forks, may process R-loops upon collisions of R-loops and replication forks | [44,175] |
| SAMHD1 | Deoxynucleoside triphosphohydrolase (dNTPase) and 3′–5′ exoribonuclease; does not resolve R-loops directly but can recruit other factors like MRE11 | [187] |
| XPG, XPF | Structure-specific nucleases involved in transcription-coupled nucleotide excision repair, may cleave and remove R-loops during transcription | [31,79] |
| RNA processing factors | ||
| SRSF1 | Splicing factor; binds the Pol II CTD and nascent RNA transcripts | [188] |
| SRSF2 | Splicing factors; mutated in myelodysplastic syndrome and cancer | [189] |
| SF3B1 | Splicing factors; mutated in myelodysplastic syndrome and cancer | [190,191] |
| SPT6 | Pol II-associated elongation factor recruits the Integrator complex to suppress R-loops generated from long non-coding RNA | [192] |
| TFIIS | Recognizes backtracked Pol II and stimulates transcript cleavage | [193] |
| U2AF1 | Splicing factors; mutated in myelodysplastic syndrome and cancer | [191] |
| SFPQ, NONO | RNA splicing; suppress telomeric R-loops | [194] |
| RNA exosome | 3′–5′ exoribonuclease; degrades non-coding RNAs and prevents R-loop accumulation | [195,196] |
| THO/TREX/TREX2 | Promote transcription termination and assembly and nuclear export of messenger ribonucleoproteins | [40,141,181,197] |
4. R-Loops as a Double-Edged Sword
4.1. Functions of R-Loops
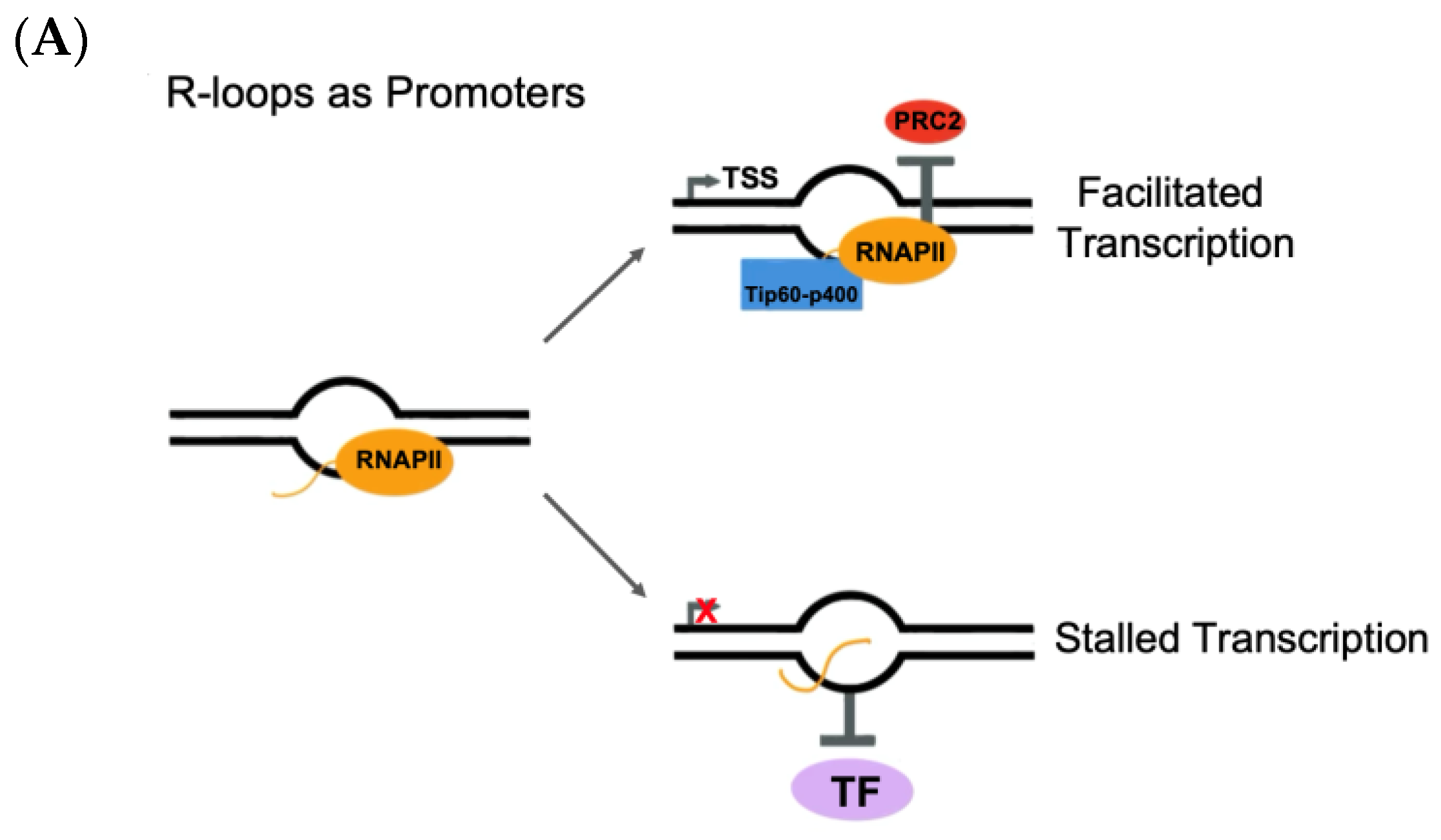
4.1.1. R-Loops and Transcriptional Regulation
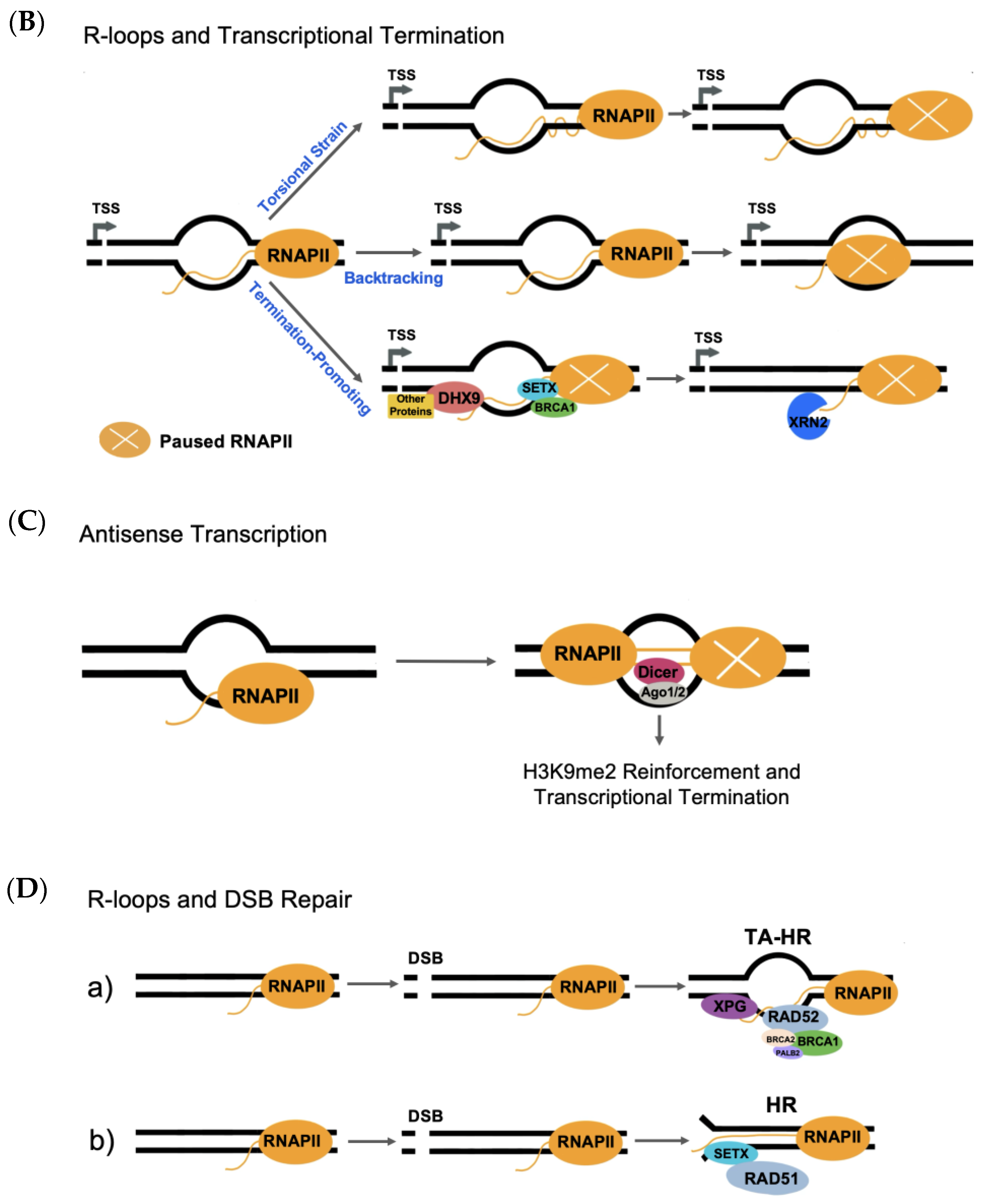
4.1.2. R-Loops and Transcriptional Termination
4.1.3. R-Loops and DNA Double-Strand Break Repair
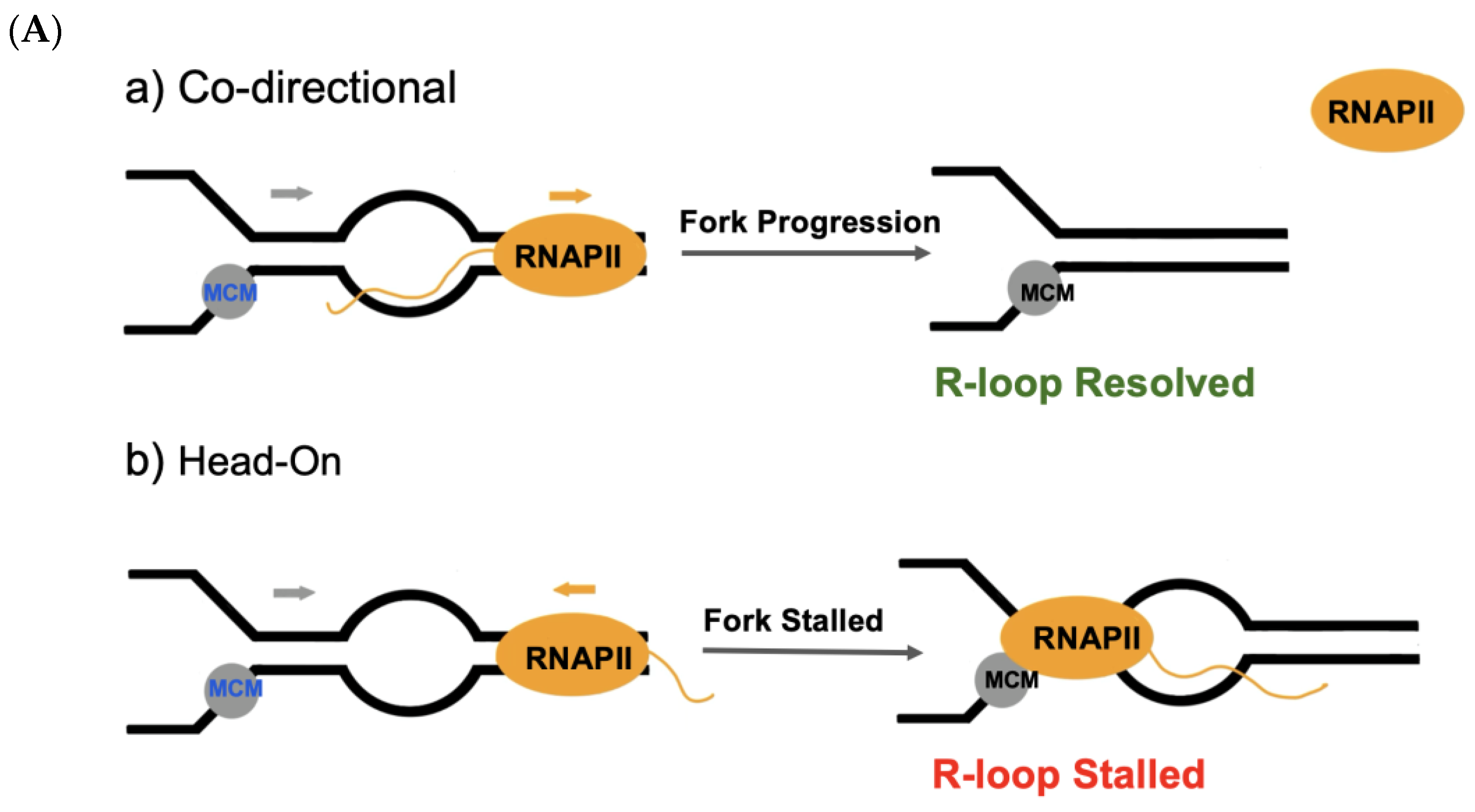
4.2. R-Loops as Threats to Genome Stability
5. R-Loops and Cancer
5.1. The Impact of Tumor Suppressor Genes on R-Loops
5.2. The Influence of Oncogenes on R-Loops
6. Technologies for Mapping of Genomic R-Loops
6.1. The S9.6 Monoclonal Antibody-Based Approach
6.2. Inactive RNase H-Based Approach
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Allison, D.F.; Wang, G.G. R-loops: Formation, function, and relevance to cell stress. Cell Stress 2019, 3, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.W.; Crothers, D.M. Stability and properties of double and triple helices: Dramatic effects of RNA or DNA backbone composition. Science 1992, 258, 1463–1466. [Google Scholar] [CrossRef]
- Thomas, M.; White, R.L.; Davis, R.W. Hybridization of RNA to double-stranded DNA: Formation of R-loops. Proc. Natl. Acad. Sci. USA 1976, 73, 2294–2298. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Lan, L.; Zou, L. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat. Rev. Mol. Cell Biol. 2022, 23, 521–540. [Google Scholar] [CrossRef]
- Marnef, A.; Legube, G. R-loops as Janus-faced modulators of DNA repair. Nat. Cell Biol. 2021, 23, 305–313. [Google Scholar] [CrossRef]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Lombraña, R.; Almeida, R.; Álvarez, A.; Gómez, M. R-loops and initiation of DNA replication in human cells: A missing link? Genet 2015, 6, 158. [Google Scholar] [CrossRef]
- Ginno, P.A.; Lim, Y.W.; Lott, P.L.; Korf, I.; Chédin, F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013, 23, 1590–1600. [Google Scholar] [CrossRef]
- Ginno, P.A.; Lott, P.L.; Christensen, H.C.; Korf, I.; Chédin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 2012, 45, 814–825. [Google Scholar] [CrossRef]
- Groh, M.; Silva, L.M.; Gromak, N. Mechanisms of transcriptional dysregulation in repeat expansion disorders. Biochem. Soc. Trans. 2014, 42, 1123–1128. [Google Scholar] [CrossRef]
- Aguilera, A.; García-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef]
- Groh, M.; Gromak, N. Out of balance: R-loops in human disease. PLoS Genet. 2014, 10, e1004630. [Google Scholar] [CrossRef]
- Yu, K.; Chedin, F.; Hsieh, C.L.; Wilson, T.E.; Lieber, M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003, 4, 442–451. [Google Scholar] [CrossRef]
- Gong, Y.; Liu, Y. R-Loops at Chromosome Ends: From Formation, Regulation, and Cellular Consequence. Cancers 2023, 15, 2178. [Google Scholar] [CrossRef] [PubMed]
- Lima, W.F.; Murray, H.M.; Damle, S.S.; Hart, C.E.; Hung, G.; De Hoyos, C.L.; Liang, X.H.; Crooke, S.T. Viable RNaseH1 knockout mice show RNaseH1 is essential for R loop processing, mitochondrial and liver function. Nucleic Acids Res. 2016, 44, 5299–5312. [Google Scholar] [CrossRef]
- Holt, I.J. The mitochondrial R-loop. Nucleic Acids Res. 2019, 47, 5480–5489. [Google Scholar] [CrossRef]
- Ujvári, A.; Luse, D.S. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 2006, 13, 49–54. [Google Scholar] [CrossRef]
- Wells, J.P.; White, J.; Stirling, P.C. R Loops and Their Composite Cancer Connections. Trends Cancer 2019, 5, 619–631. [Google Scholar] [CrossRef] [PubMed]
- El Hage, A.; French, S.L.; Beyer, A.L.; Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010, 24, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.A. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: Recent insights into factors that influence transcription elongation. Gene 2012, 493, 176–184. [Google Scholar] [CrossRef]
- Al-Hadid, Q.; Yang, Y. R-loop: An emerging regulator of chromatin dynamics. Acta Biochim. Biophys. Sin. 2016, 48, 623–631. [Google Scholar] [CrossRef]
- Dutrieux, L.; Lin, Y.L.; Lutzmann, M.; Rodriguez, R.; Cogné, M.; Pasero, P.; Moreaux, J. Transcription/Replication Conflicts in Tumorigenesis and Their Potential Role as Novel Therapeutic Targets in Multiple Myeloma. Cancers 2021, 13, 3755. [Google Scholar] [CrossRef] [PubMed]
- Gómez-González, B.; Aguilera, A. Transcription-mediated replication hindrance: A major driver of genome instability. Genes Dev. 2019, 33, 1008–1026. [Google Scholar] [CrossRef] [PubMed]
- Elsakrmy, N.; Cui, H. R-Loops and R-Loop-Binding Proteins in Cancer Progression and Drug Resistance. Int. J. Mol. Sci. 2023, 24, 7064. [Google Scholar] [CrossRef]
- Panov, K.I.; Hannan, K.; Hannan, R.D.; Hein, N. The Ribosomal Gene Loci-The Power behind the Throne. Genes 2021, 12, 763. [Google Scholar] [CrossRef] [PubMed]
- Schier, A.C.; Taatjes, D.J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef]
- Richard, P.; Manley, J.L. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009, 23, 1247–1269. [Google Scholar] [CrossRef]
- Abraham, K.J.; Khosraviani, N.; Chan, J.N.Y.; Gorthi, A.; Samman, A.; Zhao, D.Y.; Wang, M.; Bokros, M.; Vidya, E.; Ostrowski, L.A.; et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 2020, 585, 298–302. [Google Scholar] [CrossRef]
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef]
- Muniesa-Vargas, A.; Theil, A.F.; Ribeiro-Silva, C.; Vermeulen, W.; Lans, H. XPG: A multitasking genome caretaker. Cell Mol. Life Sci. 2022, 79, 166. [Google Scholar] [CrossRef] [PubMed]
- Paule, M.R.; White, R.J. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000, 28, 1283–1298. [Google Scholar] [CrossRef]
- Lata, E.; Choquet, K.; Sagliocco, F.; Brais, B.; Bernard, G.; Teichmann, M. RNA Polymerase III Subunit Mutations in Genetic Diseases. Front. Mol. Biosci. 2021, 8, 696438. [Google Scholar] [CrossRef]
- Sanchez, A.; de Vivo, A.; Tonzi, P.; Kim, J.; Huang, T.T.; Kee, Y. Transcription-replication conflicts as a source of common fragile site instability caused by BMI1-RNF2 deficiency. PLoS Genet. 2020, 16, e1008524. [Google Scholar] [CrossRef]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774–786.e719. [Google Scholar] [CrossRef]
- Haeusler, R.A.; Engelke, D.R. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 2006, 34, 4826–4836. [Google Scholar] [CrossRef]
- Kotsantis, P.; Segura-Bayona, S.; Margalef, P.; Marzec, P.; Ruis, P.; Hewitt, G.; Bellelli, R.; Patel, H.; Goldstone, R.; Poetsch, A.R.; et al. RTEL1 Regulates G4/R-Loops to Avert Replication-Transcription Collisions. Cell Rep. 2020, 33, 108546. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.S.; Maganti, R.; Tanksley, J.P.; Luo, J.; Park, J.J.H.; Balkanska-Sinclair, E.; Ling, J.; Floyd, S.R. BRD4 Prevents R-Loop Formation and Transcription-Replication Conflicts by Ensuring Efficient Transcription Elongation. Cell Rep. 2020, 32, 108166. [Google Scholar] [CrossRef] [PubMed]
- Wellinger, R.E.; Prado, F.; Aguilera, A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell Biol. 2006, 26, 3327–3334. [Google Scholar] [CrossRef]
- Kotsantis, P.; Silva, L.M.; Irmscher, S.; Jones, R.M.; Folkes, L.; Gromak, N.; Petermann, E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016, 7, 13087. [Google Scholar] [CrossRef] [PubMed]
- Stork, C.T.; Bocek, M.; Crossley, M.P.; Sollier, J.; Sanz, L.A.; Chédin, F.; Swigut, T.; Cimprich, K.A. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 2016, 5, 17548. [Google Scholar] [CrossRef] [PubMed]
- Stoy, H.; Zwicky, K.; Kuster, D.; Lang, K.S.; Krietsch, J.; Crossley, M.P.; Schmid, J.A.; Cimprich, K.A.; Merrikh, H.; Lopes, M. Direct visualization of transcription-replication conflicts reveals post-replicative DNA:RNA hybrids. Nat. Struct. Mol. Biol. 2023, 30, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhu, M.; Limbo, O.; Russell, P. RNase H eliminates R-loops that disrupt DNA replication but is nonessential for efficient DSB repair. EMBO Rep. 2018, 19, e45335. [Google Scholar] [CrossRef]
- Lang, K.S.; Hall, A.N.; Merrikh, C.N.; Ragheb, M.; Tabakh, H.; Pollock, A.J.; Woodward, J.J.; Dreifus, J.E.; Merrikh, H. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 2017, 170, 787–799.e718. [Google Scholar] [CrossRef]
- Prendergast, L.; McClurg, U.L.; Hristova, R.; Berlinguer-Palmini, R.; Greener, S.; Veitch, K.; Hernandez, I.; Pasero, P.; Rico, D.; Higgins, J.M.G.; et al. Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability. Nat. Commun. 2020, 11, 4534. [Google Scholar] [CrossRef]
- Zhang, B.; Luo, D.; Li, Y.; Perčulija, V.; Chen, J.; Lin, J.; Ye, Y.; Ouyang, S. Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1. Nat. Commun. 2021, 12, 3476. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Jiang, F.; Taylor, D.W.; Chen, J.S.; Kornfeld, J.E.; Zhou, K.; Thompson, A.J.; Nogales, E.; Doudna, J.A. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016, 351, 867–871. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Samach, A.; Mafessoni, F.; Gross, O.; Melamed-Bessudo, C.; Filler-Hayut, S.; Dahan-Meir, T.; Amsellem, Z.; Pawlowski, W.P.; Levy, A.A. CRISPR/Cas9-induced DNA breaks trigger crossover, chromosomal loss, and chromothripsis-like rearrangements. Plant Cell 2023, koad209. [Google Scholar] [CrossRef]
- Zhang, G.; Luo, Y.; Dai, X.; Dai, Z. Benchmarking deep learning methods for predicting CRISPR/Cas9 sgRNA on- and off-target activities. Brief. Bioinform. 2023, 24, bbad333. [Google Scholar] [CrossRef]
- Zeng, Y.; Cui, Y.; Zhang, Y.; Zhang, Y.; Liang, M.; Chen, H.; Lan, J.; Song, G.; Lou, J. The initiation, propagation and dynamics of CRISPR-SpyCas9 R-loop complex. Nucleic Acids Res. 2018, 46, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Pacesa, M.; Loeff, L.; Querques, I.; Muckenfuss, L.M.; Sawicka, M.; Jinek, M. R-loop formation and conformational activation mechanisms of Cas9. Nature 2022, 609, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Imaichi, S.; Kraeutler, E.; Aguilar, R.; Lee, Y.W.; Sheridan, S.D.; Lee, J.T. Site-specific R-loops induce CGG repeat contraction and fragile X gene reactivation. Cell 2023, 186, 2593–2609.e2518. [Google Scholar] [CrossRef]
- Ouyang, J.; Yadav, T.; Zhang, J.M.; Yang, H.; Rheinbay, E.; Guo, H.; Haber, D.A.; Lan, L.; Zou, L. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 2021, 594, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Rendtlew Danielsen, J.M.; Yang, Y.G.; Qi, Y. A role for small RNAs in DNA double-strand break repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef]
- Teng, Y.; Yadav, T.; Duan, M.; Tan, J.; Xiang, Y.; Gao, B.; Xu, J.; Liang, Z.; Liu, Y.; Nakajima, S.; et al. ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nat. Commun. 2018, 9, 4115. [Google Scholar] [CrossRef]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235. [Google Scholar] [CrossRef]
- Pessina, F.; Giavazzi, F.; Yin, Y.; Gioia, U.; Vitelli, V.; Galbiati, A.; Barozzi, S.; Garre, M.; Oldani, A.; Flaus, A.; et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 2019, 21, 1286–1299. [Google Scholar] [CrossRef]
- Liu, S.; Hua, Y.; Wang, J.; Li, L.; Yuan, J.; Zhang, B.; Wang, Z.; Ji, J.; Kong, D. RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination. Cell 2021, 184, 1314–1329.e1310. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kong, D. A direct role of RNA polymerase III and RNA in DNA homologous recombination. Mol. Cell Oncol. 2021, 8, 1935173. [Google Scholar] [CrossRef]
- Cohen, S.; Puget, N.; Lin, Y.L.; Clouaire, T.; Aguirrebengoa, M.; Rocher, V.; Pasero, P.; Canitrot, Y.; Legube, G. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 2018, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- Holt, I.J. R-Loops and Mitochondrial DNA Metabolism. Methods Mol. Biol. 2022, 2528, 173–202. [Google Scholar] [CrossRef]
- Falkenberg, M. Mitochondrial DNA replication in mammalian cells: Overview of the pathway. Essays Biochem. 2018, 62, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Melchionda, L.; Nasca, A.; Carrara, F.; Lamantea, E.; Zanolini, A.; Lamperti, C.; Fang, M.; Zhang, J.; Ronchi, D.; et al. RNASEH1 Mutations Impair mtDNA Replication and Cause Adult-Onset Mitochondrial Encephalomyopathy. Am. J. Hum. Genet. 2015, 97, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Wanrooij, P.H.; Uhler, J.P.; Shi, Y.; Westerlund, F.; Falkenberg, M.; Gustafsson, C.M. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012, 40, 10334–10344. [Google Scholar] [CrossRef]
- Brown, T.A.; Tkachuk, A.N.; Clayton, D.A. Native R-loops persist throughout the mouse mitochondrial DNA genome. J. Biol. Chem. 2008, 283, 36743–36751. [Google Scholar] [CrossRef]
- Silva, S.; Camino, L.P.; Aguilera, A. Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proc. Natl. Acad. Sci. USA 2018, 115, 11024–11029. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009, 276, 1494–1505. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Frolova, E.G.; Feng, C.; Grinberg, A.; Love, P.E.; Crouch, R.J. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 2003, 11, 807–815. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Barral, A.; Déjardin, J. Telomeric Chromatin and TERRA. J. Mol. Biol. 2020, 432, 4244–4256. [Google Scholar] [CrossRef] [PubMed]
- Luke, B.; Lingner, J. TERRA: Telomeric repeat-containing RNA. EMBO J. 2009, 28, 2503–2510. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef]
- Feretzaki, M.; Pospisilova, M.; Valador Fernandes, R.; Lunardi, T.; Krejci, L.; Lingner, J. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 2020, 587, 303–308. [Google Scholar] [CrossRef]
- Cusanelli, E.; Chartrand, P. Telomeric repeat-containing RNA TERRA: A noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 2015, 6, 143. [Google Scholar] [CrossRef]
- Guh, C.Y.; Shen, H.J.; Chen, L.W.; Chiu, P.C.; Liao, I.H.; Lo, C.C.; Chen, Y.; Hsieh, Y.H.; Chang, T.C.; Yen, C.P.; et al. XPF activates break-induced telomere synthesis. Nat. Commun. 2022, 13, 5781. [Google Scholar] [CrossRef]
- Pan, X.; Chen, Y.; Biju, B.; Ahmed, N.; Kong, J.; Goldenberg, M.; Huang, J.; Mohan, N.; Klosek, S.; Parsa, K.; et al. FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops. Sci. Rep. 2019, 9, 19110. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Transcribing Centromeres: Noncoding RNAs and Kinetochore Assembly. Trends Genet. 2018, 34, 587–599. [Google Scholar] [CrossRef]
- Racca, C.; Britton, S.; Hédouin, S.; Francastel, C.; Calsou, P.; Larminat, F. BRCA1 prevents R-loop-associated centromeric instability. Cell Death Dis. 2021, 12, 896. [Google Scholar] [CrossRef]
- Kabeche, L.; Nguyen, H.D.; Buisson, R.; Zou, L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 2018, 359, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Bobkov, G.O.M.; Gilbert, N.; Heun, P. Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J. Cell Biol. 2018, 217, 1957–1972. [Google Scholar] [CrossRef] [PubMed]
- McNulty, S.M.; Sullivan, L.L.; Sullivan, B.A. Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C. Dev. Cell 2017, 42, 226–240.e226. [Google Scholar] [CrossRef]
- Giunta, S.; Hervé, S.; White, R.R.; Wilhelm, T.; Dumont, M.; Scelfo, A.; Gamba, R.; Wong, C.K.; Rancati, G.; Smogorzewska, A.; et al. CENP-A chromatin prevents replication stress at centromeres to avoid structural aneuploidy. Proc. Natl. Acad. Sci. USA 2021, 118, e2015634118. [Google Scholar] [CrossRef] [PubMed]
- Mankan, A.K.; Schmidt, T.; Chauhan, D.; Goldeck, M.; Höning, K.; Gaidt, M.; Kubarenko, A.V.; Andreeva, L.; Hopfner, K.P.; Hornung, V. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014, 33, 2937–2946. [Google Scholar] [CrossRef]
- Chatzidoukaki, O.; Stratigi, K.; Goulielmaki, E.; Niotis, G.; Akalestou-Clocher, A.; Gkirtzimanaki, K.; Zafeiropoulos, A.; Altmüller, J.; Topalis, P.; Garinis, G.A. R-loops trigger the release of cytoplasmic ssDNAs leading to chronic inflammation upon DNA damage. Sci. Adv. 2021, 7, eabj5769. [Google Scholar] [CrossRef]
- Crossley, M.P.; Song, C.; Bocek, M.J.; Choi, J.H.; Kousorous, J.; Sathirachinda, A.; Lin, C.; Brickner, J.R.; Bai, G.; Lans, H.; et al. R-loop-derived cytoplasmic RNA-DNA hybrids activate an immune response. Nature 2023, 613, 187–194. [Google Scholar] [CrossRef]
- Hatchi, E.; Skourti-Stathaki, K.; Ventz, S.; Pinello, L.; Yen, A.; Kamieniarz-Gdula, K.; Dimitrov, S.; Pathania, S.; McKinney, K.M.; Eaton, M.L.; et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 2015, 57, 636–647. [Google Scholar] [CrossRef]
- Ablasser, A.; Chen, Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, aat8657. [Google Scholar] [CrossRef] [PubMed]
- Typas, D. Breaking the loop: RNA-DNA hybrids escape to the cytoplasm. Nat. Struct. Mol. Biol. 2023, 30, 134. [Google Scholar] [CrossRef]
- Koo, C.X.; Kobiyama, K.; Shen, Y.J.; LeBert, N.; Ahmad, S.; Khatoo, M.; Aoshi, T.; Gasser, S.; Ishii, K.J. RNA polymerase III regulates cytosolic RNA:DNA hybrids and intracellular microRNA expression. J. Biol. Chem. 2015, 290, 7463–7473. [Google Scholar] [CrossRef] [PubMed]
- Belotserkovskii, B.P.; Tornaletti, S.; D’Souza, A.D.; Hanawalt, P.C. R-loop generation during transcription: Formation, processing and cellular outcomes. DNA Repair 2018, 71, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro de Almeida, C.; Dhir, S.; Dhir, A.; Moghaddam, A.E.; Sattentau, Q.; Meinhart, A.; Proudfoot, N.J. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol. Cell 2018, 70, 650–662.e658. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Huang, J.T.J.; Hiom, K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 2018, 9, 4346. [Google Scholar] [CrossRef]
- Lockhart, A.; Pires, V.B.; Bento, F.; Kellner, V.; Luke-Glaser, S.; Yakoub, G.; Ulrich, H.D.; Luke, B. RNase H1 and H2 Are Differentially Regulated to Process RNA-DNA Hybrids. Cell Rep. 2019, 29, 2890–2900.e2895. [Google Scholar] [CrossRef]
- Yang, Z.; Hou, Q.; Cheng, L.; Xu, W.; Hong, Y.; Li, S.; Sun, Q. RNase H1 Cooperates with DNA Gyrases to Restrict R-Loops and Maintain Genome Integrity in Arabidopsis Chloroplasts. Plant Cell 2017, 29, 2478–2497. [Google Scholar] [CrossRef]
- Sparks, J.L.; Chon, H.; Cerritelli, S.M.; Kunkel, T.A.; Johansson, E.; Crouch, R.J.; Burgers, P.M. RNase H2-initiated ribonucleotide excision repair. Mol. Cell 2012, 47, 980–986. [Google Scholar] [CrossRef]
- Yi, C.; Yang, J.; Zhang, T.; Xie, S.; Li, W.; Qin, L.; Chen, D. A pan-cancer analysis of RNASEH1, a potential regulator of the tumor microenvironment. Clin. Transl. Oncol. 2023, 25, 2569–2586. [Google Scholar] [CrossRef]
- Kind, B.; Muster, B.; Staroske, W.; Herce, H.D.; Sachse, R.; Rapp, A.; Schmidt, F.; Koss, S.; Cardoso, M.C.; Lee-Kirsch, M.A. Altered spatio-temporal dynamics of RNase H2 complex assembly at replication and repair sites in Aicardi-Goutières syndrome. Hum. Mol. Genet. 2014, 23, 5950–5960. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.; Patrick, T.; Parmar, R.; Taylor, C.F.; Aeby, A.; Aicardi, J.; Artuch, R.; Montalto, S.A.; Bacino, C.A.; Barroso, B.; et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 2007, 81, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Crow, Y.J.; Livingston, J.H. Aicardi-Goutières syndrome: An important Mendelian mimic of congenital infection. Dev. Med. Child. Neurol. 2008, 50, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Crow, Y.J.; Leitch, A.; Hayward, B.E.; Garner, A.; Parmar, R.; Griffith, E.; Ali, M.; Semple, C.; Aicardi, J.; Babul-Hirji, R.; et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 2006, 38, 910–916. [Google Scholar] [CrossRef]
- Brosh, R.M., Jr.; Matson, S.W. History of DNA Helicases. Genes 2020, 11, 255. [Google Scholar] [CrossRef]
- Mallam, A.L.; Sidote, D.J.; Lambowitz, A.M. Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. eLife 2014, 3, e04630. [Google Scholar] [CrossRef]
- Hasanova, Z.; Klapstova, V.; Porrua, O.; Stefl, R.; Sebesta, M. Human senataxin is a bona fide R-loop resolving enzyme and transcription termination factor. Nucleic Acids Res. 2023, 51, 2818–2837. [Google Scholar] [CrossRef]
- Gatti, V.; De Domenico, S.; Melino, G.; Peschiaroli, A. Senataxin and R-loops homeostasis: Multifaced implications in carcinogenesis. Cell Death Discov. 2023, 9, 145. [Google Scholar] [CrossRef]
- Sollier, J.; Stork, C.T.; García-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef]
- Liu, Y.C.; Cheng, S.C. Functional roles of DExD/H-box RNA helicases in Pre-mRNA splicing. J. Biomed. Sci. 2015, 22, 54. [Google Scholar] [CrossRef]
- Liu, Y.; Imai, R. Function of Plant DExD/H-Box RNA Helicases Associated with Ribosomal RNA Biogenesis. Front. Plant Sci. 2018, 9, 125. [Google Scholar] [CrossRef]
- Martin, R.; Straub, A.U.; Doebele, C.; Bohnsack, M.T. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 2013, 10, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Hotz-Wagenblatt, A.; Voit, R.; Grummt, I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017, 31, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Santoriello, C.; Sporrij, A.; Yang, S.; Flynn, R.A.; Henriques, T.; Dorjsuren, B.; Custo Greig, E.; McCall, W.; Stanhope, M.E.; Fazio, M.; et al. RNA helicase DDX21 mediates nucleotide stress responses in neural crest and melanoma cells. Nat. Cell Biol. 2020, 22, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Uchida, C.; Anderson, S.F.; Lee, C.G.; Hurwitz, J.; Parvin, J.D.; Montminy, M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 1997, 90, 1107–1112. [Google Scholar] [CrossRef]
- Anderson, S.F.; Schlegel, B.P.; Nakajima, T.; Wolpin, E.S.; Parvin, J.D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998, 19, 254–256. [Google Scholar] [CrossRef]
- Chakraborty, P.; Grosse, F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair 2011, 10, 654–665. [Google Scholar] [CrossRef]
- Cristini, A.; Groh, M.; Kristiansen, M.S.; Gromak, N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 2018, 23, 1891–1905. [Google Scholar] [CrossRef]
- Acharya, S.; Kaul, Z.; Gocha, A.S.; Martinez, A.R.; Harris, J.; Parvin, J.D.; Groden, J. Association of BLM and BRCA1 during Telomere Maintenance in ALT Cells. PLoS ONE 2014, 9, e103819. [Google Scholar] [CrossRef]
- Silva, B.; Pentz, R.; Figueira, A.M.; Arora, R.; Lee, Y.W.; Hodson, C.; Wischnewski, H.; Deans, A.J.; Azzalin, C.M. FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat. Commun. 2019, 10, 2253. [Google Scholar] [CrossRef]
- Chang, E.Y.; Novoa, C.A.; Aristizabal, M.J.; Coulombe, Y.; Segovia, R.; Chaturvedi, R.; Shen, Y.; Keong, C.; Tam, A.S.; Jones, S.J.M.; et al. RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J. Cell Biol. 2017, 216, 3991–4005. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Ahmed, N.; Kong, J.; Zhang, D. Breaking the end: Target the replication stress response at the ALT telomeres for cancer therapy. Mol. Cell Oncol. 2017, 4, e1360978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; Zou, L. Alternative lengthening of telomeres: From molecular mechanisms to therapeutic outlooks. Cell Biosci. 2020, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Domingues-Silva, B.; Silva, B.; Azzalin, C.M. ALTernative Functions for Human FANCM at Telomeres. Front. Mol. Biosci. 2019, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.B.; Chen, H.V.; Acharya, D.; Rando, O.J.; Fazzio, T.G. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 2015, 22, 999–1007. [Google Scholar] [CrossRef]
- Mognato, M.; Burdak-Rothkamm, S.; Rothkamm, K. Interplay between DNA replication stress, chromatin dynamics and DNA-damage response for the maintenance of genome stability. Mutat. Res. Rev. Mutat. Res. 2021, 787, 108346. [Google Scholar] [CrossRef]
- Herrera-Moyano, E.; Mergui, X.; García-Rubio, M.L.; Barroso, S.; Aguilera, A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 2014, 28, 735–748. [Google Scholar] [CrossRef]
- Bayona-Feliu, A.; Barroso, S.; Muñoz, S.; Aguilera, A. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat. Genet. 2021, 53, 1050–1063. [Google Scholar] [CrossRef]
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Scott, C.; Mitson, M.; Taylor, S.; Higgs, D.R.; Gibbons, R.J. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015, 6, 7538. [Google Scholar] [CrossRef]
- Lam, F.C.; Kong, Y.W.; Huang, Q.; Vu Han, T.L.; Maffa, A.D.; Kasper, E.M.; Yaffe, M.B. BRD4 prevents the accumulation of R-loops and protects against transcription-replication collision events and DNA damage. Nat. Commun. 2020, 11, 4083. [Google Scholar] [CrossRef]
- Takaoka, M.; Miki, Y. BRCA1 gene: Function and deficiency. Int. J. Clin. Oncol. 2018, 23, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Block-Schmidt, A.S.; Dukowic-Schulze, S.; Wanieck, K.; Reidt, W.; Puchta, H. BRCC36A is epistatic to BRCA1 in DNA crosslink repair and homologous recombination in Arabidopsis thaliana. Nucleic Acids Res. 2011, 39, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Pecori, R.; Di Giorgio, S.; Paulo Lorenzo, J.; Nina Papavasiliou, F. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Nat. Rev. Genet. 2022, 23, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Peake, J.D.; Noguchi, E. Fanconi anemia: Current insights regarding epidemiology, cancer, and DNA repair. Hum. Genet. 2022, 141, 1811–1836. [Google Scholar] [CrossRef] [PubMed]
- García-Rubio, M.L.; Pérez-Calero, C.; Barroso, S.I.; Tumini, E.; Herrera-Moyano, E.; Rosado, I.V.; Aguilera, A. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet. 2015, 11, e1005674. [Google Scholar] [CrossRef]
- Schwab, R.A.; Nieminuszczy, J.; Shah, F.; Langton, J.; Lopez Martinez, D.; Liang, C.C.; Cohn, M.A.; Gibbons, R.J.; Deans, A.J.; Niedzwiedz, W. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol. Cell 2015, 60, 351–361. [Google Scholar] [CrossRef]
- Liang, Z.; Liang, F.; Teng, Y.; Chen, X.; Liu, J.; Longerich, S.; Rao, T.; Green, A.M.; Collins, N.B.; Xiong, Y.; et al. Binding of FANCI-FANCD2 Complex to RNA and R-Loops Stimulates Robust FANCD2 Monoubiquitination. Cell Rep. 2019, 26, 564–572.e565. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Yadav, T.; Giri, S.; Saez, B.; Graubert, T.A.; Zou, L. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol. Cell 2017, 65, 832–847.e834. [Google Scholar] [CrossRef]
- Shivji, M.K.K.; Renaudin, X.; Williams, Ç.H.; Venkitaraman, A.R. BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation. Cell Rep. 2018, 22, 1031–1039. [Google Scholar] [CrossRef]
- Sessa, G.; Gómez-González, B.; Silva, S.; Pérez-Calero, C.; Beaurepere, R.; Barroso, S.; Martineau, S.; Martin, C.; Ehlén, Å.; Martínez, J.S.; et al. BRCA2 promotes DNA-RNA hybrid resolution by DDX5 helicase at DNA breaks to facilitate their repair‡. EMBO J. 2021, 40, e106018. [Google Scholar] [CrossRef]
- Bhatia, V.; Barroso, S.I.; García-Rubio, M.L.; Tumini, E.; Herrera-Moyano, E.; Aguilera, A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014, 511, 362–365. [Google Scholar] [CrossRef]
- D’Alessandro, G.; Whelan, D.R.; Howard, S.M.; Vitelli, V.; Renaudin, X.; Adamowicz, M.; Iannelli, F.; Jones-Weinert, C.W.; Lee, M.; Matti, V.; et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 2018, 9, 5376. [Google Scholar] [CrossRef]
- Wilson-Sali, T.; Hsieh, T.S. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc. Natl. Acad. Sci. USA 2002, 99, 7974–7979. [Google Scholar] [CrossRef] [PubMed]
- Tuduri, S.; Crabbé, L.; Conti, C.; Tourrière, H.; Holtgreve-Grez, H.; Jauch, A.; Pantesco, V.; De Vos, J.; Thomas, A.; Theillet, C.; et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009, 11, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Yang, X.; Huang, S.N.; Agama, K.; Baechler, S.A.; Sun, Y.; Zhang, H.; Saha, L.K.; Su, S.; Jenkins, L.M.; et al. Resolution of R-loops by topoisomerase III-β (TOP3B) in coordination with the DEAD-box helicase DDX5. Cell Rep. 2022, 40, 111067. [Google Scholar] [CrossRef] [PubMed]
- Rychlik, M.P.; Chon, H.; Cerritelli, S.M.; Klimek, P.; Crouch, R.J.; Nowotny, M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol. Cell 2010, 40, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, K.J.; Carroll, P.; Lettice, L.; Tarnauskaitė, Ž.; Reddy, K.; Dix, F.; Revuelta, A.; Abbondati, E.; Rigby, R.E.; Rabe, B.; et al. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 2016, 35, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, N.; Park, S.H.; Wells, J.; Hwang, T.; Ryu, E.; Kim, B.G.; Hwang, S.; Kim, S.J.; Kang, S.; et al. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020, 48, 7218–7238. [Google Scholar] [CrossRef]
- Schütz, P.; Karlberg, T.; van den Berg, S.; Collins, R.; Lehtiö, L.; Högbom, M.; Holmberg-Schiavone, L.; Tempel, W.; Park, H.W.; Hammarström, M.; et al. Comparative structural analysis of human DEAD-box RNA helicases. PLoS ONE 2010, 5, e12791. [Google Scholar] [CrossRef]
- Topisirovic, I.; Siddiqui, N.; Lapointe, V.L.; Trost, M.; Thibault, P.; Bangeranye, C.; Piñol-Roma, S.; Borden, K.L. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009, 28, 1087–1098. [Google Scholar] [CrossRef]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Mersaoui, S.Y.; Yu, Z.; Coulombe, Y.; Karam, M.; Busatto, F.F.; Masson, J.Y.; Richard, S. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J. 2019, 38, e100986. [Google Scholar] [CrossRef]
- Hodroj, D.; Recolin, B.; Serhal, K.; Martinez, S.; Tsanov, N.; Abou Merhi, R.; Maiorano, D. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017, 36, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Cao, Y.; Ling, T.; Li, P.; Wu, S.; Peng, D.; Wang, Y.; Jia, X.; Chen, S.; Xu, A.; et al. DDX23, an Evolutionary Conserved dsRNA Sensor, Participates in Innate Antiviral Responses by Pairing With TRIF or MAVS. Front. Immunol. 2019, 10, 2202. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, Y.; Qiu, C.; Chen, J.; Wu, H.; Wang, Q.; Ma, X.; Song, K.; Kong, B. Splicing Factor DDX23, Transcriptionally Activated by E2F1, Promotes Ovarian Cancer Progression by Regulating FOXM1. Front. Oncol. 2021, 11, 749144. [Google Scholar] [CrossRef]
- Matson, J.P.; Zou, L. A genome-wide and cotranscriptional suppressor of R loops. Genes Dev. 2020, 34, 863–864. [Google Scholar] [CrossRef]
- Pérez-Calero, C.; Bayona-Feliu, A.; Xue, X.; Barroso, S.I.; Muñoz, S.; González-Basallote, V.M.; Sung, P.; Aguilera, A. UAP56/DDX39B is a major cotranscriptional RNA-DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 2020, 34, 898–912. [Google Scholar] [CrossRef]
- Shen, H. UAP56- a key player with surprisingly diverse roles in pre-mRNA splicing and nuclear export. BMB Rep. 2009, 42, 185–188. [Google Scholar] [CrossRef]
- Kota, K.P.; Wagner, S.R.; Huerta, E.; Underwood, J.M.; Nickerson, J.A. Binding of ATP to UAP56 is necessary for mRNA export. J. Cell Sci. 2008, 121, 1526–1537. [Google Scholar] [CrossRef]
- Mosler, T.; Conte, F.; Longo, G.M.C.; Mikicic, I.; Kreim, N.; Möckel, M.M.; Petrosino, G.; Flach, J.; Barau, J.; Luke, B.; et al. R-loop proximity proteomics identifies a role of DDX41 in transcription-associated genomic instability. Nat. Commun. 2021, 12, 7314. [Google Scholar] [CrossRef]
- Qin, K.; Jian, D.; Xue, Y.; Cheng, Y.; Zhang, P.; Wei, Y.; Zhang, J.; Xiong, H.; Zhang, Y.; Yuan, X. DDX41 regulates the expression and alternative splicing of genes involved in tumorigenesis and immune response. Oncol. Rep. 2021, 45, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.S.; Arna, A.B.; Dong, H.; Yadav, M.; Aggarwal, A.; Wu, Y. Structure-function analysis of DEAD-box helicase DDX43. Methods 2022, 204, 286–299. [Google Scholar] [CrossRef]
- Wu, H.; Zhai, L.T.; Chen, P.Y.; Xi, X.G. DDX43 prefers single strand substrate and its full binding activity requires physical connection of all domains. Biochem. Biophys. Res. Commun. 2019, 520, 594–599. [Google Scholar] [CrossRef]
- Chakraborty, P.; Hiom, K. DHX9-dependent recruitment of BRCA1 to RNA promotes DNA end resection in homologous recombination. Nat. Commun. 2021, 12, 4126. [Google Scholar] [CrossRef] [PubMed]
- Dhote, V.; Sweeney, T.R.; Kim, N.; Hellen, C.U.; Pestova, T.V. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc. Natl. Acad. Sci. USA 2012, 109, E3150–E3159. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.R.; Dhote, V.; Guca, E.; Hellen, C.U.T.; Hashem, Y.; Pestova, T.V. Functional role and ribosomal position of the unique N-terminal region of DHX29, a factor required for initiation on structured mammalian mRNAs. Nucleic Acids Res. 2021, 49, 12955–12969. [Google Scholar] [CrossRef]
- Chen, X.; Yuan, J.; Xue, G.; Campanario, S.; Wang, D.; Wang, W.; Mou, X.; Liew, S.W.; Umar, M.I.; Isern, J.; et al. Translational control by DHX36 binding to 5′UTR G-quadruplex is essential for muscle stem-cell regenerative functions. Nat. Commun. 2021, 12, 5043. [Google Scholar] [CrossRef]
- Marabitti, V.; Valenzisi, P.; Lillo, G.; Malacaria, E.; Palermo, V.; Pichierri, P.; Franchitto, A. R-Loop-Associated Genomic Instability and Implication of WRN and WRNIP1. Int. J. Mol. Sci. 2022, 23, 1547. [Google Scholar] [CrossRef]
- Drosopoulos, W.C.; Kosiyatrakul, S.T.; Schildkraut, C.L. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J. Cell Biol. 2015, 210, 191–208. [Google Scholar] [CrossRef]
- Cohen, S.; Guenolé, A.; Lazar, I.; Marnef, A.; Clouaire, T.; Vernekar, D.V.; Puget, N.; Rocher, V.; Arnould, C.; Aguirrebengoa, M.; et al. A POLD3/BLM dependent pathway handles DSBs in transcribed chromatin upon excessive RNA:DNA hybrid accumulation. Nat. Commun. 2022, 13, 2012. [Google Scholar] [CrossRef]
- Yeo, A.J.; Becherel, O.J.; Luff, J.E.; Graham, M.E.; Richard, D.; Lavin, M.F. Senataxin controls meiotic silencing through ATR activation and chromatin remodeling. Cell Discov. 2015, 1, 15025. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.T.; Bernstein, K.A. Role and Regulation of the RECQL4 Family during Genomic Integrity Maintenance. Genes 2021, 12, 1919. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Shamanna, R.A.; Keijzers, G.; Anand, R.; Rasmussen, L.J.; Cejka, P.; Croteau, D.L.; Bohr, V.A. RECQL4 Promotes DNA End Resection in Repair of DNA Double-Strand Breaks. Cell Rep. 2016, 16, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Ahn, B.; Bohr, V.A. Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology 2009, 10, 235–252. [Google Scholar] [CrossRef]
- Matos, D.A.; Zhang, J.M.; Ouyang, J.; Nguyen, H.D.; Genois, M.M.; Zou, L. ATR Protects the Genome against R Loops through a MUS81-Triggered Feedback Loop. Mol. Cell 2020, 77, 514–527.e514. [Google Scholar] [CrossRef]
- Tresini, M.; Warmerdam, D.O.; Kolovos, P.; Snijder, L.; Vrouwe, M.G.; Demmers, J.A.; van IJcken, W.F.; Grosveld, F.G.; Medema, R.H.; Hoeijmakers, J.H.; et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature 2015, 523, 53–58. [Google Scholar] [CrossRef]
- Walker, C.; Herranz-Martin, S.; Karyka, E.; Liao, C.; Lewis, K.; Elsayed, W.; Lukashchuk, V.; Chiang, S.C.; Ray, S.; Mulcahy, P.J.; et al. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat. Neurosci. 2017, 20, 1225–1235. [Google Scholar] [CrossRef]
- McBride, M.J.; Pulice, J.L.; Beird, H.C.; Ingram, D.R.; D’Avino, A.R.; Shern, J.F.; Charville, G.W.; Hornick, J.L.; Nakayama, R.T.; Garcia-Rivera, E.M.; et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 2018, 33, 1128–1141.e1127. [Google Scholar] [CrossRef]
- Klusmann, I.; Wohlberedt, K.; Magerhans, A.; Teloni, F.; Korbel, J.O.; Altmeyer, M.; Dobbelstein, M. Chromatin modifiers Mdm2 and RNF2 prevent RNA:DNA hybrids that impair DNA replication. Proc. Natl. Acad. Sci. USA 2018, 115, E11311–E11320. [Google Scholar] [CrossRef]
- Kim, J.J.; Lee, S.Y.; Gong, F.; Battenhouse, A.M.; Boutz, D.R.; Bashyal, A.; Refvik, S.T.; Chiang, C.M.; Xhemalce, B.; Paull, T.T.; et al. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev. 2019, 33, 1751–1774. [Google Scholar] [CrossRef]
- Salas-Armenteros, I.; Pérez-Calero, C.; Bayona-Feliu, A.; Tumini, E.; Luna, R.; Aguilera, A. Human THO-Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J. 2017, 36, 3532–3547. [Google Scholar] [CrossRef] [PubMed]
- Vohhodina, J.; Goehring, L.J.; Liu, B.; Kong, Q.; Botchkarev, V.V., Jr.; Huynh, M.; Liu, Z.; Abderazzaq, F.O.; Clark, A.P.; Ficarro, S.B.; et al. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat. Commun. 2021, 12, 3542. [Google Scholar] [CrossRef] [PubMed]
- Hatchi, E.; Goehring, L.; Landini, S.; Skourti-Stathaki, K.; DeConti, D.K.; Abderazzaq, F.O.; Banerjee, P.; Demers, T.M.; Wang, Y.E.; Quackenbush, J.; et al. BRCA1 and RNAi factors promote repair mediated by small RNAs and PALB2-RAD52. Nature 2021, 591, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Makharashvili, N.; Arora, S.; Yin, Y.; Fu, Q.; Wen, X.; Lee, J.H.; Kao, C.H.; Leung, J.W.; Miller, K.M.; Paull, T.T. Sae2/CtIP prevents R-loop accumulation in eukaryotic cells. eLife 2018, 7, e42733. [Google Scholar] [CrossRef] [PubMed]
- Yeo, A.J.; Becherel, O.J.; Luff, J.E.; Cullen, J.K.; Wongsurawat, T.; Jenjaroenpun, P.; Kuznetsov, V.A.; McKinnon, P.J.; Lavin, M.F. R-loops in proliferating cells but not in the brain: Implications for AOA2 and other autosomal recessive ataxias. PLoS ONE 2014, 9, e90219. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Anand, R.; Zhang, X.; Francia, S.; Michelini, F.; Galbiati, A.; Williams, H.; Ronato, D.A.; Masson, J.Y.; Rothenberg, E.; et al. MRE11-RAD50-NBS1 Complex Is Sufficient to Promote Transcription by RNA Polymerase II at Double-Strand Breaks by Melting DNA Ends. Cell Rep. 2021, 34, 108565. [Google Scholar] [CrossRef]
- Park, K.; Ryoo, J.; Jeong, H.; Kim, M.; Lee, S.; Hwang, S.Y.; Ahn, J.; Kim, D.; Moon, H.C.; Baek, D.; et al. Aicardi-Goutières syndrome-associated gene SAMHD1 preserves genome integrity by preventing R-loop formation at transcription-replication conflict regions. PLoS Genet. 2021, 17, e1009523. [Google Scholar] [CrossRef]
- Li, X.; Manley, J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005, 122, 365–378. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Leong, W.Y.; Li, W.; Reddy, P.N.G.; Sullivan, J.D.; Walter, M.J.; Zou, L.; Graubert, T.A. Spliceosome Mutations Induce R Loop-Associated Sensitivity to ATR Inhibition in Myelodysplastic Syndromes. Cancer Res. 2018, 78, 5363–5374. [Google Scholar] [CrossRef]
- Sorrells, S.; Nik, S.; Casey, M.J.; Cameron, R.C.; Truong, H.; Toruno, C.; Gulfo, M.; Lowe, A.; Jette, C.; Stewart, R.A.; et al. Spliceosomal components protect embryonic neurons from R-loop-mediated DNA damage and apoptosis. Dis. Model. Mech. 2018, 11, dmm031583. [Google Scholar] [CrossRef]
- Cheruiyot, A.; Li, S.; Nonavinkere Srivatsan, S.; Ahmed, T.; Chen, Y.; Lemacon, D.S.; Li, Y.; Yang, Z.; Wadugu, B.A.; Warner, W.A.; et al. Nonsense-Mediated RNA Decay Is a Unique Vulnerability of Cancer Cells Harboring SF3B1 or U2AF1 Mutations. Cancer Res. 2021, 81, 4499–4513. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Tellier, M.; Foxwell, J.; Ribeiro de Almeida, C.; Tan-Wong, S.M.; Dhir, S.; Dujardin, G.; Dhir, A.; Murphy, S.; Proudfoot, N.J. Deregulated Expression of Mammalian lncRNA through Loss of SPT6 Induces R-Loop Formation, Replication Stress, and Cellular Senescence. Mol. Cell 2018, 72, 970–984.e977. [Google Scholar] [CrossRef]
- Zatreanu, D.; Han, Z.; Mitter, R.; Tumini, E.; Williams, H.; Gregersen, L.; Dirac-Svejstrup, A.B.; Roma, S.; Stewart, A.; Aguilera, A.; et al. Elongation Factor TFIIS Prevents Transcription Stress and R-Loop Accumulation to Maintain Genome Stability. Mol. Cell 2019, 76, 57–69.e59. [Google Scholar] [CrossRef]
- Petti, E.; Buemi, V.; Zappone, A.; Schillaci, O.; Broccia, P.V.; Dinami, R.; Matteoni, S.; Benetti, R.; Schoeftner, S. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat. Commun. 2019, 10, 1001. [Google Scholar] [CrossRef] [PubMed]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.P.; et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Prim, J.; Bonath, F.; Visa, N. RNA at DNA Double-Strand Breaks: The Challenge of Dealing with DNA:RNA Hybrids. Bioessays 2020, 42, e1900225. [Google Scholar] [CrossRef] [PubMed]
- Gómez-González, B.; García-Rubio, M.; Bermejo, R.; Gaillard, H.; Shirahige, K.; Marín, A.; Foiani, M.; Aguilera, A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011, 30, 3106–3119. [Google Scholar] [CrossRef]
- Dumelie, J.G.; Jaffrey, S.R. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. eLife 2017, 6, e28306. [Google Scholar] [CrossRef]
- van Kruijsbergen, I.; Hontelez, S.; Veenstra, G.J. Recruiting polycomb to chromatin. Int. J. Biochem. Cell Biol. 2015, 67, 177–187. [Google Scholar] [CrossRef]
- Shi, J.; Xu, J.; Chen, Y.E.; Li, J.S.; Cui, Y.; Shen, L.; Li, J.J.; Li, W. The concurrence of DNA methylation and demethylation is associated with transcription regulation. Nat. Commun. 2021, 12, 5285. [Google Scholar] [CrossRef]
- Boque-Sastre, R.; Soler, M.; Oliveira-Mateos, C.; Portela, A.; Moutinho, C.; Sayols, S.; Villanueva, A.; Esteller, M.; Guil, S. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. USA 2015, 112, 5785–5790. [Google Scholar] [CrossRef]
- Cloutier, S.C.; Wang, S.; Ma, W.K.; Al Husini, N.; Dhoondia, Z.; Ansari, A.; Pascuzzi, P.E.; Tran, E.J. Regulated Formation of lncRNA-DNA Hybrids Enables Faster Transcriptional Induction and Environmental Adaptation. Mol. Cell 2016, 61, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, N.J. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 2016, 352, aad9926. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Kamieniarz-Gdula, K.; Proudfoot, N.J. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 2014, 516, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Belotserkovskii, B.P.; Hanawalt, P.C. Anchoring nascent RNA to the DNA template could interfere with transcription. Biophys. J. 2011, 100, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E. RNA polymerase backtracking in gene regulation and genome instability. Cell 2012, 149, 1438–1445. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Patidar, P.L.; Motea, E.A.; Dang, T.T.; Manley, J.L.; Boothman, D.A. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016, 12, e1006107. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.S.; Bushell, M. DNA:RNA hybrids form at DNA double-strand breaks in transcriptionally active loci. Cell Death Dis. 2020, 11, 280. [Google Scholar] [CrossRef]
- Yasuhara, T.; Kato, R.; Hagiwara, Y.; Shiotani, B.; Yamauchi, M.; Nakada, S.; Shibata, A.; Miyagawa, K. Human Rad52 Promotes XPG-Mediated R-loop Processing to Initiate Transcription-Associated Homologous Recombination Repair. Cell 2018, 175, 558–570.e511. [Google Scholar] [CrossRef]
- Meers, C.; Keskin, H.; Banyai, G.; Mazina, O.; Yang, T.; Gombolay, A.L.; Mukherjee, K.; Kaparos, E.I.; Newnam, G.; Mazin, A.; et al. Genetic Characterization of Three Distinct Mechanisms Supporting RNA-Driven DNA Repair and Modification Reveals Major Role of DNA Polymerase ζ. Mol. Cell 2020, 79, 1037–1050.e1035. [Google Scholar] [CrossRef]
- Keskin, H.; Shen, Y.; Huang, F.; Patel, M.; Yang, T.; Ashley, K.; Mazin, A.V.; Storici, F. Transcript-RNA-templated DNA recombination and repair. Nature 2014, 515, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.T.; Hawley, B.R.; Skalka, G.L.; Baldock, R.A.; Smith, E.M.; Bader, A.S.; Malewicz, M.; Watts, F.Z.; Wilczynska, A.; Bushell, M. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 2018, 9, 532. [Google Scholar] [CrossRef] [PubMed]
- Wahba, L.; Gore, S.K.; Koshland, D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife 2013, 2, e00505. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Cimprich, K.A. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015, 25, 514–522. [Google Scholar] [CrossRef]
- Symington, L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002, 66, 630–670, table of contents. [Google Scholar] [CrossRef] [PubMed]
- Costantino, L.; Koshland, D. Genome-wide Map of R-Loop-Induced Damage Reveals How a Subset of R-Loops Contributes to Genomic Instability. Mol. Cell 2018, 71, 487–497.e483. [Google Scholar] [CrossRef]
- Jalan, M.; Olsen, K.S.; Powell, S.N. Emerging Roles of RAD52 in Genome Maintenance. Cancers 2019, 11, 1038. [Google Scholar] [CrossRef]
- San Martin Alonso, M.; Noordermeer, S.M. Untangling the crosstalk between BRCA1 and R-loops during DNA repair. Nucleic Acids Res. 2021, 49, 4848–4863. [Google Scholar] [CrossRef]
- Prado, F.; Aguilera, A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005, 24, 1267–1276. [Google Scholar] [CrossRef]
- Srivatsan, A.; Tehranchi, A.; MacAlpine, D.M.; Wang, J.D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010, 6, e1000810. [Google Scholar] [CrossRef]
- Mackay, R.P.; Xu, Q.; Weinberger, P.M. R-Loop Physiology and Pathology: A Brief Review. DNA Cell Biol. 2020, 39, 1914–1925. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Khan, E.S.; Danckwardt, S. Pathophysiological Role and Diagnostic Potential of R-Loops in Cancer and Beyond. Genes 2022, 13, 2181. [Google Scholar] [CrossRef] [PubMed]
- Brambati, A.; Zardoni, L.; Achar, Y.J.; Piccini, D.; Galanti, L.; Colosio, A.; Foiani, M.; Liberi, G. Dormant origins and fork protection mechanisms rescue sister forks arrested by transcription. Nucleic Acids Res. 2018, 46, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Marabitti, V.; Lillo, G.; Malacaria, E.; Palermo, V.; Sanchez, M.; Pichierri, P.; Franchitto, A. ATM pathway activation limits R-loop-associated genomic instability in Werner syndrome cells. Nucleic Acids Res. 2019, 47, 3485–3502. [Google Scholar] [CrossRef]
- Roy, S.; Tomaszowski, K.H.; Luzwick, J.W.; Park, S.; Li, J.; Murphy, M.; Schlacher, K. p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and POLθ pathways. eLife 2018, 7, e31723. [Google Scholar] [CrossRef]
- Barlow, J.H.; Faryabi, R.B.; Callén, E.; Wong, N.; Malhowski, A.; Chen, H.T.; Gutierrez-Cruz, G.; Sun, H.W.; McKinnon, P.; Wright, G.; et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 2013, 152, 620–632. [Google Scholar] [CrossRef]
- Gorthi, A.; Romero, J.C.; Loranc, E.; Cao, L.; Lawrence, L.A.; Goodale, E.; Iniguez, A.B.; Bernard, X.; Masamsetti, V.P.; Roston, S.; et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 2018, 555, 387–391. [Google Scholar] [CrossRef]
- Jones, S.E.; Fleuren, E.D.G.; Frankum, J.; Konde, A.; Williamson, C.T.; Krastev, D.B.; Pemberton, H.N.; Campbell, J.; Gulati, A.; Elliott, R.; et al. ATR Is a Therapeutic Target in Synovial Sarcoma. Cancer Res. 2017, 77, 7014–7026. [Google Scholar] [CrossRef]
- Jones, R.M.; Mortusewicz, O.; Afzal, I.; Lorvellec, M.; García, P.; Helleday, T.; Petermann, E. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene 2013, 32, 3744–3753. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, J.Y.; Huang, Y.J.; Gu, Y.; Qiu, J.; Qian, H.; Shao, C.; Zhang, X.; Hu, J.; Li, H.; et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol. Cell 2018, 69, 412–425.e416. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.A.; Hieter, P.; Stirling, P.C. Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 2014, 30, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Vanoosthuyse, V. Strengths and Weaknesses of the Current Strategies to Map and Characterize R-Loops. Noncoding RNA 2018, 4, 9. [Google Scholar] [CrossRef]
- Chédin, F.; Hartono, S.R.; Sanz, L.A.; Vanoosthuyse, V. Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J. 2021, 40, e106394. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.J.; Hamperl, S.; Swigut, T.; Cimprich, K.A. qDRIP: A method to quantitatively assess RNA-DNA hybrid formation genome-wide. Nucleic Acids Res. 2020, 48, e84. [Google Scholar] [CrossRef]
- Nadel, J.; Athanasiadou, R.; Lemetre, C.; Wijetunga, N.A.; Broin, P.Ó.; Sato, H.; Zhang, Z.; Jeddeloh, J.; Montagna, C.; Golden, A.; et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics. Chromatin 2015, 8, 46. [Google Scholar] [CrossRef]
- Wahba, L.; Costantino, L.; Tan, F.J.; Zimmer, A.; Koshland, D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016, 30, 1327–1338. [Google Scholar] [CrossRef]
- Sanz, L.A.; Chédin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2019, 14, 1734–1755. [Google Scholar] [CrossRef]
- Xu, W.; Xu, H.; Li, K.; Fan, Y.; Liu, Y.; Yang, X.; Sun, Q. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 2017, 3, 704–714. [Google Scholar] [CrossRef]
- Hartono, S.R.; Malapert, A.; Legros, P.; Bernard, P.; Chédin, F.; Vanoosthuyse, V. The Affinity of the S9.6 Antibody for Double-Stranded RNAs Impacts the Accurate Mapping of R-Loops in Fission Yeast. J. Mol. Biol. 2018, 430, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.D.; Garboczi, D.N.; Singh, K.; Hu, Z.; Leppla, S.H.; Leysath, C.E. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit. 2013, 26, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Legros, P.; Malapert, A.; Niinuma, S.; Bernard, P.; Vanoosthuyse, V. RNA processing factors Swd2.2 and Sen1 antagonize RNA Pol III-dependent transcription and the localization of condensin at Pol III Genes. PLoS Genet. 2014, 10, e1004794. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, J.Y.; Zhang, X.; Fu, X.D.; Chen, L. R-ChIP for genome-wide mapping of R-loops by using catalytically inactive RNASEH1. Nat. Protoc. 2019, 14, 1661–1685. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Zafar, A.; Luo, L.; Denning, A.M.; Gu, J.; Bennett, A.; Yuan, F.; Zhang, Y. R-Loops in Genome Instability and Cancer. Cancers 2023, 15, 4986. https://doi.org/10.3390/cancers15204986
Li F, Zafar A, Luo L, Denning AM, Gu J, Bennett A, Yuan F, Zhang Y. R-Loops in Genome Instability and Cancer. Cancers. 2023; 15(20):4986. https://doi.org/10.3390/cancers15204986
Chicago/Turabian StyleLi, Fang, Alyan Zafar, Liang Luo, Ariana Maria Denning, Jun Gu, Ansley Bennett, Fenghua Yuan, and Yanbin Zhang. 2023. "R-Loops in Genome Instability and Cancer" Cancers 15, no. 20: 4986. https://doi.org/10.3390/cancers15204986
APA StyleLi, F., Zafar, A., Luo, L., Denning, A. M., Gu, J., Bennett, A., Yuan, F., & Zhang, Y. (2023). R-Loops in Genome Instability and Cancer. Cancers, 15(20), 4986. https://doi.org/10.3390/cancers15204986






